

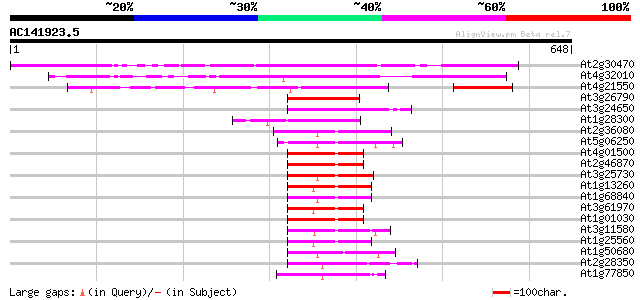
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141923.5 - phase: 0 /pseudo
(648 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g30470 putative VP1/ABI3 family regulatory protein 422 e-118
At4g32010 predicted protein 315 4e-86
At4g21550 VP1 like protein 160 3e-39
At3g26790 FUSCA3 (FUS3) 84 2e-16
At3g24650 abscisic acid-insensitive protein 3 80 5e-15
At1g28300 VP1 protein, putative 79 6e-15
At2g36080 putative RAV2-like DNA binding protein 77 4e-14
At5g06250 putative protein 75 1e-13
At4g01500 putative DNA-binding protein 74 2e-13
At2g46870 RAV-like B3 domain DNA binding protein 73 4e-13
At3g25730 AP2 domain transcription factor 72 7e-13
At1g13260 unknown protein 72 7e-13
At1g68840 putative DNA-binding protein protein RAV2 72 1e-12
At3g61970 RAV-like protein 70 5e-12
At1g01030 DNA-binding protein, putative 68 1e-11
At3g11580 DNA binding protein like 66 7e-11
At1g25560 unknown protein 62 8e-10
At1g50680 hypothetical protein 54 2e-07
At2g28350 unknown protein 54 3e-07
At1g77850 putative auxin response factor protein (T32E8.16) 52 1e-06
>At2g30470 putative VP1/ABI3 family regulatory protein
Length = 790
Score = 422 bits (1086), Expect = e-118
Identities = 256/591 (43%), Positives = 335/591 (56%), Gaps = 61/591 (10%)
Query: 1 MGVEVCMNNYCKEGSTGEWKKGWSLISGGFAKLCNKCGLAYENSLFCDKFHRHETGWRKC 60
MG ++CMN C ST EWKKGW L SG A LC +CG AYE+SLFC++FH+ ++GWR+C
Sbjct: 6 MGSKMCMNASCGTTSTVEWKKGWPLRSGLLADLCYRCGSAYESSLFCEQFHKDQSGWREC 65
Query: 61 SNCSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNT--ENHNRFSQTTKNNASDQ 118
CSK +H GCI SK + E +D+GG+ C TC QL LNT EN FS+ +D+
Sbjct: 66 YLCSKRLHCGCIASKVTIELMDYGGVGCSTCACCHQLNLNTRGENPGVFSRLPMKTLADR 125
Query: 119 YGEHIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIGPKTEEVKCQFNKE 178
+H++G E G+ G+ P G K EE
Sbjct: 126 --QHVNG----ESGGRNE----------GDLFSQPLVMG-------GDKREEFM-----P 157
Query: 179 DTRFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSRNSVLPLATEIGEG 238
F +M S T L+ E+ + +L++ +A+ S + ATE EG
Sbjct: 158 HRGFGKLMSPESTTTGHR-LDAAGEMHESSPLQPSLNMGLAVNPFSPS----FATEAVEG 212
Query: 239 KLEGKASSHFQQGQTSQSILAQLSKTGIAMNLETNKGMISHPPRRPPADVKGKNQLLSRY 298
S ++ +IL + S+ I+ +K + R PP + +G+ LL RY
Sbjct: 213 MKHISPSQSNMVHCSASNILQKPSRPAISTPPVASKSAQARIGR-PPVEGRGRGHLLPRY 271
Query: 299 WPRITDQELEKLSGDLKSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVP 358
WP+ TD+E++++SG+L +VPLFEK LS SDAGRIGRLVLPKACAEA+ P I QSEG+P
Sbjct: 272 WPKYTDKEVQQISGNLNLNIVPLFEKTLSASDAGRIGRLVLPKACAEAYFPPISQSEGIP 331
Query: 359 LQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFG 418
L+ QD+ G EWTFQFR+WPNNNSRMYVLEGVTPCIQS+ L AGDTVTFSR+DPG K + G
Sbjct: 332 LKIQDVRGREWTFQFRYWPNNNSRMYVLEGVTPCIQSMMLQAGDTVTFSRVDPGGKLIMG 391
Query: 419 FRRSLTSIVTQDASTSSHSNGILIKDTNFSGAPQNLNSLSSFSNLLQSMKGNGEPYLNGH 478
R++ + D +NG +DT+ SG +N S++ S + K LNG
Sbjct: 392 SRKAANA---GDMQGCGLTNGTSTEDTSSSGVTENPPSINGSSCISLIPK-----ELNGM 443
Query: 479 SEHLRLGNGTADWLKTANSEEEMNNGPL-QRLVSVSEKKRSRNIGTKTKRLHIHSEDAME 537
E+L E N G + V EKKR+R IG K KRL +HSE++ME
Sbjct: 444 PENL---------------NSETNGGRIGDDPTRVKEKKRTRTIGAKNKRLLLHSEESME 488
Query: 538 LRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDEPPVFGK-IKTNASPSG 587
LRLTWEEAQ+ L P PSV+P V IE+Q EEYDEPPVFGK PSG
Sbjct: 489 LRLTWEEAQDLLRPSPSVKPTIVVIEEQEIEEYDEPPVFGKRTIVTTKPSG 539
>At4g32010 predicted protein
Length = 675
Score = 315 bits (808), Expect = 4e-86
Identities = 210/545 (38%), Positives = 277/545 (50%), Gaps = 108/545 (19%)
Query: 46 FCDKFHRHETGWRKCSNCSKPIHSGCIVSKSSFEYLDFGGITCVTCVKPSQLCLNTENHN 105
F DKF R K +H GCI S+ E L+ GG+TC++C K S L +H
Sbjct: 9 FLDKFLR------------KRLHCGCIASRFMMELLENGGVTCISCAKKSGLISMNVSHE 56
Query: 106 RFSQTTKNNASDQYGEHIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIG 165
+ + AS EH+ ++E+ +L+ RI D S +
Sbjct: 57 SNGKDFPSFAS---AEHVGS--VLERTNLKHLLHFQRI--------------DPTHSSLQ 97
Query: 166 PKTEEVKCQFNKEDTRFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSR 225
K EE + L+ ++H + E K + +L ++LG +
Sbjct: 98 MKQEESLLPSS------LDALRHKT---------------ERKELSAQPNLSISLGPTLM 136
Query: 226 NSVLPLATEIGEGKLEGKASSHFQQGQTSQSILAQLSKTG-IAMNLETNKGMISH-PPRR 283
S P + + K +S FQ S+ +L + + + IA +E + ++S R
Sbjct: 137 TS--PFHDAAVDDR--SKTNSIFQLAPRSRQLLPKPANSAPIAAGMEPSGSLVSQIHVAR 192
Query: 284 PPADVKGKNQLLSRYWPRITDQELEKLSGDL--------------KSTVVPLFEKVLSPS 329
PP + +GK QLL RYWPRITDQEL +LSG S ++PLFEKVLS S
Sbjct: 193 PPPEGRGKTQLLPRYWPRITDQELLQLSGQYPHLYESLTVYFPSSNSKIIPLFEKVLSAS 252
Query: 330 DAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYVLEGV 389
DAGRIGRLVLPKACAEA+ P I EG+PL+ QDI G EW FQFRFWPNNNSRMYVLEGV
Sbjct: 253 DAGRIGRLVLPKACAEAYFPPISLPEGLPLKIQDIKGKEWVFQFRFWPNNNSRMYVLEGV 312
Query: 390 TPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSNGILIKDTNFSG 449
TPCIQS+QL AGDTVTFSR +P K + G+R++ S T
Sbjct: 313 TPCIQSMQLQAGDTVTFSRTEPEGKLVMGYRKATNSTAT--------------------- 351
Query: 450 APQNLNSLSSFSNLLQSMKGNGEPYLNGHSEHLRLGNGTADWLKTANSEEEMNNGPLQRL 509
Q KG+ EP LN S L G G +W K SE+ + +
Sbjct: 352 ---------------QMFKGSSEPNLNMFSNSLNPGCGDINWSKLEKSEDMAKDNLFLQS 396
Query: 510 VSVSEKKRSRNIGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEE 569
S +KR RNIGTK+KRL I S D +EL++TWEEAQE L PP S +P+ T+E+Q FEE
Sbjct: 397 SLTSARKRVRNIGTKSKRLLIDSVDVLELKITWEEAQELLRPPQSTKPSIFTLENQDFEE 456
Query: 570 YDEPP 574
YD+ P
Sbjct: 457 YDKLP 461
>At4g21550 VP1 like protein
Length = 739
Score = 160 bits (404), Expect = 3e-39
Identities = 121/387 (31%), Positives = 183/387 (47%), Gaps = 47/387 (12%)
Query: 67 IHSGCIVSKSSFEYLDFGGITCVTCVK------PSQLCLNTENHNRFSQTTKNNASDQYG 120
IH GCI S S++ +D GGI C+ C + P + N S T N S Q
Sbjct: 80 IHCGCIASASAYTLMDAGGIECLACARKKFALSPISEKFKDLSINWSSSTRSNQISYQPP 139
Query: 121 EHIDGRLLVEQAGKGNLMQLCRIGEVGESSRWPQAQRDAMVSCIGPKTEEVKCQFNKEDT 180
+D +L Q G + A ++ + +C T E K N
Sbjct: 140 SCLDPSVL----------QFDFRNRGGNNEFSQPASKERVTAC----TMEKKRGMNDMIG 185
Query: 181 RFLNVMKHSSHLTAFTTLENNRPSWETKSIDETLSLKMALGTSSRNSVLPLATEI---GE 237
+ ++ ++ F + P +SLK + +P+ T I G
Sbjct: 186 KLMSENSKHYRVSPFPNVNVYHP---------LISLKEGPCGTQLAFPVPITTPIEKTGH 236
Query: 238 GKLEGKASSHFQQGQTSQSILAQLSKTGIAMNLETNKGMISHPPRRPPADVKGKNQLLSR 297
+L+G H + + L+ + ++ +++H + GK Q++ R
Sbjct: 237 SRLDGSNLWHTRNSSPLSRLHNDLNGGADSPFESKSRNVMAH------LETPGKYQVVPR 290
Query: 298 YWPRIT--DQELEKLSGDLKSTVVPLFE----KVLSPSDAGRIGRLVLPKACAEAFLPRI 351
+WP+++ +Q L+ S + S+++ K+LS +D G+ RLVLPK AEAFLP++
Sbjct: 291 FWPKVSYKNQVLQNQSKEYPSSLIDTTLEYNFKILSATDTGK--RLVLPKKYAEAFLPQL 348
Query: 352 LQSEGVPLQFQDIMGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDP 411
++GVPL QD MG EW FQFRFWP++ R+YVLEGVTP IQ+LQL AGDTV FSR+DP
Sbjct: 349 SHTKGVPLTVQDPMGKEWRFQFRFWPSSKGRIYVLEGVTPFIQTLQLQAGDTVIFSRLDP 408
Query: 412 GEKFLFGFRR-SLTSIVTQDASTSSHS 437
K + GFR+ S+T Q HS
Sbjct: 409 ERKLILGFRKASITQSSDQADPADMHS 435
Score = 63.5 bits (153), Expect = 3e-10
Identities = 31/69 (44%), Positives = 45/69 (64%)
Query: 513 SEKKRSRNIGTKTKRLHIHSEDAMELRLTWEEAQEFLCPPPSVEPNFVTIEDQVFEEYDE 572
S KK+S + T++KR + D L+LTWEEAQ FL PPP++ P+ V IED FEEY++
Sbjct: 456 SGKKKSSMMITRSKRQKVEKGDDNLLKLTWEEAQGFLLPPPNLTPSRVVIEDYEFEEYED 515
Query: 573 PPVFGKIKT 581
V ++ +
Sbjct: 516 LAVVSQVSS 524
>At3g26790 FUSCA3 (FUS3)
Length = 313
Score = 84.3 bits (207), Expect = 2e-16
Identities = 39/85 (45%), Positives = 56/85 (65%), Gaps = 1/85 (1%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMG-NEWTFQFRFWPNN 379
LF+K L SD + R++LPK AEA LP + EG+P++ +D+ G + WTF++R+WPNN
Sbjct: 91 LFQKELKNSDVSSLRRMILPKKAAEAHLPALECKEGIPIRMEDLDGFHVWTFKYRYWPNN 150
Query: 380 NSRMYVLEGVTPCIQSLQLNAGDTV 404
NSRMYVLE + + L GD +
Sbjct: 151 NSRMYVLENTGDFVNAHGLQLGDFI 175
>At3g24650 abscisic acid-insensitive protein 3
Length = 720
Score = 79.7 bits (195), Expect = 5e-15
Identities = 52/146 (35%), Positives = 78/146 (52%), Gaps = 8/146 (5%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDI-MGNEWTFQFRFWPNN 379
L +KVL SD G +GR+VLPK AE LP + +G+ L +DI W ++RFWPNN
Sbjct: 571 LLQKVLKQSDVGNLGRIVLPKKEAETHLPELEARDGISLAMEDIGTSRVWNMRYRFWPNN 630
Query: 380 NSRMYVLEGVTPCIQSLQLNAGD-TVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSN 438
SRMY+LE +++ L GD V +S + G+ + G + S +A SS +
Sbjct: 631 KSRMYLLENTGDFVKTNGLQEGDFIVIYSDVKCGKYLIRGVKVRQPSGQKPEAPPSSAAT 690
Query: 439 GILIKDTNFSGAPQNLNSLSSFSNLL 464
K N S +N+N+ S +N++
Sbjct: 691 ----KRQNKS--QRNINNNSPSANVV 710
>At1g28300 VP1 protein, putative
Length = 363
Score = 79.3 bits (194), Expect = 6e-15
Identities = 53/154 (34%), Positives = 82/154 (52%), Gaps = 11/154 (7%)
Query: 258 LAQLSKTGIAMNLETNKGMISHPPRRPPADVKGKNQLLS---RYWPRITDQE--LEKLSG 312
+A++++ M N S P P V K QL+ + +I+D++ ++ +
Sbjct: 107 MARINRKNAMMRSRNN----SSPNSSPSELVDSKRQLMMLNLKNNVQISDKKDSYQQSTF 162
Query: 313 DLKSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMG-NEWTF 371
D K V L EK L SD G +GR+VLPK AEA LP++ EG+ +Q +D+ W+F
Sbjct: 163 DNKKLRV-LCEKELKNSDVGSLGRIVLPKRDAEANLPKLSDKEGIVVQMRDVFSMQSWSF 221
Query: 372 QFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVT 405
+++FW NN SRMYVLE ++ GD +T
Sbjct: 222 KYKFWSNNKSRMYVLENTGEFVKQNGAEIGDFLT 255
>At2g36080 putative RAV2-like DNA binding protein
Length = 158
Score = 76.6 bits (187), Expect = 4e-14
Identities = 50/144 (34%), Positives = 77/144 (52%), Gaps = 9/144 (6%)
Query: 305 QELEKLSGDLKSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQS-----EGVPL 359
Q+ ++ D+ LFEK L+PSD G++ RLV+PK AE + P + +G+ L
Sbjct: 6 QQQQQHQNDVVEEKEALFEKPLTPSDVGKLNRLVIPKQHAERYFPLAAAAADAVEKGLLL 65
Query: 360 QFQDIMGNEWTFQFRFWPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSR-IDPGEKFLF 417
F+D G W F++ +W N+S+ YVL +G + ++ L+AGD V F R G +F
Sbjct: 66 CFEDEEGKPWRFRYSYW--NSSQSYVLTKGWSRYVKEKHLDAGDVVLFHRHRSDGGRFFI 123
Query: 418 GFRRSLTSIVTQDASTSSHSNGIL 441
G+RR S + D+ SN L
Sbjct: 124 GWRRRGDSSSSSDSYRHVQSNASL 147
>At5g06250 putative protein
Length = 282
Score = 74.7 bits (182), Expect = 1e-13
Identities = 57/167 (34%), Positives = 89/167 (53%), Gaps = 28/167 (16%)
Query: 310 LSGDLKSTVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLP--RILQS----------EGV 357
L DLK + LFEK L+PSD G++ RLV+PK AE + P +L S +G+
Sbjct: 37 LHDDLKES---LFEKSLTPSDVGKLNRLVIPKQHAEKYFPLNAVLVSSAAADTSSSEKGM 93
Query: 358 PLQFQDIMGNEWTFQFRFWPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSRIDPGEKFL 416
L F+D G W F++ +W N+S+ YVL +G + ++ QL+ GD V F R + L
Sbjct: 94 LLSFEDESGKSWRFRYSYW--NSSQSYVLTKGWSRFVKDKQLDPGDVVFFQRHRSDSRRL 151
Query: 417 F-GFRR----SLTSIVTQDASTSSHSNGILI-----KDTNFSGAPQN 453
F G+RR S +S+ +++ ++ S G L +N+S P +
Sbjct: 152 FIGWRRRGQGSSSSVAATNSAVNTSSMGALSYHQIHATSNYSNPPSH 198
>At4g01500 putative DNA-binding protein
Length = 328
Score = 74.3 bits (181), Expect = 2e-13
Identities = 39/89 (43%), Positives = 57/89 (63%), Gaps = 3/89 (3%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQFQDIMGNEWTFQFRFWPNNN 380
+F+KVL+PSD G++ RLV+PK AE F P G L FQD G W F++ +W N+
Sbjct: 30 MFDKVLTPSDVGKLNRLVIPKQHAENFFPLEDNQNGTVLDFQDKNGKMWRFRYSYW--NS 87
Query: 381 SRMYVL-EGVTPCIQSLQLNAGDTVTFSR 408
S+ YV+ +G + ++ +L AGDTV+F R
Sbjct: 88 SQSYVMTKGWSRFVKEKKLFAGDTVSFYR 116
>At2g46870 RAV-like B3 domain DNA binding protein
Length = 310
Score = 73.2 bits (178), Expect = 4e-13
Identities = 37/91 (40%), Positives = 61/91 (66%), Gaps = 5/91 (5%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQS--EGVPLQFQDIMGNEWTFQFRFWPN 378
+F+KV++PSD G++ RLV+PK AE F P S +G+ L F+D+ G W F++ +W
Sbjct: 34 MFDKVVTPSDVGKLNRLVIPKQHAERFFPLDSSSNEKGLLLNFEDLTGKSWRFRYSYW-- 91
Query: 379 NNSRMYVL-EGVTPCIQSLQLNAGDTVTFSR 408
N+S+ YV+ +G + ++ +L+AGD V+F R
Sbjct: 92 NSSQSYVMTKGWSRFVKDKKLDAGDIVSFQR 122
>At3g25730 AP2 domain transcription factor
Length = 333
Score = 72.4 bits (176), Expect = 7e-13
Identities = 41/107 (38%), Positives = 66/107 (61%), Gaps = 9/107 (8%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQS-----EGVPLQFQDIMGNEWTFQFRF 375
LFEK ++PSD G++ RLV+PK AE P L + +G+ L F+D+ G W F++ +
Sbjct: 182 LFEKTVTPSDVGKLNRLVIPKHQAEKHFPLPLGNNNVSVKGMLLNFEDVNGKVWRFRYSY 241
Query: 376 WPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSRI-DPGEKFLFGFR 420
W N+S+ YVL +G + ++ +L AGD ++F R D +KF G++
Sbjct: 242 W--NSSQSYVLTKGWSRFVKEKRLCAGDLISFKRSNDQDQKFFIGWK 286
>At1g13260 unknown protein
Length = 344
Score = 72.4 bits (176), Expect = 7e-13
Identities = 40/101 (39%), Positives = 63/101 (61%), Gaps = 6/101 (5%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLP---RILQSEGVPLQFQDIMGNEWTFQFRFWP 377
LFEK ++PSD G++ RLV+PK AE P + +GV L F+D+ G W F++ +W
Sbjct: 187 LFEKAVTPSDVGKLNRLVIPKHHAEKHFPLPSSNVSVKGVLLNFEDVNGKVWRFRYSYW- 245
Query: 378 NNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSRIDPGEKFLF 417
N+S+ YVL +G + ++ L AGD V+FSR + ++ L+
Sbjct: 246 -NSSQSYVLTKGWSRFVKEKNLRAGDVVSFSRSNGQDQQLY 285
>At1g68840 putative DNA-binding protein protein RAV2
Length = 352
Score = 72.0 bits (175), Expect = 1e-12
Identities = 41/102 (40%), Positives = 61/102 (59%), Gaps = 7/102 (6%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQS----EGVPLQFQDIMGNEWTFQFRFW 376
LFEK ++PSD G++ RLV+PK AE P S +GV + F+D+ G W F++ +W
Sbjct: 187 LFEKAVTPSDVGKLNRLVIPKQHAEKHFPLPSPSPAVTKGVLINFEDVNGKVWRFRYSYW 246
Query: 377 PNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSRIDPGEKFLF 417
N+S+ YVL +G + ++ L AGD VTF R E+ L+
Sbjct: 247 --NSSQSYVLTKGWSRFVKEKNLRAGDVVTFERSTGLERQLY 286
>At3g61970 RAV-like protein
Length = 299
Score = 69.7 bits (169), Expect = 5e-12
Identities = 36/94 (38%), Positives = 61/94 (64%), Gaps = 8/94 (8%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLP-----RILQSEGVPLQFQDIMGNEWTFQFRF 375
+F+KV++PSD G++ RLV+PK AE + P ++G+ L F+D GN W F++ +
Sbjct: 22 MFDKVVTPSDVGKLNRLVIPKQHAERYFPLDNSTTNDSNKGLLLNFEDRSGNSWRFRYSY 81
Query: 376 WPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSR 408
W N+S+ YV+ +G + ++ +L+AGD V+F R
Sbjct: 82 W--NSSQSYVMTKGWSRFVKDKKLDAGDIVSFQR 113
>At1g01030 DNA-binding protein, putative
Length = 358
Score = 68.2 bits (165), Expect = 1e-11
Identities = 36/91 (39%), Positives = 57/91 (62%), Gaps = 5/91 (5%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLP--RILQSEGVPLQFQDIMGNEWTFQFRFWPN 378
+F+KV++PSD G++ RLV+PK AE + P G L FQD G W F++ +W
Sbjct: 55 MFDKVVTPSDVGKLNRLVIPKQHAERYFPLDSSNNQNGTLLNFQDRNGKMWRFRYSYW-- 112
Query: 379 NNSRMYVL-EGVTPCIQSLQLNAGDTVTFSR 408
N+S+ YV+ +G + ++ +L+AGD V+F R
Sbjct: 113 NSSQSYVMTKGWSRFVKEKKLDAGDIVSFQR 143
>At3g11580 DNA binding protein like
Length = 267
Score = 65.9 bits (159), Expect = 7e-11
Identities = 49/145 (33%), Positives = 78/145 (53%), Gaps = 28/145 (19%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPR-------------ILQSEGVPLQFQDIMGN 367
LFEK L+PSD G++ RLV+PK AE + P +G+ L F+D G
Sbjct: 28 LFEKSLTPSDVGKLNRLVIPKQHAEKYFPLNNNNNNGGSGDDVATTEKGMLLSFEDESGK 87
Query: 368 EWTFQFRFWPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFS--RIDPGEKFLFGFRR--- 421
W F++ +W N+S+ YVL +G + ++ L+AGD V F R D F+ G+RR
Sbjct: 88 CWKFRYSYW--NSSQSYVLTKGWSRYVKDKHLDAGDVVFFQRHRFDLHRLFI-GWRRRGE 144
Query: 422 ----SLTSIVTQDA--STSSHSNGI 440
S+V+Q+A +T+++ +G+
Sbjct: 145 ASSSPAVSVVSQEALVNTTAYWSGL 169
>At1g25560 unknown protein
Length = 361
Score = 62.4 bits (150), Expect = 8e-10
Identities = 37/108 (34%), Positives = 58/108 (53%), Gaps = 13/108 (12%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLP----------RILQSEGVPLQFQDIMGNEWT 370
LFEK ++PSD G++ RLV+PK AE P ++GV + +D G W
Sbjct: 194 LFEKTVTPSDVGKLNRLVIPKQHAEKHFPLPAMTTAMGMNPSPTKGVLINLEDRTGKVWR 253
Query: 371 FQFRFWPNNNSRMYVL-EGVTPCIQSLQLNAGDTVTFSRIDPGEKFLF 417
F++ +W N+S+ YVL +G + ++ L AGD V F R ++ L+
Sbjct: 254 FRYSYW--NSSQSYVLTKGWSRFVKEKNLRAGDVVCFERSTGPDRQLY 299
>At1g50680 hypothetical protein
Length = 337
Score = 54.3 bits (129), Expect = 2e-07
Identities = 39/151 (25%), Positives = 67/151 (43%), Gaps = 27/151 (17%)
Query: 321 LFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQS-------------EGVPLQFQDIMGN 367
LF+K L+PSD G++ RLV+PK A ++P I E V + F D
Sbjct: 156 LFQKELTPSDVGKLNRLVIPKKYAVKYMPFISADQSEKEEGEIVGSVEDVEVVFYDRAMR 215
Query: 368 EWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRID--PGEKFLFGFRRSLT- 424
+W F++ +W ++ S ++ G ++ L D + F D K L G R++
Sbjct: 216 QWKFRYCYWKSSQSFVFT-RGWNSFVKEKNLKEKDVIAFYTCDVPNNVKTLEGQRKNFLM 274
Query: 425 ----------SIVTQDASTSSHSNGILIKDT 445
S+V ++ S + H + + +K T
Sbjct: 275 IDVHCFSDNGSVVAEEVSMTVHDSSVQVKKT 305
>At2g28350 unknown protein
Length = 693
Score = 53.9 bits (128), Expect = 3e-07
Identities = 45/153 (29%), Positives = 71/153 (45%), Gaps = 13/153 (8%)
Query: 322 FEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQ---FQDIMGNEWTFQFRFWPN 378
F K L+ SDA G +P+ CAE PR+ S P+Q +DI G W F+ +
Sbjct: 115 FAKTLTQSDANNGGGFSVPRYCAETIFPRLDYSAEPPVQTVIAKDIHGETWKFR-HIYRG 173
Query: 379 NNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRRSLTSIVTQDASTSSHSN 438
R + G + + +L AGD++ F R + G+ G RR+ + +A + +
Sbjct: 174 TPRRHLLTTGWSTFVNQKKLIAGDSIVFLRSESGD-LCVGIRRAKRGGLGSNAGSDNPYP 232
Query: 439 GILIKDTNFSGAPQNLNSLSSFSNLLQSMKGNG 471
G FSG ++ S ++ S L+ MK NG
Sbjct: 233 G-------FSGFLRDDESTTTTSKLMM-MKRNG 257
>At1g77850 putative auxin response factor protein (T32E8.16)
Length = 585
Score = 51.6 bits (122), Expect = 1e-06
Identities = 41/130 (31%), Positives = 62/130 (47%), Gaps = 8/130 (6%)
Query: 309 KLSGDLK-STVVPLFEKVLSPSDAGRIGRLVLPKACAEAFLPRILQSEGVPLQ---FQDI 364
+ GD+ + V F K+L+PSDA G +P+ CA++ P + P+Q DI
Sbjct: 105 RFDGDVDDNNKVTTFAKILTPSDANNGGGFSVPRFCADSVFPLLNFQIDPPVQKLYVTDI 164
Query: 365 MGNEWTFQFRFWPNNNSRMYVLEGVTPCIQSLQLNAGDTVTFSRIDPGEKFLFGFRRSLT 424
G W F+ + R + G + + S +L AGD+V F R E F+ G RR T
Sbjct: 165 HGAVWDFR-HIYRGTPRRHLLTTGWSKFVNSKKLIAGDSVVFMRKSADEMFI-GVRR--T 220
Query: 425 SIVTQDASTS 434
I + D +S
Sbjct: 221 PISSSDGGSS 230
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,106,022
Number of Sequences: 26719
Number of extensions: 670526
Number of successful extensions: 1925
Number of sequences better than 10.0: 57
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 1858
Number of HSP's gapped (non-prelim): 64
length of query: 648
length of database: 11,318,596
effective HSP length: 106
effective length of query: 542
effective length of database: 8,486,382
effective search space: 4599619044
effective search space used: 4599619044
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC141923.5


