

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141862.21 - phase: 0 /pseudo
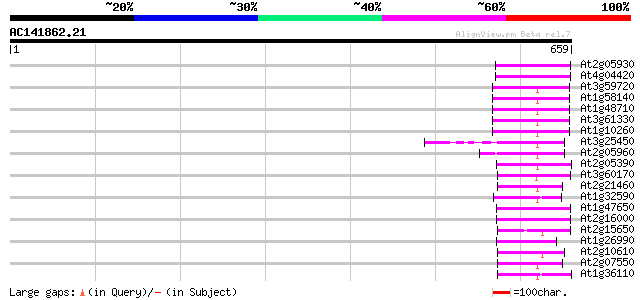
(659 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g05930 copia-like retroelement pol polyprotein 77 3e-14
At4g04420 putative transposon protein 72 1e-12
At3g59720 copia-type reverse transcriptase-like protein 64 2e-10
At1g58140 hypothetical protein 64 2e-10
At1g48710 hypothetical protein 64 2e-10
At3g61330 copia-type polyprotein 63 6e-10
At1g10260 putative lectin receptor kinase 62 1e-09
At3g25450 hypothetical protein 59 9e-09
At2g05960 putative retroelement pol polyprotein 59 9e-09
At2g05390 putative retroelement pol polyprotein 56 6e-08
At3g60170 putative protein 56 7e-08
At2g21460 putative retroelement pol polyprotein 54 4e-07
At1g32590 hypothetical protein, 5' partial 54 4e-07
At1g47650 hypothetical protein 53 5e-07
At2g16000 putative retroelement pol polyprotein 53 6e-07
At2g15650 putative retroelement pol polyprotein 53 6e-07
At1g26990 polyprotein, putative 53 6e-07
At2g10610 pseudogene 52 8e-07
At2g07550 putative retroelement pol polyprotein 52 1e-06
At1g36110 hypothetical protein 50 4e-06
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 77.0 bits (188), Expect = 3e-14
Identities = 38/89 (42%), Positives = 50/89 (55%), Gaps = 1/89 (1%)
Query: 571 QRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNS-SISINNVW 629
++ WY DSG SRHMTG +S T +V FGG GKI G G + + + NV+
Sbjct: 514 KKPWYFDSGASRHMTGSQSNLENYTSVKESKVTFGGGDKGKIKGKGDLTKAEKPQLTNVY 573
Query: 630 LVDGLKHNLLSISQFCDNGYDVMFSKTNC 658
V+GL NL+S+SQ CD G V F+ C
Sbjct: 574 FVEGLTANLISVSQLCDEGLTVSFNSVKC 602
>At4g04420 putative transposon protein
Length = 1008
Score = 72.0 bits (175), Expect = 1e-12
Identities = 36/89 (40%), Positives = 48/89 (53%), Gaps = 1/89 (1%)
Query: 571 QRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNS-SISINNVW 629
++ WY DSG SRHM G +S T +V FGG KI G G + + + NV+
Sbjct: 606 KKPWYFDSGASRHMVGSQSNLENYTSVKESKVTFGGGDKRKIKGKGDLTKAEKPQLTNVY 665
Query: 630 LVDGLKHNLLSISQFCDNGYDVMFSKTNC 658
V+GL NL+S+SQ CD G V F+ C
Sbjct: 666 FVEGLTANLISVSQLCDEGLTVSFNSVKC 694
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 64.3 bits (155), Expect = 2e-10
Identities = 33/95 (34%), Positives = 46/95 (47%), Gaps = 5/95 (5%)
Query: 568 REKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSS 622
+E+ WYLDSG S HM G KS+F L G V G ++ G G I
Sbjct: 329 QEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDH 388
Query: 623 ISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKTN 657
I+NV+ + +K N+LS+ Q + GYD+ N
Sbjct: 389 QFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNN 423
>At1g58140 hypothetical protein
Length = 1320
Score = 64.3 bits (155), Expect = 2e-10
Identities = 33/95 (34%), Positives = 46/95 (47%), Gaps = 5/95 (5%)
Query: 568 REKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSS 622
+E+ WYLDSG S HM G KS+F L G V G ++ G G I
Sbjct: 329 QEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDH 388
Query: 623 ISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKTN 657
I+NV+ + +K N+LS+ Q + GYD+ N
Sbjct: 389 QFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNN 423
>At1g48710 hypothetical protein
Length = 1352
Score = 64.3 bits (155), Expect = 2e-10
Identities = 33/95 (34%), Positives = 46/95 (47%), Gaps = 5/95 (5%)
Query: 568 REKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSS 622
+E+ WYLDSG S HM G KS+F L G V G ++ G G I
Sbjct: 329 QEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDH 388
Query: 623 ISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKTN 657
I+NV+ + +K N+LS+ Q + GYD+ N
Sbjct: 389 QFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNN 423
>At3g61330 copia-type polyprotein
Length = 1352
Score = 62.8 bits (151), Expect = 6e-10
Identities = 32/95 (33%), Positives = 46/95 (47%), Gaps = 5/95 (5%)
Query: 568 REKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSS 622
+++ WYLDSG S HM G KS+F L G V G ++ G G I
Sbjct: 329 QKENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDH 388
Query: 623 ISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKTN 657
I+NV+ + +K N+LS+ Q + GYD+ N
Sbjct: 389 QFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNN 423
>At1g10260 putative lectin receptor kinase
Length = 194
Score = 61.6 bits (148), Expect = 1e-09
Identities = 32/95 (33%), Positives = 45/95 (46%), Gaps = 5/95 (5%)
Query: 568 REKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSS 622
+E+ WYLDSG S HM G KS+F L G V G ++ G G I
Sbjct: 12 QEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKGKGNILIRLKNGDH 71
Query: 623 ISINNVWLVDGLKHNLLSISQFCDNGYDVMFSKTN 657
I+N + + +K N+LS+ Q + GYD+ N
Sbjct: 72 QFISNGYYIPSMKTNILSLGQLLEKGYDIRLKDNN 106
>At3g25450 hypothetical protein
Length = 1343
Score = 58.9 bits (141), Expect = 9e-09
Identities = 47/169 (27%), Positives = 82/169 (47%), Gaps = 28/169 (16%)
Query: 488 LNQNLKSQSQRIRRTKQLLLLRKQYPKVLNLKY*MIRSHSAFTLRYKGGKVKPPELTQKD 547
+N+ +K++S I R +L+ L++Q K + +H A +L + L +K
Sbjct: 229 INKEIKAKSHVIDRLLKLIRLQEQKEKEED------DTHEAESLMMH----EVVYLNEK- 277
Query: 548 P*RYGYLNLNWLKLQMCLRAREKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGN 607
N+ +L+ C+ +WYLD+G S HMTG ++ F L G+V+FG +
Sbjct: 278 -------NIRPTELESCIN-----NAWYLDNGASNHMTGNRAWFCKLDEMITGKVRFGDD 325
Query: 608 QTGKIIGTGTI-----GNSSISINNVWLVDGLKHNLLSISQFCDNGYDV 651
I G G+I G + +V+ + LK N+LS+ Q ++G D+
Sbjct: 326 SCINIKGKGSIPFISKGGERKILFDVYYIPDLKSNILSLGQATESGCDI 374
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 58.9 bits (141), Expect = 9e-09
Identities = 36/104 (34%), Positives = 54/104 (51%), Gaps = 7/104 (6%)
Query: 553 YLNLNWLKLQMCLRAREKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKI 612
YLN + L++ +K +WYLD+G S HMTG + F L G+VKFG + I
Sbjct: 238 YLNEGNMNLEIYEACSDK--AWYLDNGASNHMTGNRDWFCKLDEMVTGKVKFGDDSRIDI 295
Query: 613 IGTGTI-----GNSSISINNVWLVDGLKHNLLSISQFCDNGYDV 651
G G+I ++ NV+ + LK N++S+ Q + G DV
Sbjct: 296 RGKGSILFLTKNGEPKTLANVYYIPDLKSNIISLGQATEAGCDV 339
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 56.2 bits (134), Expect = 6e-08
Identities = 32/92 (34%), Positives = 46/92 (49%), Gaps = 5/92 (5%)
Query: 573 SWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSSISINN 627
SWYLD+G S HMTG F L G+V+FG + I G G+I G ++ +
Sbjct: 279 SWYLDNGASNHMTGNLQWFSKLNEMITGKVRFGDDSRIDIKGKGSIVLITKGGIRKTLTD 338
Query: 628 VWLVDGLKHNLLSISQFCDNGYDVMFSKTNCT 659
V+ + LK N++S+ Q + G DV T
Sbjct: 339 VYFIPDLKSNIISLGQATEAGCDVRMKDDQLT 370
>At3g60170 putative protein
Length = 1339
Score = 55.8 bits (133), Expect = 7e-08
Identities = 30/89 (33%), Positives = 44/89 (48%), Gaps = 4/89 (4%)
Query: 574 WYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGT----IGNSSISINNVW 629
W+LDSGCS HMTG K F L VK G + ++G G+ + + I V+
Sbjct: 300 WFLDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGSVKVKVNGVTQVIPEVY 359
Query: 630 LVDGLKHNLLSISQFCDNGYDVMFSKTNC 658
V L++NLLS+ Q + G ++ C
Sbjct: 360 YVPELRNNLLSLGQLQERGLAILIRDGTC 388
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 53.5 bits (127), Expect = 4e-07
Identities = 28/81 (34%), Positives = 40/81 (48%), Gaps = 5/81 (6%)
Query: 574 WYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSSISINNV 628
W +D+GCS HMT ++ F L GG V+ G K+ G GTI + + NV
Sbjct: 287 WVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNV 346
Query: 629 WLVDGLKHNLLSISQFCDNGY 649
+ + NLLS+ F +GY
Sbjct: 347 RYIPEMDRNLLSLGTFEKSGY 367
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 53.5 bits (127), Expect = 4e-07
Identities = 30/85 (35%), Positives = 46/85 (53%), Gaps = 6/85 (7%)
Query: 569 EKQRSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSSI 623
E+++ W+LDSGCS HM G + FL L V+ G ++ + G G + G +
Sbjct: 253 EEKQIWFLDSGCSNHMCGTREWFLELDSGFKQNVRLGDDRRMAVEGKGKLRLEVDGRIQV 312
Query: 624 SINNVWLVDGLKHNLLSISQFCDNG 648
I++V+ V GLK+NL S+ Q G
Sbjct: 313 -ISDVYFVPGLKNNLFSVGQLQQKG 336
>At1g47650 hypothetical protein
Length = 1409
Score = 53.1 bits (126), Expect = 5e-07
Identities = 32/89 (35%), Positives = 47/89 (51%), Gaps = 2/89 (2%)
Query: 572 RSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTIG-NSSISINNVWL 630
+S +DSG H++ +KSLFL L V T +I G GT+ N I + N+
Sbjct: 330 KSQVIDSGAIHHVSHDKSLFLNLDTSVVSAVNLPAGPTVRISGVGTLRLNDDILLKNILF 389
Query: 631 VDGLKHNLLSISQFCDN-GYDVMFSKTNC 658
+ + NL+SIS D+ G V+F KT+C
Sbjct: 390 IPEFRLNLISISSLTDDIGSRVIFDKTSC 418
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 52.8 bits (125), Expect = 6e-07
Identities = 31/88 (35%), Positives = 47/88 (53%), Gaps = 2/88 (2%)
Query: 573 SWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTIG-NSSISINNVWLV 631
+W +DSG + H++ ++SLF +L V T KI G GT+ N I + NV +
Sbjct: 430 TWVIDSGATHHVSHDRSLFSSLDTSVLSAVNLPTGPTVKISGVGTLKLNDDILLKNVLFI 489
Query: 632 DGLKHNLLSISQFCDN-GYDVMFSKTNC 658
+ NL+SIS D+ G V+F K +C
Sbjct: 490 PEFRLNLISISSLTDDIGSRVIFDKNSC 517
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 52.8 bits (125), Expect = 6e-07
Identities = 31/92 (33%), Positives = 46/92 (49%), Gaps = 10/92 (10%)
Query: 574 WYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSIS-------IN 626
W +DSGC+ HMT E+ F + ++ + G I+ T G+ ++ I
Sbjct: 326 WLVDSGCTNHMTKEERYFSNINKSIKVPIRV---RNGDIVMTAGKGDITVMTRHGKRIIK 382
Query: 627 NVWLVDGLKHNLLSISQFCDNGYDVMFSKTNC 658
NV+LV GL+ NLLS+ Q +GY V F C
Sbjct: 383 NVFLVPGLEKNLLSVPQIISSGYWVRFQDKRC 414
>At1g26990 polyprotein, putative
Length = 1436
Score = 52.8 bits (125), Expect = 6e-07
Identities = 29/72 (40%), Positives = 42/72 (58%), Gaps = 1/72 (1%)
Query: 572 RSWYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTIG-NSSISINNVWL 630
R+W +DSG S H+T E++L+ T D V+ T KI GTG I ++S++NV
Sbjct: 419 RAWVIDSGASHHVTHERNLYHTYKALDRTFVRLPNGHTVKIEGTGFIQLTDALSLHNVLF 478
Query: 631 VDGLKHNLLSIS 642
+ K NLLS+S
Sbjct: 479 IPEFKFNLLSVS 490
>At2g10610 pseudogene
Length = 531
Score = 52.4 bits (124), Expect = 8e-07
Identities = 26/83 (31%), Positives = 47/83 (56%), Gaps = 5/83 (6%)
Query: 574 WYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTIGNSSIS-----INNV 628
WYLD+G S HMTG++ F + G+++FG + I G GTI ++ + +V
Sbjct: 172 WYLDNGASNHMTGDRRYFDKMDHSITGKIRFGDDSRIDIKGKGTIAFIDLNGKPRVMLDV 231
Query: 629 WLVDGLKHNLLSISQFCDNGYDV 651
+ + LK N++S+ Q ++G ++
Sbjct: 232 YFIPELKSNIISLGQATESGCEI 254
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 51.6 bits (122), Expect = 1e-06
Identities = 28/81 (34%), Positives = 39/81 (47%), Gaps = 5/81 (6%)
Query: 574 WYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI-----GNSSISINNV 628
W LD+GCS HMT ++ F GG V+ G ++ G GTI +I + NV
Sbjct: 309 WILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGTIRVKNSDGLTIVLTNV 368
Query: 629 WLVDGLKHNLLSISQFCDNGY 649
+ + NLLS+ F GY
Sbjct: 369 RYIPDMDRNLLSLGTFEKAGY 389
>At1g36110 hypothetical protein
Length = 745
Score = 50.1 bits (118), Expect = 4e-06
Identities = 27/92 (29%), Positives = 47/92 (50%), Gaps = 7/92 (7%)
Query: 574 WYLDSGCSRHMTGEKSLFLTLTMKDGGEVKFGGNQTGKIIGTGTI------GNSSISINN 627
WYLD+G S HMTG ++ F + G+V+FG + I G G+I G + +
Sbjct: 280 WYLDNGASNHMTGNRAYFSKIDESITGKVRFGDDSCIDIKGKGSILFMSKNGEKKV-LAE 338
Query: 628 VWLVDGLKHNLLSISQFCDNGYDVMFSKTNCT 659
V+ + +K N++S+ Q + G ++ + T
Sbjct: 339 VYFIPDVKSNIISLGQATEAGCNIRMKENYLT 370
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.351 0.154 0.514
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,201,931
Number of Sequences: 26719
Number of extensions: 453888
Number of successful extensions: 1603
Number of sequences better than 10.0: 61
Number of HSP's better than 10.0 without gapping: 41
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 1522
Number of HSP's gapped (non-prelim): 71
length of query: 659
length of database: 11,318,596
effective HSP length: 106
effective length of query: 553
effective length of database: 8,486,382
effective search space: 4692969246
effective search space used: 4692969246
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC141862.21


