

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141863.4 + phase: 0 /partial
(1196 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
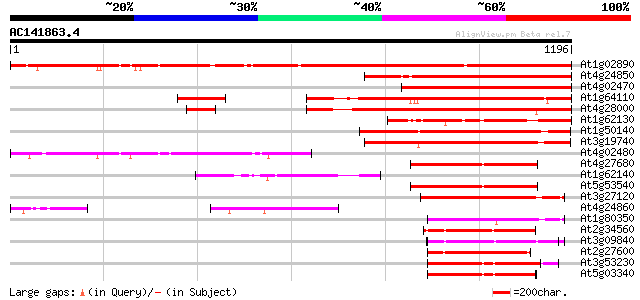
Score E
Sequences producing significant alignments: (bits) Value
At1g02890 hypothetical protein 1057 0.0
At4g24850 unknown protein (At4g24850) 623 e-178
At4g02470 602 e-172
At1g64110 unknown protein 479 e-135
At4g28000 putative protein 475 e-134
At1g62130 hypothetical protein 451 e-127
At1g50140 hypothetical protein, 5' partial 431 e-120
At3g19740 ATPase, putative 422 e-118
At4g02480 326 6e-89
At4g27680 unknown protein 280 3e-75
At1g62140 hypothetical protein 279 8e-75
At5g53540 26S proteasome regulatory particle chain RPT6-like pro... 276 7e-74
At3g27120 spastin protein, putative 240 3e-63
At4g24860 putative protein 226 5e-59
At1g80350 CAD ATPase (AAA1) 215 1e-55
At2g34560 katanin like protein 204 3e-52
At3g09840 putative transitional endoplasmic reticulum ATPase 183 4e-46
At2g27600 putative AAA-type ATPase 179 8e-45
At3g53230 CDC48 - like protein 179 1e-44
At5g03340 transitional endoplasmic reticulum ATPase 178 1e-44
>At1g02890 hypothetical protein
Length = 1240
Score = 1057 bits (2734), Expect = 0.0
Identities = 629/1244 (50%), Positives = 807/1244 (64%), Gaps = 86/1244 (6%)
Query: 1 PDNGAASSEKPPENSNPEPSADPGKCA-QPDAQIDEPVAAADDDKADTTPPIADASTPT- 58
P + +++SE P +N P +DPG + P+ + +P + D ++ TT + T T
Sbjct: 34 PASSSSASEVPIDNQ--APVSDPGSISGDPELRTSDP-QSNDAERPVTTTDVPAMETDTN 90
Query: 59 -----LVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNFTIGSSR 113
LV P + K + + APW +LLSQ Q+P++ + FT+G R
Sbjct: 91 PELEGLVTPTPAGEVVVEAEKSKSSKKRIAKAPWAKLLSQFPQNPHLVMRGSVFTVGR-R 149
Query: 114 NCNFHLKDHTISGNLCKIKHTQREGSDVAVLESTGSKGSVIVNGTLVKKSTCCTLNSGDE 173
C+ ++DH++ LC+++ ++ G VA LE G+ V VNG + ++STC L GDE
Sbjct: 150 ACDLCIRDHSMPNVLCELRQSEHGGPSVASLEIIGNGVLVQVNGKIYQRSTCVHLRGGDE 209
Query: 174 VVFGLHGNHSY-----------IFQQVNTE--------VAVKGAEVQSGIGKFMQLERRS 214
++F G H+Y IFQ + E ++ E QS K + +E R+
Sbjct: 210 IIFTTPGKHAYVSFYKFFENVQIFQPLKDENLAAPDRTSSLSLFEAQSAPLKGLHVETRA 269
Query: 215 GDPSAVAG-ASILASLSNLRQDLTRWKSPSQTASKPHQGADVSIHTVLPDGTE-----IE 268
D S+V G AS+LAS+S L+ + P+ + K Q ++V VLP + ++
Sbjct: 270 RDSSSVDGTASLLASISKLQN--VPFLPPTAKSVKRQQNSEVP---VLPSSCDDFILDVD 324
Query: 269 LDGLGNSTP-----SMGTDKAADAEASNKNTPMDCDPEDAGAEP--GNVKYSGVNDLLRP 321
L+ ++ SM A+ + A+N + D + D EP GN+ +RP
Sbjct: 325 LNDADSNNDHAAIASMEKTVASTSCAANDDHDADGNGMDPFQEPEAGNIPDPAYE--IRP 382
Query: 322 FFRILAGSTTCKLK--LSKSICKQVLEERNGAEDTQAASTSGTSVRCAVFKEDAHAAILD 379
+L + L+ +SK + + E R ++ + S S + R A K+ IL+
Sbjct: 383 ILSLLGDPSEFDLRGSISKILVDERREVREMPKEYERPSASVLTRRQA-HKDSLRGGILN 441
Query: 380 GKEQEVSFDNFPYYLSENTKNVLIAACFIHLKH-KEHAKYTADLPTVNPRILLSGPAGSE 438
++ EVSF+NFPY+LS TK+VL+ + + H+K+ KE+A+Y +DLPT PR SE
Sbjct: 442 PQDIEVSFENFPYFLSGTTKDVLMISTYAHIKYGKEYAEYASDLPTACPR--------SE 493
Query: 439 IYSEMLVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDM 498
IY EML KALAK GAKL+I DS LL GG + KEA+ K+ E+ K++ A
Sbjct: 494 IYQEMLAKALAKQCGAKLMIVDSLLLPGGSTPKEADTTKESSRRERLSVLAKRAVQAAQA 553
Query: 499 AKSTDPPASETDTPSSSNVPTPLGLESQAKLETDSVPS---TSGTAKNCLFKLGDRVKYS 555
A + P SS G+ + L + +V ++ T+K+ FK GDRV++
Sbjct: 554 A------VLQHKKPISS---VEAGITGGSTLSSQAVRRQEVSTATSKSYTFKAGDRVRFL 604
Query: 556 SSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEAGQG 615
S + + +GP+ G +GKV+L F+ N SKIGVRFD+ IPDG DLG CE
Sbjct: 605 GPSTSSLASLRAPPRGPATGFQGKVLLAFEGNGSSKIGVRFDRSIPDGNDLGGLCEEDHA 664
Query: 616 FFCNITDLRLENSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPY-SF 674
+ LRLE+S D+ DK IN +FEV +ES ILF+K+ EKS+ GN D Y +
Sbjct: 665 -----SSLRLESSSSDDADKLAINEIFEVAFNESERGSLILFLKDIEKSVSGNTDVYITL 719
Query: 675 KSKLEKLPDNVVVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFG-RLHDR 733
KSKLE LP+N+VVI S T D+RKEKSH GG LFTKFGSNQTALLDLAFPD+FG RL DR
Sbjct: 720 KSKLENLPENIVVIASQTQLDNRKEKSHPGGFLFTKFGSNQTALLDLAFPDTFGGRLQDR 779
Query: 734 GKEVPKPNKTLTKLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRS 793
E+PK K +T+LFPNKVTI +P+DEA L WK +L+RD E LK + N+ +R VLS++
Sbjct: 780 NTEMPKAVKQITRLFPNKVTIQLPEDEASLVDWKDKLERDTEILKAQANITSIRAVLSKN 839
Query: 794 GMESDGLESLCVKDLTLTNENSEKILGWALSHHLMQNPEADA-DAKLVLSSESIQYGIGI 852
+ +E LC+KD TL +++ EK++G+A +HHLM E D KL++S+ESI YG+ +
Sbjct: 840 QLVCPDIEILCIKDQTLPSDSVEKVVGFAFNHHLMNCSEPTVKDNKLIISAESITYGLQL 899
Query: 853 FQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVM 912
IQNE+KS KKSLKDVVTENEFEK+LL DVIPP+DIGV+F DIGALENVKDTLKELVM
Sbjct: 900 LHEIQNENKSTKKSLKDVVTENEFEKKLLSDVIPPSDIGVSFSDIGALENVKDTLKELVM 959
Query: 913 LPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFG 972
LPLQRPELF KGQLTKP KGILLFGPPGTGKTMLAKAVAT+AGANFINISMSSITSK
Sbjct: 960 LPLQRPELFGKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSK--- 1016
Query: 973 EGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKD 1032
GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFM+NWDGLRTKD
Sbjct: 1017 -GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKD 1075
Query: 1033 TERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAI 1092
ERV+VLAATNRP+DLDEAVIRRLPRRLMVNLPD+ NR+KIL VILAKE+++ DVDL AI
Sbjct: 1076 KERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDSANRSKILSVILAKEEMAEDVDLEAI 1135
Query: 1093 ANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNME 1152
ANMTDGYSGSDLKNLCVTAAH PI+EILEKEKKE + A AE R P L S D+R LNM
Sbjct: 1136 ANMTDGYSGSDLKNLCVTAAHLPIREILEKEKKERSVAQAENRAMPQLYSSTDVRPLNMN 1195
Query: 1153 DFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
DFK AH QVCASV+S+S NM EL QWNELYGEGGSR K +LSYF
Sbjct: 1196 DFKTAHDQVCASVASDSSNMNELQQWNELYGEGGSRKKTSLSYF 1239
>At4g24850 unknown protein (At4g24850)
Length = 442
Score = 623 bits (1607), Expect = e-178
Identities = 316/445 (71%), Positives = 371/445 (83%), Gaps = 8/445 (1%)
Query: 756 MPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLES----LCVKDLTLT 811
MPQDE L WK Q+DRD ET K+K N +HLR VL R G+ +GLE+ +C+KDLTL
Sbjct: 1 MPQDEKRLTLWKHQMDRDAETSKVKSNFNHLRMVLRRRGLGCEGLETTWSRMCLKDLTLQ 60
Query: 812 NENSEKILGWALSHHLMQNPEADADAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVV 871
++ EKI+GWA +H+ +NP+ D AK+ LS ESI++GIG+ +QN+ K S KD+V
Sbjct: 61 RDSVEKIIGWAFGNHISKNPDTDP-AKVTLSRESIEFGIGL---LQNDLKGSTSSKKDIV 116
Query: 872 TENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCK 931
EN FEKRLL DVI P+DI VTFDDIGALE VKD LKELVMLPLQRPELFCKG+LTKPCK
Sbjct: 117 VENVFEKRLLSDVILPSDIDVTFDDIGALEKVKDILKELVMLPLQRPELFCKGELTKPCK 176
Query: 932 GILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPS 991
GILLFGPPGTGKTMLAKAVA +A ANFINISMSSITSKWFGEGEKYVKAVFSLASK++PS
Sbjct: 177 GILLFGPPGTGKTMLAKAVAKEADANFINISMSSITSKWFGEGEKYVKAVFSLASKMSPS 236
Query: 992 VIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEA 1051
VIFVDEVDSMLGRRE+P EHEA RK+KNEFM++WDGL T++ ERV+VLAATNRP+DLDEA
Sbjct: 237 VIFVDEVDSMLGRREHPREHEASRKIKNEFMMHWDGLTTQERERVLVLAATNRPFDLDEA 296
Query: 1052 VIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTA 1111
VIRRLPRRLMV LPD NRA ILKVILAKEDLS D+D+G IA+MT+GYSGSDLKNLCVTA
Sbjct: 297 VIRRLPRRLMVGLPDTSNRAFILKVILAKEDLSPDLDIGEIASMTNGYSGSDLKNLCVTA 356
Query: 1112 AHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVN 1171
AHRPIKEILEKEK+E AA+A+G+ P L GS D+R+LN+EDF+ AH+ V ASVSSES
Sbjct: 357 AHRPIKEILEKEKRERDAALAQGKVPPPLSGSSDLRALNVEDFRDAHKWVSASVSSESAT 416
Query: 1172 MTELVQWNELYGEGGSRVKKALSYF 1196
MT L QWN+L+GEGGS +++ S++
Sbjct: 417 MTALQQWNKLHGEGGSGKQQSFSFY 441
>At4g02470
Length = 371
Score = 602 bits (1551), Expect = e-172
Identities = 307/361 (85%), Positives = 326/361 (90%)
Query: 836 DAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFD 895
D KLV+S+ESI YG+ IQNE+KSLKKSLKDVVTENEFEK+LL DVIPP+DIGV+FD
Sbjct: 10 DNKLVISAESISYGLQTLHDIQNENKSLKKSLKDVVTENEFEKKLLSDVIPPSDIGVSFD 69
Query: 896 DIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAG 955
DIGALENVK+TLKELVMLPLQRPELF KGQLTKP KGILLFGPPGTGKTMLAKAVAT+AG
Sbjct: 70 DIGALENVKETLKELVMLPLQRPELFDKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAG 129
Query: 956 ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR 1015
ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR
Sbjct: 130 ANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMR 189
Query: 1016 KMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILK 1075
KMKNEFMVNWDGLRTKD ERV+VLAATNRP+DLDEAVIRRLPRRLMVNLPDA NR+KIL
Sbjct: 190 KMKNEFMVNWDGLRTKDRERVLVLAATNRPFDLDEAVIRRLPRRLMVNLPDATNRSKILS 249
Query: 1076 VILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGR 1135
VILAKE+++ DVDL AIANMTDGYSGSDLKNLCVTAAH PI+EILEKEKKE AA AE R
Sbjct: 250 VILAKEEIAPDVDLEAIANMTDGYSGSDLKNLCVTAAHFPIREILEKEKKEKTAAQAENR 309
Query: 1136 PAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSY 1195
P P L D+RSL M DFK AH QVCASVSS+S NM EL QWNELYGEGGSR K +LSY
Sbjct: 310 PTPPLYSCTDVRSLTMNDFKAAHDQVCASVSSDSSNMNELQQWNELYGEGGSRKKTSLSY 369
Query: 1196 F 1196
F
Sbjct: 370 F 370
>At1g64110 unknown protein
Length = 824
Score = 479 bits (1234), Expect = e-135
Identities = 271/619 (43%), Positives = 388/619 (61%), Gaps = 96/619 (15%)
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHT 692
+K L+ +L++V+ S+ +P +L++++ E + + Y+ F+ L+KL V+++GS
Sbjct: 245 EKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQKLLQKLSGPVLILGSR- 303
Query: 693 HSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKV 752
++DL+ D+ +E+ ++ L+ +FP +
Sbjct: 304 -------------------------IVDLSSEDA--------QEI---DEKLSAVFPYNI 327
Query: 753 TIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTN 812
I P+DE L SWK QL+RD+ ++ + N +H+ VLS + + D LES+ +D + +
Sbjct: 328 DIRPPEDETHLVSWKSQLERDMNMIQTQDNRNHIMEVLSENDLICDDLESISFEDTKVLS 387
Query: 813 ENSEKILGWALSHHLMQNPEAD-ADAKLVLSSESIQYGIGIF------------QAIQNE 859
E+I+ ALS+HLM N + + + KLV+SS S+ +G +F Q + E
Sbjct: 388 NYIEEIVVSALSYHLMNNKDPEYRNGKLVISSISLSHGFSLFREGKAGGREKLKQKTKEE 447
Query: 860 S------KSLKKSLK---------------------------DVVTENEFEKRLLGDVIP 886
S +S+K K +V +NEFEKR+ +VIP
Sbjct: 448 SSKEVKAESIKPETKTESVTTVSSKEEPEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIP 507
Query: 887 PNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTML 946
+I VTF DIGAL+ +K++L+ELVMLPL+RP+LF G L KPC+GILLFGPPGTGKTML
Sbjct: 508 AEEINVTFKDIGALDEIKESLQELVMLPLRRPDLFTGG-LLKPCRGILLFGPPGTGKTML 566
Query: 947 AKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRE 1006
AKA+A +AGA+FIN+SMS+ITSKWFGE EK V+A+F+LASK++P++IFVDEVDSMLG+R
Sbjct: 567 AKAIAKEAGASFINVSMSTITSKWFGEDEKNVRALFTLASKVSPTIIFVDEVDSMLGQRT 626
Query: 1007 NPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPD 1066
GEHEAMRK+KNEFM +WDGL TK ER++VLAATNRP+DLDEA+IRR RR+MV LP
Sbjct: 627 RVGEHEAMRKIKNEFMSHWDGLMTKPGERILVLAATNRPFDLDEAIIRRFERRIMVGLPA 686
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE 1126
NR KIL+ +LAKE + ++D +A MT+GY+GSDLKNLC TAA+RP++E++++E+ +
Sbjct: 687 VENREKILRTLLAKEKVDENLDYKELAMMTEGYTGSDLKNLCTTAAYRPVRELIQQERIK 746
Query: 1127 LAAAVAEGRPAPALRGSDD---------IRSLNMEDFKHAHQQVCASVSSESVNMTELVQ 1177
+ P A G +D +R LN +DFK A QV AS ++E M EL Q
Sbjct: 747 DTEKKKQREPTKA--GEEDEGKEERVITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQ 804
Query: 1178 WNELYGEGGSRVKKALSYF 1196
WNELYGEGGSR K+ L+YF
Sbjct: 805 WNELYGEGGSRKKEQLTYF 823
Score = 97.1 bits (240), Expect = 6e-20
Identities = 47/103 (45%), Positives = 71/103 (68%), Gaps = 2/103 (1%)
Query: 358 STSGTSVRCAVFKEDAHAAILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAK 417
S+S +V +++ ++DG+E +++FD FPYYLSE T+ +L +A ++HLKH + +K
Sbjct: 32 SSSNNAVTADKMEKEILRQVVDGRESKITFDEFPYYLSEQTRVLLTSAAYVHLKHFDASK 91
Query: 418 YTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFGAKLLIFD 460
YT +L + ILLSGPA E+Y +ML KALA +F AKLL+ D
Sbjct: 92 YTRNLSPASRAILLSGPA--ELYQQMLAKALAHFFDAKLLLLD 132
Score = 32.3 bits (72), Expect = 1.7
Identities = 32/139 (23%), Positives = 60/139 (43%), Gaps = 8/139 (5%)
Query: 839 LVLSSESIQYGIGIFQAIQNESKSLK-----KSLKDVVTENEFEKRLLGDVIPPNDIGVT 893
++LS+ + G+G+ + + K S + VT ++ EK +L V+ + +T
Sbjct: 1 MLLSALGVGVGVGVGLGLASGQAVGKWAGGNSSSNNAVTADKMEKEILRQVVDGRESKIT 60
Query: 894 FDDIGAL--ENVKDTLKELVMLPLQRPELFCKGQLTKPC-KGILLFGPPGTGKTMLAKAV 950
FD+ E + L + L+ + + P + ILL GP + MLAKA+
Sbjct: 61 FDEFPYYLSEQTRVLLTSAAYVHLKHFDASKYTRNLSPASRAILLSGPAELYQQMLAKAL 120
Query: 951 ATDAGANFINISMSSITSK 969
A A + + ++ K
Sbjct: 121 AHFFDAKLLLLDVNDFALK 139
>At4g28000 putative protein
Length = 726
Score = 475 bits (1223), Expect = e-134
Identities = 261/581 (44%), Positives = 375/581 (63%), Gaps = 58/581 (9%)
Query: 634 DKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYSFKSKLEKLPDNVVVIGSHTH 693
++ + +L++V+ S S +P I+++++ EK F+ L KL V+V+GS
Sbjct: 185 ERLFLQSLYKVLVSISETNPIIIYLRDVEKLCQSERFYKLFQRLLTKLSGPVLVLGS--- 241
Query: 694 SDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLTKLFPNKVT 753
RL + + + + ++ LFP +
Sbjct: 242 -----------------------------------RLLEPEDDCQEVGEGISALFPYNIE 266
Query: 754 IHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNE 813
I P+DE L SWK + + D++ ++ + N +H+ VL+ + +E D L S+C D +
Sbjct: 267 IRPPEDENQLMSWKTRFEDDMKVIQFQDNKNHIAEVLAANDLECDDLGSICHADTMFLSS 326
Query: 814 NSEKILGWALSHHLMQNPEAD-ADAKLVLSSESIQYGIGIFQAIQN---ESKSLKKSL-- 867
+ E+I+ A+S+HLM N E + + +LV+SS S+ +G+ I Q Q +S L ++
Sbjct: 327 HIEEIVVSAISYHLMNNKEPEYKNGRLVISSNSLSHGLNILQEGQGCFEDSLKLDTNIDS 386
Query: 868 KDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLT 927
K+V +NEFEKR+ +VIP N+IGVTF DIG+L+ K++L+ELVMLPL+RP+LF KG L
Sbjct: 387 KEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPLRRPDLF-KGGLL 445
Query: 928 KPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASK 987
KPC+GILLFGPPGTGKTM+AKA+A +AGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K
Sbjct: 446 KPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 505
Query: 988 IAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYD 1047
++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM +WDGL + +R++VLAATNRP+D
Sbjct: 506 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRILVLAATNRPFD 565
Query: 1048 LDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNL 1107
LDEA+IRR RR+MV LP +R KIL+ +L+KE + ++D +A MTDGYSGSDLKN
Sbjct: 566 LDEAIIRRFERRIMVGLPSVESREKILRTLLSKEK-TENLDFQELAQMTDGYSGSDLKNF 624
Query: 1108 CVTAAHRPIKEIL--------EKEKKELAAAVAEGRPAPALRGSDD----IRSLNMEDFK 1155
C TAA+RP++E++ E+ K+E A +E S++ +R L+MED K
Sbjct: 625 CTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGITLRPLSMEDMK 684
Query: 1156 HAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
A QV AS ++E M EL QWN+LYGEGGSR K+ LSYF
Sbjct: 685 VAKSQVAASFAAEGAGMNELKQWNDLYGEGGSRKKEQLSYF 725
Score = 68.2 bits (165), Expect = 3e-11
Identities = 31/62 (50%), Positives = 45/62 (72%)
Query: 377 ILDGKEQEVSFDNFPYYLSENTKNVLIAACFIHLKHKEHAKYTADLPTVNPRILLSGPAG 436
I+DG+E V+FD FPYYLSE T+ +L +A ++HLK + +K+T +L + ILLSGPA
Sbjct: 54 IVDGRESSVTFDEFPYYLSEKTRLLLTSAAYVHLKQSDISKHTRNLAPGSKAILLSGPAD 113
Query: 437 SE 438
+E
Sbjct: 114 TE 115
>At1g62130 hypothetical protein
Length = 372
Score = 451 bits (1161), Expect = e-127
Identities = 249/412 (60%), Positives = 297/412 (71%), Gaps = 61/412 (14%)
Query: 805 VKDLTLTNENSEKILGWALSHHLMQNPEADADAKLVLSSESIQYGIGIFQAIQNESKSLK 864
+KDLTL +++EKI+GWALSHH+ NP AD D +++LS ES++ GI + ++ ESK
Sbjct: 1 MKDLTLRRDSAEKIIGWALSHHIKSNPGADPDVRVILSLESLKCGI---ELLEIESK--- 54
Query: 865 KSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKG 924
KSLKD+VTEN FE + D+IPP++IGVTFDDIGALENVKDTLKELVMLP Q PELFCKG
Sbjct: 55 KSLKDIVTENTFE---ISDIIPPSEIGVTFDDIGALENVKDTLKELVMLPFQWPELFCKG 111
Query: 925 QLTK--------------------PCKGILLFGPPGTGKTMLAKAVATDAGANFINISMS 964
QLTK PC GILLFGP GTGKTMLAKAVAT+AGAN IN+SMS
Sbjct: 112 QLTKMLTLWIGGFLISLLLYFSTQPCNGILLFGPSGTGKTMLAKAVATEAGANLINMSMS 171
Query: 965 SITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVN 1024
+WF EGEKYVKAVFSLASKI+PS+IF+DEV+SML H K KNEF++N
Sbjct: 172 ----RWFSEGEKYVKAVFSLASKISPSIIFLDEVESML--------HRYRLKTKNEFIIN 219
Query: 1025 WDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLS 1084
WDGLRT + ERV+VLAATNRP+DLDEAVIRRLP RLMV LPDA +R+KILKVIL+KEDLS
Sbjct: 220 WDGLRTNEKERVLVLAATNRPFDLDEAVIRRLPHRLMVGLPDARSRSKILKVILSKEDLS 279
Query: 1085 SDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSD 1144
D D+ +A+MT+GYSG+DL KE AAVAEGR PA G
Sbjct: 280 PDFDIDEVASMTNGYSGNDL--------------------KERDAAVAEGRVPPAGSGGS 319
Query: 1145 DIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELYGEGGSRVKKALSYF 1196
D+R L MEDF++A + V S+SS+SVNMT L QWNE YGEGGSR ++ S +
Sbjct: 320 DLRVLKMEDFRNALELVSMSISSKSVNMTALRQWNEDYGEGGSRRNESFSQY 371
>At1g50140 hypothetical protein, 5' partial
Length = 601
Score = 431 bits (1108), Expect = e-120
Identities = 216/451 (47%), Positives = 309/451 (67%), Gaps = 13/451 (2%)
Query: 746 KLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCV 805
KLF N + +H P++E L +K+QL D + + N++ L L + L +
Sbjct: 163 KLFTNVMRLHPPKEEDTLRLFKKQLGEDRRIVISRSNINELLKALEEHELLCTDLYQVNT 222
Query: 806 KDLTLTNENSEKILGWALSHHLMQNP-EADADAKLVLSSESIQYGIGIFQAIQNESKSLK 864
+ LT + +EK +GWA +H+L P +L L ES++ I + +++ S
Sbjct: 223 DGVILTKQKAEKAIGWAKNHYLASCPVPLVKGGRLSLPRESLEISIARLRKLEDNSLKPS 282
Query: 865 KSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKG 924
++LK++ ++E+E+ + V+ P +IGV F+DIGALE+VK L ELV+LP++RPELF +G
Sbjct: 283 QNLKNIA-KDEYERNFVSAVVAPGEIGVKFEDIGALEDVKKALNELVILPMRRPELFARG 341
Query: 925 QLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSL 984
L +PCKGILLFGPPGTGKT+LAKA+AT+AGANFI+I+ S++TSKWFG+ EK KA+FS
Sbjct: 342 NLLRPCKGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSF 401
Query: 985 ASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNR 1044
A+K+AP +IFVDE+DS+LG R EHEA R+M+NEFM WDGLR+KD++R+++L ATNR
Sbjct: 402 ATKLAPVIIFVDEIDSLLGARGGSSEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNR 461
Query: 1045 PYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDL 1104
P+DLD+AVIRRLPRR+ V+LPDA NR KILK+ L E+L SD +A T+GYSGSDL
Sbjct: 462 PFDLDDAVIRRLPRRIYVDLPDAENRLKILKIFLTPENLESDFQFEKLAKETEGYSGSDL 521
Query: 1105 KNLCVTAAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCAS 1164
KNLC+ AA+RP++E+L++E+K A + G +RSL+++DF + +V S
Sbjct: 522 KNLCIAAAYRPVQELLQEEQKGARAEASPG-----------LRSLSLDDFIQSKAKVSPS 570
Query: 1165 VSSESVNMTELVQWNELYGEGGSRVKKALSY 1195
V+ ++ M EL +WNE YGEGGSR K +
Sbjct: 571 VAYDATTMNELRKWNEQYGEGGSRTKSPFGF 601
>At3g19740 ATPase, putative
Length = 439
Score = 422 bits (1086), Expect = e-118
Identities = 213/445 (47%), Positives = 303/445 (67%), Gaps = 17/445 (3%)
Query: 757 PQDEALLASWKQQLDRDVETLKIKGNLHHLRTVLSRSGMESDGLESLCVKDLTLTNENSE 816
P++E L + +QL D + + NL+ L L + + L + + LT + +E
Sbjct: 6 PKEEENLIVFNKQLGEDRRIVMSRSNLNELLKALEENELLCTDLYQVNTDGVILTKQRAE 65
Query: 817 KILGWALSHHLMQNPEADA-DAKLVLSSESIQYGIGIFQAIQNESKSLKKSLKDV----- 870
K++GWA +H+L P + +L+L ESI+ + +A ++ S+ ++LK
Sbjct: 66 KVIGWARNHYLSSCPSPSIKEGRLILPRESIEISVKRLKAQEDISRKPTQNLKRFLFLQN 125
Query: 871 VTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPC 930
+ ++EFE + V+ P +IGV FDDIGALE+VK TL ELV+LP++RPELF +G L +PC
Sbjct: 126 IAKDEFETNFVSAVVAPGEIGVKFDDIGALEHVKKTLNELVILPMRRPELFTRGNLLRPC 185
Query: 931 KGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAP 990
KGILLFGPPGTGKT+LAKA+AT+AGANFI+I+ S++TSKWFG+ EK KA+FS ASK+AP
Sbjct: 186 KGILLFGPPGTGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFASKLAP 245
Query: 991 SVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDE 1050
+IFVDEVDS+LG R EHEA R+M+NEFM WDGLR+KD++R+++L ATNRP+DLD+
Sbjct: 246 VIIFVDEVDSLLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRPFDLDD 305
Query: 1051 AVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVT 1110
AVIRRLPRR+ V+LPDA NR KILK+ L E+L + + +A T+GYSGSDLKNLC+
Sbjct: 306 AVIRRLPRRIYVDLPDAENRLKILKIFLTPENLETGFEFDKLAKETEGYSGSDLKNLCIA 365
Query: 1111 AAHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESV 1170
AA+RP++E+L++E K+ S D+R L+++DF + +V SV+ ++
Sbjct: 366 AAYRPVQELLQEENKD-----------SVTNASPDLRPLSLDDFIQSKAKVSPSVAYDAT 414
Query: 1171 NMTELVQWNELYGEGGSRVKKALSY 1195
M EL +WNE YGEGG+R K +
Sbjct: 415 TMNELRKWNEQYGEGGTRTKSPFGF 439
>At4g02480
Length = 731
Score = 326 bits (835), Expect = 6e-89
Identities = 239/691 (34%), Positives = 351/691 (50%), Gaps = 70/691 (10%)
Query: 1 PDNGAASSEKPPENSNP--EPSADPGK----CAQPDAQ-IDEPVAAAD------DDKADT 47
P +++SE P EN P +P ++ G+ + P A ++PV D + D
Sbjct: 51 PAGSSSASEVPIENQGPASDPGSESGEPELGSSDPQAMDAEKPVVTTDVPVMENSPETDA 110
Query: 48 TPPIADASTPTLVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSAQHPNVSICIPNF 107
P + +TPT+ + + S K APW +LLSQ +Q+P+ I P F
Sbjct: 111 NPEVEVLATPTVAGEAVADADKS-----KAAKKRALKAPWAKLLSQYSQNPHRVIRGPVF 165
Query: 108 TIGSSRNCNFHLKDHTISGNLCKIKHT----QREGSDVAVLESTGSKGSVIVNGTLVKKS 163
T+G R C+ ++D + LC++K + Q G VA LE G+ V VNG +KS
Sbjct: 166 TVGR-RGCDLSIRDQAMPSTLCELKQSESLLQHGGPSVASLEILGNGVIVHVNGKCYQKS 224
Query: 164 TCCTLNSGDEVVFGLHGNHSYIF-----QQVNTEVAVKGAEVQSGIGKFMQLERRSGD-- 216
TC L GDEV+F L+G H+Y+ + + ++ E + K + +E R+GD
Sbjct: 225 TCVHLRGGDEVIFSLNGKHAYVSFLELRETPDRASSLSICEARGAPLKGVHVETRAGDVD 284
Query: 217 -PSAVAGASILASLSNLRQDLTRWKSPSQTASKPHQGADV-----SIHTVLPDGTEIELD 270
S V GASILASLS LR P A K Q V S + + D + D
Sbjct: 285 GASDVDGASILASLSKLRS--FHLLPPIAKAGKRQQNPAVPVVPSSFNDCISDTDMNDAD 342
Query: 271 GLGNSTPSMGTDKAADAEA---SNKNTPMDCDPEDA--GAEPGNVKYSGVNDLLRPFFRI 325
+ +K A A +N+N +D D A+ GNV +G +RP +
Sbjct: 343 SNNDHAAVASVEKIAAASTPGTANENLNVDGSGLDPFQEADGGNVPAAGYE--IRPIVHL 400
Query: 326 LAGSTTCKLKLSKSICKQVLEERNGAED--TQAASTSGTSVRCAVFKEDAHAAILDGKEQ 383
L S++ ++ S S ++L+ER ++ + +S S R FK+ +L+ +
Sbjct: 401 LGESSSFDIRGSIS---RLLDERREVKEFLREFDLSSTISTRRQAFKDSLRGGVLNAQNI 457
Query: 384 EVSFDNFPYYLSENTKNVLIAACFIHLKH-KEHAKYTADLPTVNPRILLSGPAGSEIYSE 442
++SF+NFPYYLS TK VL+ + ++H+ ++A + DL T PRILLSGP+GSEIY E
Sbjct: 458 DISFENFPYYLSATTKGVLMISMYVHMNGGSKYANFATDLTTACPRILLSGPSGSEIYQE 517
Query: 443 MLVKALAKYFGAKLLIFDSQLLLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKST 502
ML KALAK FGAKL+I DS LL GG ++EAE K+G E+ K++ A + +
Sbjct: 518 MLAKALAKQFGAKLMIVDSLLLPGGSPAREAESSKEGSRRERLSMLAKRAVQAAQVLQHK 577
Query: 503 DPPAS-ETDTPSSSNVPTPLGLESQAKLETDSVPSTSGTAKNCLFKLG---------DRV 552
P +S + D S L SQA + + ++ T+K+ FK G DRV
Sbjct: 578 KPTSSVDADITGGST------LSSQALPKQEV---STATSKSYTFKAGMMFFFSSDCDRV 628
Query: 553 KYSSSSACLYQTSSSRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEA 612
K+ SA + + +GP+ GS+GKV L F+DN SKIG+RFD+P+ DG DLG CE
Sbjct: 629 KFVGPSASAISSLQGQLRGPAIGSQGKVALAFEDNCASKIGIRFDRPVQDGNDLGGLCEE 688
Query: 613 GQGFFCNITDLRLENSGIDELDKSLINTLFE 643
GFFC + LRLE S D+ DK +N +FE
Sbjct: 689 DHGFFCAASSLRLEGSSSDDADKLAVNEIFE 719
Score = 30.4 bits (67), Expect = 6.3
Identities = 35/138 (25%), Positives = 60/138 (43%), Gaps = 24/138 (17%)
Query: 841 LSSESIQYGI-GIFQAIQNESKSLKKSLKDV-------VTENEFEKRLLGDVIPPNDIGV 892
L ES + I G + +E + +K+ L++ F+ L G V+ +I +
Sbjct: 400 LLGESSSFDIRGSISRLLDERREVKEFLREFDLSSTISTRRQAFKDSLRGGVLNAQNIDI 459
Query: 893 TFDDIGALENVKDTLKELVMLPLQRPELFCKG---------QLTKPCKGILLFGPPGTG- 942
+F++ + T K ++M+ + + G LT C ILL GP G+
Sbjct: 460 SFENFPYY--LSATTKGVLMISMY---VHMNGGSKYANFATDLTTACPRILLSGPSGSEI 514
Query: 943 -KTMLAKAVATDAGANFI 959
+ MLAKA+A GA +
Sbjct: 515 YQEMLAKALAKQFGAKLM 532
>At4g27680 unknown protein
Length = 398
Score = 280 bits (717), Expect = 3e-75
Identities = 141/272 (51%), Positives = 194/272 (70%), Gaps = 1/272 (0%)
Query: 854 QAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVML 913
+A++++ + K+ + +V N +E + DVI P+ I V F IG LE +K L ELV+L
Sbjct: 43 KALEHKKEISKRLGRPLVQTNPYEDVIACDVINPDHIDVEFGSIGGLETIKQALYELVIL 102
Query: 914 PLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGE 973
PL+RPELF G+L P KG+LL+GPPGTGKTMLAKA+A ++GA FIN+ +S++ SKWFG+
Sbjct: 103 PLKRPELFAYGKLLGPQKGVLLYGPPGTGKTMLAKAIAKESGAVFINVRVSNLMSKWFGD 162
Query: 974 GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDT 1033
+K V AVFSLA K+ P++IF+DEV+S LG+R + +HEAM MK EFM WDG T
Sbjct: 163 AQKLVSAVFSLAYKLQPAIIFIDEVESFLGQRRST-DHEAMANMKTEFMALWDGFSTDPH 221
Query: 1034 ERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIA 1093
RV+VLAATNRP +LDEA++RRLP+ + +PD RA+ILKV L E + D+D IA
Sbjct: 222 ARVMVLAATNRPSELDEAILRRLPQAFEIGIPDRRERAEILKVTLKGERVEPDIDFDHIA 281
Query: 1094 NMTDGYSGSDLKNLCVTAAHRPIKEILEKEKK 1125
+ +GY+GSD+ LC AA+ PI+EIL+ E+K
Sbjct: 282 RLCEGYTGSDIFELCKKAAYFPIREILDAERK 313
>At1g62140 hypothetical protein
Length = 556
Score = 279 bits (713), Expect = 8e-75
Identities = 177/404 (43%), Positives = 226/404 (55%), Gaps = 80/404 (19%)
Query: 396 ENTKNVLIAACFIHLKHKE--HAKYTADLPTVNPRILLSGPAGSEIYSEMLVKALAKYFG 453
E+TK VL+A +HL +A Y +DL +NPRILLSGPAGSEIY E+L KALA F
Sbjct: 217 EHTKYVLLAVSDMHLNKMNIGYAPYASDLTILNPRILLSGPAGSEIYQEILAKALANSFN 276
Query: 454 AKLLIFDSQLLLGGLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTDPPASETDTPS 513
AKLLIFDS +LG +++KE E L +G P D KS D + + D
Sbjct: 277 AKLLIFDSNPILGVMTAKEFESLMNG-------------PALIDRGKSLDLSSGQGD--- 320
Query: 514 SSNVPTPLGLESQAKLETDSVPSTSGTAKNCLFKL-------GDRVKYSSSSACLYQTSS 566
S++P+P + P + GT + L L GDRV++ C +S
Sbjct: 321 -SSIPSPA-----------TSPRSFGTPISGLLILHWGKTLAGDRVRFFGDELCPGLPTS 368
Query: 567 SRYKGPSNGSRGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEAGQGFFCNITDLRLE 626
+GP G GKV+L+FD+NP +K+GVRF+ P+PDGVDLG CE G GFFC+ TDL+ E
Sbjct: 369 ---RGPPYGFIGKVLLVFDENPSAKVGVRFENPVPDGVDLGQLCEMGHGFFCSATDLQFE 425
Query: 627 NSGIDELDKSLINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNV 685
+S D+L++ L+ LFEV +SR P I+F+K+AEK VGN S FKSKLE + DN+
Sbjct: 426 SSASDDLNELLVTKLFEVAHDQSRTCPVIIFLKDAEKYFVGNSHFCSAFKSKLEVISDNL 485
Query: 686 VVIGSHTHSDSRKEKSHAGGLLFTKFGSNQTALLDLAFPDSFGRLHDRGKEVPKPNKTLT 745
+VI S THSD+ KEK GR LT
Sbjct: 486 IVICSQTHSDNPKEK-------------------------GIGR--------------LT 506
Query: 746 KLFPNKVTIHMPQDEALLASWKQQLDRDVETLKIKGNLHHLRTV 789
LF NKVTI+MPQ E LL SWK LDRD ETLK+K N +HLR V
Sbjct: 507 DLFVNKVTIYMPQGEELLKSWKYHLDRDAETLKMKANYNHLRMV 550
Score = 35.0 bits (79), Expect = 0.26
Identities = 31/101 (30%), Positives = 44/101 (42%), Gaps = 37/101 (36%)
Query: 135 QREGSDVAVLESTGSKGSVIVNGTLVKKSTCCTLNSGDEVVFGLHGNHSYIFQQVNTEVA 194
+R G+ VAVL+ TG+ G + +N V K+ L+SG
Sbjct: 84 KRNGNVVAVLDITGTGGPLRINKAFVIKNVSHELHSG----------------------- 120
Query: 195 VKGAEVQSGIGKFMQLERRSGDPSAVAGASILASLSNLRQD 235
KF+QLER + DPS V S+LASL R++
Sbjct: 121 -----------KFLQLEREARDPSRV---SMLASLEISREN 147
>At5g53540 26S proteasome regulatory particle chain RPT6-like protein
Length = 403
Score = 276 bits (705), Expect = 7e-74
Identities = 137/272 (50%), Positives = 194/272 (70%), Gaps = 1/272 (0%)
Query: 854 QAIQNESKSLKKSLKDVVTENEFEKRLLGDVIPPNDIGVTFDDIGALENVKDTLKELVML 913
++++++ + K+ + ++ N++E + DVI P I V F IG LE++K L ELV+L
Sbjct: 46 KSLEHKREIAKRLGRPLIQTNQYEDVIACDVINPLHIDVEFGSIGGLESIKQALYELVIL 105
Query: 914 PLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGE 973
PL+RPELF G+L P KG+LL+GPPGTGKTMLAKA+A ++ A FIN+ +S++ SKWFG+
Sbjct: 106 PLKRPELFAYGKLLGPQKGVLLYGPPGTGKTMLAKAIARESEAVFINVKVSNLMSKWFGD 165
Query: 974 GEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDT 1033
+K V AVFSLA K+ P++IF+DEVDS LG+R + ++EAM MK EFM WDG T
Sbjct: 166 AQKLVSAVFSLAYKLQPAIIFIDEVDSFLGQRRST-DNEAMSNMKTEFMALWDGFTTDQN 224
Query: 1034 ERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRAKILKVILAKEDLSSDVDLGAIA 1093
RV+VLAATNRP +LDEA++RR P+ + +PD RA+ILKV+L E + SD++ IA
Sbjct: 225 ARVMVLAATNRPSELDEAILRRFPQSFEIGMPDCQERAQILKVVLKGESVESDINYDRIA 284
Query: 1094 NMTDGYSGSDLKNLCVTAAHRPIKEILEKEKK 1125
+ + Y+GSD+ LC AA+ PI+EILE EK+
Sbjct: 285 RLCEDYTGSDIFELCKKAAYFPIREILEAEKE 316
>At3g27120 spastin protein, putative
Length = 327
Score = 240 bits (613), Expect = 3e-63
Identities = 140/312 (44%), Positives = 199/312 (62%), Gaps = 22/312 (7%)
Query: 877 EKRLLGDV---IPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGI 933
E RL+ V I D V +DDI LE+ K + E+V+ PL RP++F KG P KG+
Sbjct: 29 EPRLIEHVSNEIMDRDPNVRWDDIAGLEHAKKCVTEMVIWPLLRPDIF-KG-CRSPGKGL 86
Query: 934 LLFGPPGTGKTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVI 993
LLFGPPGTGKTM+ KA+A +A A F IS SS+TSKW GEGEK V+A+F +AS P+VI
Sbjct: 87 LLFGPPGTGKTMIGKAIAGEAKATFFYISASSLTSKWIGEGEKLVRALFGVASCRQPAVI 146
Query: 994 FVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVI 1053
FVDE+DS+L +R++ GEHE+ R++K +F++ +G +E+++++ ATNRP +LDEA
Sbjct: 147 FVDEIDSLLSQRKSDGEHESSRRLKTQFLIEMEGF-DSGSEQILLIGATNRPQELDEAAR 205
Query: 1054 RRLPRRLMVNLPDAPNRAKILKVILAKEDL--SSDVDLGAIANMTDGYSGSDLKNLCVTA 1111
RRL +RL + LP + RA I++ +L K+ L SD D+ I N+T+GYSGSD+KNL A
Sbjct: 206 RRLTKRLYIPLPSSEARAWIIQNLLKKDGLFTLSDDDMNIICNLTEGYSGSDMKNLVKDA 265
Query: 1112 AHRPIKEILEKEKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVN 1171
P++E L++ G L DD+R + ++DFK A Q+V SVS +
Sbjct: 266 TMGPLREALKR-----------GIDITNLT-KDDMRLVTLQDFKDALQEVRPSVSQNELG 313
Query: 1172 MTELVQWNELYG 1183
+ E WN +G
Sbjct: 314 IYE--NWNNQFG 323
>At4g24860 putative protein
Length = 566
Score = 226 bits (577), Expect = 5e-59
Identities = 139/305 (45%), Positives = 181/305 (58%), Gaps = 33/305 (10%)
Query: 428 RILLSGPAGSEIYSEMLVKALAKYFGAKLLIFDSQLLLG--------------------- 466
R L+ +GSEIY E L KALA+ AKLLIFDS +LG
Sbjct: 213 RFLVGVVSGSEIYQETLAKALARDLEAKLLIFDSYPILGFTRGKFLHLHLFVYFPDYGYE 272
Query: 467 --GLSSKEAELLKDGFNAEKSCSCPKQSPTATDMAKSTDPPASETDTPSSSNVPTPLGLE 524
L++KE E L+DG + KSC P QS D KS+D A SS + +
Sbjct: 273 ITALTAKEVESLRDGLASNKSCKLPNQSIELIDQGKSSDLSAGG-GVASSLSPAASSDSD 331
Query: 525 SQAKLETDSVP-STSGTAK------NCLF-KLGDRVKYSSSSACLYQTSSSRYKGPSNGS 576
SQ +LE +++P S + T K +CL K+ + + S L+ + +GP NG+
Sbjct: 332 SQLQLEPETLPRSVNHTLKKGMPPLHCLQQKILLQSSWISGLRILHLEEKNTCRGPPNGT 391
Query: 577 RGKVVLIFDDNPLSKIGVRFDKPIPDGVDLGSACEAGQGFFCNITDLRLENSGIDELDKS 636
GKV+L+FD+NP +K+GVRFDKPIPDGVDLG CE+G GFFC TDL ++S +L +
Sbjct: 392 TGKVILVFDENPSAKVGVRFDKPIPDGVDLGELCESGHGFFCKATDLPFKSSSFKDLVRL 451
Query: 637 LINTLFEVVTSESRDSPFILFMKEAEKSIVGNGDPYS-FKSKLEKLPDNVVVIGSHTHSD 695
L+NTLFEVV SESR PFILF+K+AEKS+ GN D YS F+ +LE LP+NV+VI S THSD
Sbjct: 452 LVNTLFEVVHSESRTCPFILFLKDAEKSVAGNFDLYSAFQIRLEYLPENVIVICSQTHSD 511
Query: 696 SRKEK 700
K K
Sbjct: 512 HLKVK 516
Score = 47.4 bits (111), Expect = 5e-05
Identities = 45/178 (25%), Positives = 74/178 (41%), Gaps = 28/178 (15%)
Query: 2 DNGAASSEKPPENSNPEPSADPGKCA-------QPDAQIDEPVAAADDDKADTTPPIADA 54
D + +K + + P +D KC D+QID AAA + PP+A A
Sbjct: 32 DKSPSKRQKLEDGGDTLPPSDSSKCVLGDTTPTSGDSQIDASAAAATTSQP---PPVAQA 88
Query: 55 STPTLVADKPRASFSSWSLYQKQNPNLESSAPWCRLLSQSA----QHPNVSICIPNFTIG 110
+ +ASF W+ + N PWCRLLSQSA Q P V+ + +
Sbjct: 89 IL------QEKASFERWTYVHSRFEN-----PWCRLLSQSAQVYQQMPIVAAKPGSVQVP 137
Query: 111 SSRNCNF-HLKDHTISGNLCKIKHTQREGSDV--AVLESTGSKGSVIVNGTLVKKSTC 165
+ + + + H+I +L ++ H + ++++ G +G VN K C
Sbjct: 138 AGKFLDLERMTGHSIISSLERLIHASSKSHQAPESMVQVDGMEGIFSVNNQDSKMEVC 195
>At1g80350 CAD ATPase (AAA1)
Length = 523
Score = 215 bits (548), Expect = 1e-55
Identities = 123/302 (40%), Positives = 182/302 (59%), Gaps = 24/302 (7%)
Query: 891 GVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAV 950
GV +DD+ L K L+E V+LPL PE F + +P KG+L+FGPPGTGKT+LAKAV
Sbjct: 235 GVRWDDVAGLSEAKRLLEEAVVLPLWMPEYF--QGIRRPWKGVLMFGPPGTGKTLLAKAV 292
Query: 951 ATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGE 1010
AT+ G F N+S +++ SKW GE E+ V+ +F LA APS IF+DE+DS+ R GE
Sbjct: 293 ATECGTTFFNVSSATLASKWRGESERMVRCLFDLARAYAPSTIFIDEIDSLCNSRGGSGE 352
Query: 1011 HEAMRKMKNEFMVNWDGLRTKDTER------VIVLAATNRPYDLDEAVIRRLPRRLMVNL 1064
HE+ R++K+E +V DG+ T V+VLAATN P+D+DEA+ RRL +R+ + L
Sbjct: 353 HESSRRVKSELLVQVDGVSNTATNEDGSRKIVMVLAATNFPWDIDEALRRRLEKRIYIPL 412
Query: 1065 PDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPI-KEILEKE 1123
PD +R ++ + L +++SDV++ +A T+GYSG DL N+C A+ + ++I K
Sbjct: 413 PDFESRKALININLRTVEVASDVNIEDVARRTEGYSGDDLTNVCRDASMNGMRRKIAGKT 472
Query: 1124 KKELAAAVAEGRPAPALRGSDDIRS--LNMEDFKHAHQQVCASVSSESVNMTELVQWNEL 1181
+ E+ DDI + + M DF+ A ++V SVSS + E +W
Sbjct: 473 RDEIKN-----------MSKDDISNDPVAMCDFEEAIRKVQPSVSSSDIEKHE--KWLSE 519
Query: 1182 YG 1183
+G
Sbjct: 520 FG 521
>At2g34560 katanin like protein
Length = 384
Score = 204 bits (518), Expect = 3e-52
Identities = 110/240 (45%), Positives = 157/240 (64%), Gaps = 6/240 (2%)
Query: 883 DVIPPNDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTG 942
D+I N + ++ I LEN K LKE V++P++ P F G LT P KGILLFGPPGTG
Sbjct: 92 DIIRGNP-NIKWESIKGLENAKKLLKEAVVMPIKYPTYF-NGLLT-PWKGILLFGPPGTG 148
Query: 943 KTMLAKAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSML 1002
KTMLAKAVAT+ F NIS SS+ SKW G+ EK ++ +F LA APS IF+DE+D+++
Sbjct: 149 KTMLAKAVATECNTTFFNISASSVVSKWRGDSEKLIRVLFDLARHHAPSTIFLDEIDAII 208
Query: 1003 GRR--ENPGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRL 1060
+R E EHEA R++K E ++ DGL+ K E V VLAATN P++LD A++RRL +R+
Sbjct: 209 SQRGGEGRSEHEASRRLKTELLIQMDGLQ-KTNELVFVLAATNLPWELDAAMLRRLEKRI 267
Query: 1061 MVNLPDAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEIL 1120
+V LPD R + ++++ + + + ++GYSGSD++ LC AA +P++ L
Sbjct: 268 LVPLPDPEARRGMFEMLIPSQPGDEPLPHDVLVEKSEGYSGSDIRILCKEAAMQPLRRTL 327
>At3g09840 putative transitional endoplasmic reticulum ATPase
Length = 809
Score = 183 bits (465), Expect = 4e-46
Identities = 105/284 (36%), Positives = 163/284 (56%), Gaps = 9/284 (3%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V+++DIG LENVK L+E V P++ PE F K ++ P KG+L +GPPG GKT+LAKA+A
Sbjct: 477 VSWNDIGGLENVKRELQETVQYPVEHPEKFEKFGMS-PSKGVLFYGPPGCGKTLLAKAIA 535
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+ ANFI++ + + WFGE E V+ +F A + AP V+F DE+DS+ +R
Sbjct: 536 NECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGGGSGG 595
Query: 1012 E---AMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPD 1066
+ A ++ N+ + DG+ K T V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 596 DGGGAADRVLNQLLTEMDGMNAKKT--VFIIGATNRPDIIDSALLRPGRLDQLIYIPLPD 653
Query: 1067 APNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKEKKE 1126
+R I K L K ++ DVD+GA+A T G+SG+D+ +C A I+E +EK+ E
Sbjct: 654 EDSRLNIFKAALRKSPIAKDVDIGALAKYTQGFSGADITEICQRACKYAIRENIEKD-IE 712
Query: 1127 LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESV 1170
+E A G D++ + F+ + + SVS +
Sbjct: 713 KEKRRSENPEAMEEDGVDEVSEIKAAHFEESMKYARRSVSDADI 756
Score = 168 bits (425), Expect = 2e-41
Identities = 109/301 (36%), Positives = 165/301 (54%), Gaps = 23/301 (7%)
Query: 888 NDIGVTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLA 947
+D+G +DD+G + ++ELV LPL+ P+LF K KP KGILL+GPPG+GKT++A
Sbjct: 202 DDVG--YDDVGGVRKQMAQIRELVELPLRHPQLF-KSIGVKPPKGILLYGPPGSGKTLIA 258
Query: 948 KAVATDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRREN 1007
+AVA + GA F I+ I SK GE E ++ F A K APS+IF+DE+DS+ +RE
Sbjct: 259 RAVANETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREK 318
Query: 1008 PGEHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLP 1065
E R++ ++ + DGL+++ VIV+ ATNRP +D A+ R R R + + +P
Sbjct: 319 T-NGEVERRIVSQLLTLMDGLKSR--AHVIVMGATNRPNSIDPALRRFGRFDREIDIGVP 375
Query: 1066 DAPNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE---ILEK 1122
D R ++L++ L+ DVDL I+ T GY G+DL LC AA + I+E +++
Sbjct: 376 DEIGRLEVLRIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVIDL 435
Query: 1123 EKKELAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESVNMTELVQWNELY 1182
E + A + ++ E F A S E+V V WN++
Sbjct: 436 EDDSIDAEILNS------------MAVTNEHFHTALGNSNPSALRETVVEVPNVSWNDIG 483
Query: 1183 G 1183
G
Sbjct: 484 G 484
>At2g27600 putative AAA-type ATPase
Length = 435
Score = 179 bits (454), Expect = 8e-45
Identities = 95/219 (43%), Positives = 142/219 (64%), Gaps = 5/219 (2%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
+ + D+ LE+ K L+E V+LP++ P+ F + +P + LL+GPPGTGK+ LAKAVA
Sbjct: 129 IKWSDVAGLESAKQALQEAVILPVKFPQFFTGKR--RPWRAFLLYGPPGTGKSYLAKAVA 186
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
T+A + F ++S S + SKW GE EK V +F +A + APS+IFVDE+DS+ G R E
Sbjct: 187 TEADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESAPSIIFVDEIDSLCGTRGEGNES 246
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIRRLPRRLMVNLPDAPNRA 1071
EA R++K E +V G+ D E+V+VLAATN PY LD+A+ RR +R+ + LP+A R
Sbjct: 247 EASRRIKTELLVQMQGVGHND-EKVLVLAATNTPYALDQAIRRRFDKRIYIPLPEAKARQ 305
Query: 1072 KILKVILAKEDLS-SDVDLGAIANMTDGYSGSDLKNLCV 1109
+ KV L + ++ D + T+G+SGSD+ ++CV
Sbjct: 306 HMFKVHLGDTPHNLTEPDFEYLGQKTEGFSGSDV-SVCV 343
>At3g53230 CDC48 - like protein
Length = 815
Score = 179 bits (453), Expect = 1e-44
Identities = 106/284 (37%), Positives = 164/284 (57%), Gaps = 11/284 (3%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V+++DIG LENVK L+E V P++ PE F K ++ P KG+L +GPPG GKT+LAKA+A
Sbjct: 478 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMS-PSKGVLFYGPPGCGKTLLAKAIA 536
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENP--G 1009
+ ANFI+I + + WFGE E V+ +F A + AP V+F DE+DS+ +R N
Sbjct: 537 NECQANFISIKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSVGD 596
Query: 1010 EHEAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
A ++ N+ + DG+ K T V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 597 AGGAADRVLNQLLTEMDGMNAKKT--VFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDE 654
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKE-KKE 1126
+R +I K L K ++ DVDL A+A T G+SG+D+ +C + I+E +EK+ +KE
Sbjct: 655 ESRYQIFKSCLRKSPVAKDVDLRALAKYTQGFSGADITEICQRSCKYAIRENIEKDIEKE 714
Query: 1127 LAAAVAEGRPAPALRGSDDIRSLNMEDFKHAHQQVCASVSSESV 1170
A + P ++I + F+ + + SVS +
Sbjct: 715 RKRAES---PEAMEEDEEEIAEIKAGHFEESMKYARRSVSDADI 755
Score = 167 bits (422), Expect = 4e-41
Identities = 97/245 (39%), Positives = 149/245 (60%), Gaps = 9/245 (3%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V +DD+G + ++ELV LPL+ P+LF K KP KGILL+GPPG+GKT++A+AVA
Sbjct: 205 VGYDDVGGVRKQMAQIRELVELPLRHPQLF-KSIGVKPPKGILLYGPPGSGKTLIARAVA 263
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+ GA F I+ I SK GE E ++ F A K APS+IF+DE+DS+ +RE
Sbjct: 264 NETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HG 322
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPN 1069
E R++ ++ + DGL+++ VIV+ ATNRP +D A+ R R R + + +PD
Sbjct: 323 EVERRIVSQLLTLMDGLKSR--AHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIG 380
Query: 1070 RAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKE---ILEKEKKE 1126
R ++L++ L+ DVDL ++ T GY G+DL LC AA + I+E +++ + +E
Sbjct: 381 RLEVLRIHTKNMKLAEDVDLERVSKDTHGYVGADLAALCTEAALQCIREKMDVIDLDDEE 440
Query: 1127 LAAAV 1131
+ A +
Sbjct: 441 IDAEI 445
>At5g03340 transitional endoplasmic reticulum ATPase
Length = 810
Score = 178 bits (452), Expect = 1e-44
Identities = 96/236 (40%), Positives = 145/236 (60%), Gaps = 7/236 (2%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V+++DIG LENVK L+E V P++ PE F K ++ P KG+L +GPPG GKT+LAKA+A
Sbjct: 477 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMS-PSKGVLFYGPPGCGKTLLAKAIA 535
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+ ANFI++ + + WFGE E V+ +F A + AP V+F DE+DS+ +R N
Sbjct: 536 NECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSAGD 595
Query: 1012 E--AMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDA 1067
A ++ N+ + DG+ K T V ++ ATNRP +D A++R RL + + + LPD
Sbjct: 596 AGGAADRVLNQLLTEMDGMNAKKT--VFIIGATNRPDIIDSALLRPGRLDQLIYIPLPDE 653
Query: 1068 PNRAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILEKE 1123
+R I K L K ++ DVD+ A+A T G+SG+D+ +C A I+E +EK+
Sbjct: 654 DSRLNIFKACLRKSPVAKDVDVTALAKYTQGFSGADITEICQRACKYAIRENIEKD 709
Score = 166 bits (420), Expect = 7e-41
Identities = 96/232 (41%), Positives = 142/232 (60%), Gaps = 6/232 (2%)
Query: 892 VTFDDIGALENVKDTLKELVMLPLQRPELFCKGQLTKPCKGILLFGPPGTGKTMLAKAVA 951
V +DD+G + ++ELV LPL+ P+LF K KP KGILL+GPPG+GKT++A+AVA
Sbjct: 204 VGYDDVGGVRKQMAQIRELVELPLRHPQLF-KSIGVKPPKGILLYGPPGSGKTLIARAVA 262
Query: 952 TDAGANFINISMSSITSKWFGEGEKYVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEH 1011
+ GA F I+ I SK GE E ++ F A K APS+IF+DE+DS+ +RE
Sbjct: 263 NETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-NG 321
Query: 1012 EAMRKMKNEFMVNWDGLRTKDTERVIVLAATNRPYDLDEAVIR--RLPRRLMVNLPDAPN 1069
E R++ ++ + DGL+++ VIV+ ATNRP +D A+ R R R + + +PD
Sbjct: 322 EVERRIVSQLLTLMDGLKSR--AHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIG 379
Query: 1070 RAKILKVILAKEDLSSDVDLGAIANMTDGYSGSDLKNLCVTAAHRPIKEILE 1121
R ++L++ L+ DVDL I+ T GY G+DL LC AA + I+E ++
Sbjct: 380 RLEVLRIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMD 431
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.131 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,600,113
Number of Sequences: 26719
Number of extensions: 1242720
Number of successful extensions: 3632
Number of sequences better than 10.0: 153
Number of HSP's better than 10.0 without gapping: 109
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 3234
Number of HSP's gapped (non-prelim): 258
length of query: 1196
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1086
effective length of database: 8,379,506
effective search space: 9100143516
effective search space used: 9100143516
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC141863.4


