

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.6 - phase: 0
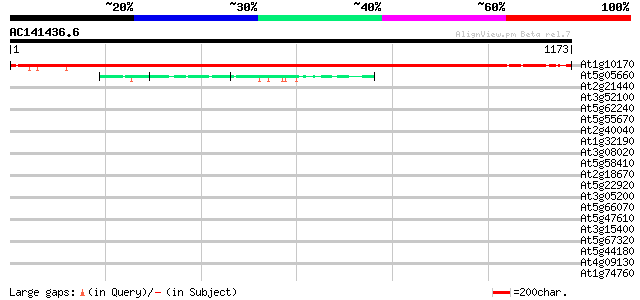
(1173 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g10170 hypothetical protein 1426 0.0
At5g05660 putative protein 288 1e-77
At2g21440 unknown protein (At2g21440) 42 0.003
At3g52100 putative protein 39 0.013
At5g62240 putative protein 39 0.017
At5g55670 unknown protein 39 0.017
At2g40040 unknown protein 38 0.039
At1g32190 unknown protein 37 0.066
At3g08020 unknown protein 36 0.11
At5g58410 unknown protein 36 0.15
At2g18670 putative RING zinc finger protein 36 0.15
At5g22920 PGPD14 protein 35 0.19
At3g05200 RING-H2 zinc finger protein ATL6 (ATL6) 35 0.25
At5g66070 putative protein 35 0.33
At5g47610 unknown protein 35 0.33
At3g15400 anther development protein, ATA20 35 0.33
At5g67320 unknown protein 34 0.43
At5g44180 unknown protein 34 0.43
At4g09130 putative protein 34 0.43
At1g74760 34 0.43
>At1g10170 hypothetical protein
Length = 1188
Score = 1426 bits (3692), Expect = 0.0
Identities = 691/1212 (57%), Positives = 830/1212 (68%), Gaps = 63/1212 (5%)
Query: 1 MSLQQRRERREGSRFPSHRPPRQE-WIPKGAGASSSASTT-----STTTTASTTTAAAAT 54
MS Q RR+R + SHR Q+ WIP+ + SS +T + T A +A+
Sbjct: 1 MSFQVRRDRSDDR---SHRFNHQQTWIPRNSSTSSVVVNEPLLPPNTDRNSETLDAGSAS 57
Query: 55 TTV--------DQPTSLHSHEKNTNSDGGSSNQGAVVAPTFARHRSNHHVAHRVEREHVA 106
V P S + H++++N NQ +H+ + ++ +H
Sbjct: 58 RPVYLQRQHNASGPPSYNHHQRSSNIGPPPPNQHRRYNAPDNQHQRSDNIGPPQPNQHRR 117
Query: 107 HRT-DRDHVAH------RVERERVAHRVEREHVAHRMEREHVAHRVEREHVAHRV--ERE 157
+ D H + R R + E +H +R R + + + R+
Sbjct: 118 YNAPDNQHQRSDNSGPPQPYRHRRNNAPENQHQRSDNIGPPPPNRQRRNNASGTLPDNRQ 177
Query: 158 HVAHRVERGRGRSGNMAGRQYGSRDSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIW 217
VA R D +LPQLVQE+QEKL K ++ECMICYD V RSA IW
Sbjct: 178 RVASRTRPVNQGKRVAKEENVVLTDPNLPQLVQELQEKLVKSSIECMICYDKVGRSANIW 237
Query: 218 SCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNWRCPGCQSVQHTSSKDIKYACFCGK 277
SCSSCYSIFH+NCIK+WARAPTSVDL AEKN G NWRCPGCQSVQ TSSK+I Y CFCGK
Sbjct: 238 SCSSCYSIFHINCIKRWARAPTSVDLLAEKNQGDNWRCPGCQSVQLTSSKEISYRCFCGK 297
Query: 278 RVDPPSDLYLTPHSCGEPCGKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPP 337
R DPPSD YLTPHSCGEPCGKPLEKE E +++LCPH CVLQCHPGPCPPCKAFAPP
Sbjct: 298 RRDPPSDPYLTPHSCGEPCGKPLEKEFAPAETTEEDLCPHVCVLQCHPGPCPPCKAFAPP 357
Query: 338 RLCPCGKKRIATRCSDRQSDLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFC 397
R CPCGKK + TRCS+R+SDL CGQRCDKLL CGRH CE CHVGPCDPCQVL+ A+CFC
Sbjct: 358 RSCPCGKKMVTTRCSERRSDLVCGQRCDKLLSCGRHQCERTCHVGPCDPCQVLVNATCFC 417
Query: 398 SKMTQVLFCGEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSR 457
K + + CG+M +KGE +AE GV+SC NCG LGC NH C EVCHPG CG+C+ LPSR
Sbjct: 418 KKKVETVICGDMNVKGELKAEDGVYSCSFNCGKPLGCGNHFCSEVCHPGPCGDCDLLPSR 477
Query: 458 VKACCCGKTKLEDE-RKSCVDPIPTCSKVCSKTLRCGVHACKETCHVGECPPCKVLISQK 516
VK C CG T+LE++ R+SC+DPIP+CS VC K L C +H C E CH G+CPPC V ++QK
Sbjct: 478 VKTCYCGNTRLEEQIRQSCLDPIPSCSNVCRKLLPCRLHTCNEMCHAGDCPPCLVQVNQK 537
Query: 517 CRCGSTSRTVECYKTT--ENQKFTCQKPCGAKKNCGRHRCSEKCCPLSGPNNGLTTPDWD 574
CRCGSTSR VECY TT E +KF C KPCG KKNCGRHRCSE+CCPL + DWD
Sbjct: 538 CRCGSTSRAVECYITTSSEAEKFVCAKPCGRKKNCGRHRCSERCCPLLNGKKNDLSGDWD 597
Query: 575 PHFCSMLCGKKLRCGQHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPL 634
PH C + C KKLRCGQH CE+LCHSGHCPPCLE IFTDL CACG TSIPPPL CGT P
Sbjct: 598 PHVCQIPCQKKLRCGQHSCESLCHSGHCPPCLEMIFTDLTCACGRTSIPPPLSCGTPVPS 657
Query: 635 CQLPCSVPQPCGHSGSHSCHFGDCPPCSVPVSKECVGGHVILRNIPCGSNNIKCNNPCGR 694
CQLPC +PQPCGHS +H CHFGDCPPCS PV K+CVGGHV+LRNIPCG +I+C CG+
Sbjct: 658 CQLPCPIPQPCGHSDTHGCHFGDCPPCSTPVEKKCVGGHVVLRNIPCGLKDIRCTKICGK 717
Query: 695 TRQCGLHACGRSCHSPPCDILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPGCPCPDA 754
TR+CG+HAC R+CH PCD G+R C Q CGAPR+ CRH C ALCHP PCPD
Sbjct: 718 TRRCGMHACARTCHPEPCDSFNESEAGMRVTCRQKCGAPRTDCRHTCAALCHPSAPCPDL 777
Query: 755 RCEFPVTITCSCGRISANVPCDVGG---NNSNYNADAIFEASIIQKLPMPLQPVDANGQK 811
RCEF VTITCSCGRI+A VPCD GG N SN A EAS++QKLP PLQPV+++G +
Sbjct: 778 RCEFSVTITCSCGRITATVPCDAGGRSANGSNVYCAAYDEASVLQKLPAPLQPVESSGNR 837
Query: 812 VPLGQRKLMCDEECAKLERKRVLADAFDIT-PSLDALHFGENSSY-ELLSDTFRRDPKWV 869
+PLGQRKL CD+ECAKLERKRVL DAFDIT P+L+ALHF ENS+ E++SD +RRDPKWV
Sbjct: 838 IPLGQRKLSCDDECAKLERKRVLQDAFDITPPNLEALHFSENSAMTEIISDLYRRDPKWV 897
Query: 870 LAIEERCKILVLGKSKGTTHGLKVHVFCPMIKDKRDAVRMIAERWKLAVNAAGWEPKRFI 929
LA+EERCK LVLGK++G+T LKVH+FCPM KDKRD VR+IAERWKL V+ AGWEPKRF
Sbjct: 898 LAVEERCKFLVLGKARGSTSALKVHIFCPMQKDKRDTVRLIAERWKLGVSNAGWEPKRFT 957
Query: 930 VISATQKSKAPARVLGVK-GTTTLNAPLPTAFDPLVDMDPRLVVSFPDLPRDADISALVL 988
V+ T KSK P R++G + G ++ P P +D LVDMDP LVVSF DLPR+A+ISALVL
Sbjct: 958 VVHVTAKSKPPTRIIGARGGAISIGGPHPPFYDSLVDMDPGLVVSFLDLPREANISALVL 1017
Query: 989 RFGGECELVWLNDRNALAVFHDPARAATAMRRLDHGTVYQGAVSFVQNAGASAASSVTSA 1048
RFGGECELVWLND+NALAVFHD ARAATAMRRL+HG+VY GAV VQ+ G S S+ +
Sbjct: 1018 RFGGECELVWLNDKNALAVFHDHARAATAMRRLEHGSVYHGAV-VVQSGGQS--PSLNNV 1074
Query: 1049 WGGTKEGAL----RSNPWKKAAVLDPGWKEDSWGDEQWTTAGDSANIQPSAL---KKEAP 1101
WG + + NPW++A + + +DSWG E G S + Q SAL K +P
Sbjct: 1075 WGKLPGSSAWDVDKGNPWRRAVIQE---SDDSWGAEDSPIGGSSTDAQASALRSAKSNSP 1131
Query: 1102 IPASLNPWNVLNHESSSSSSPATVIRSVASGKQTESGNVSTKVEPSAGGADGGNSDATEA 1161
I S+N W+VL + +S+S+ + +A +++ S + K +P G +
Sbjct: 1132 IVTSVNRWSVLEPKKASTST----LEPIAQIEESSSSKTTGK-QPVEGSGE--------- 1177
Query: 1162 AEVVDDWEKAFE 1173
EVVDDWEK E
Sbjct: 1178 -EVVDDWEKVCE 1188
>At5g05660 putative protein
Length = 820
Score = 288 bits (738), Expect = 1e-77
Identities = 194/661 (29%), Positives = 255/661 (38%), Gaps = 178/661 (26%)
Query: 189 VQEIQEKLTKGTVECMICYDMVRRSAPIWSC-SSCYSIFHLNCIKKWARAPTSVDLSAEK 247
+Q + G V C+IC + ++R+ P WSC SSC+++FHL CI+ WAR +DL A +
Sbjct: 73 IQSFLASSSSGAVSCLICLERIKRTDPTWSCTSSCFAVFHLFCIQSWARQ--CLDLQAAR 130
Query: 248 NLGFN----------WRCPGCQSVQHTSSKDIKYACFCGKRVDPPSDL-YLTPHSCGEPC 296
+ W CP C+S S +Y C+CGK DPP+D ++ PHSCGE C
Sbjct: 131 AVTRPSSNPTEPEAVWNCPKCRSSYQKSKIPRRYLCYCGKEEDPPADNPWILPHSCGEVC 190
Query: 297 GKPLEKEVFVTEERKDELCPHACVLQCHPGPCPPCKAFAPPRLCPCGKKRIATRCSDRQS 356
+PL C H C+L CHPGPC C + C CG RC +Q
Sbjct: 191 ERPLSNN-----------CGHCCLLLCHPGPCASCPKLVKAK-CFCGGVEDVRRCGHKQ- 237
Query: 357 DLTCGQRCDKLLDCGRHHCENACHVGPCDPCQVLIEASCFCSKMTQVLFCGEMAMKGEFE 416
+CG C+++LDC H+C CH G C PC+ C C K+ + C E
Sbjct: 238 -FSCGDVCERVLDCNIHNCREICHDGECPPCRERAVYKCSCGKVKEEKDCCER------- 289
Query: 417 AEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVKACCCGKTKLEDERKSCV 476
VF C ++C N+L C H+C CH G CG C + R +C CGK + SC
Sbjct: 290 ----VFRCEASCENMLNCGKHVCERGCHAGECGLCPYQGKR--SCPCGKRFYQG--LSCD 341
Query: 477 DPIPTCSKVCSKTLRCGVHACKETCHVGEC-PPCKVLISQKCRCG---------STSRTV 526
P C C K L CG H C E CH G C C++++++ CRCG T++ V
Sbjct: 342 VVAPLCGGTCDKVLGCGYHRCPERCHRGPCLETCRIVVTKSCRCGVTKKQIWDDMTNKIV 401
Query: 527 ECYKTTENQKFTC-------QKPCGAKKNCGRHRCSEKCCPLSGPNNGL----------- 568
C +T + T + C N C P PN
Sbjct: 402 YCKISTHARGMTLFVACSLQRSMCSLSNNGNNILCLWCHGPRPPPNREFILKPTKKMLHI 461
Query: 569 -------------TTPDWDP-------------------HFCSMLCGKKLRCGQHVCETL 596
P W P C LCG L CG H C
Sbjct: 462 QAESTPGSPCPRCPEPVWRPCVGHHLAAEKRMICSDRTQFACDNLCGNPLPCGNHYCSYF 521
Query: 597 CH------------SGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQP 644
CH S C C DL C T P CQ PC
Sbjct: 522 CHALDIRSSSLDKRSESCEKC------DLRCQKERT------------PRCQHPC----- 558
Query: 645 CGHSGSHSCHFGDCPPCSVPVSKECVGGHVILRNIPCGSNNIKC--NNPCGRTRQCGLHA 702
CH DCPPC V + C G ++ + +C N Q +
Sbjct: 559 -----PRRCHPEDCPPCKTLVKRSCHCGAMV--------HAFECIYYNTMSEKDQMKARS 605
Query: 703 CGRSCHSPPCDILPGIVKGLRAACGQTCGAPRSGCRHMCMALCHPG-CPCPDARCEFPVT 761
C CH LP C H+C +CHPG CP P+ +C V+
Sbjct: 606 CRGPCHRK----LP-------------------NCTHLCPEICHPGQCPLPE-KCGKKVS 641
Query: 762 I 762
I
Sbjct: 642 I 642
Score = 70.1 bits (170), Expect = 7e-12
Identities = 54/182 (29%), Positives = 72/182 (38%), Gaps = 24/182 (13%)
Query: 292 CGEPCGKPLEKEVFVTEERK--DELCPHACVLQC-HPGPCPP--CKAFAPPRLCPCGKKR 346
C EP +P E+R + AC C +P PC C F C
Sbjct: 474 CPEPVWRPCVGHHLAAEKRMICSDRTQFACDNLCGNPLPCGNHYCSYF-------CHALD 526
Query: 347 IATRCSDRQSDLT--CGQRCDKLLD--CGRHHCENACHVGPCDPCQVLIEASCFCSKMTQ 402
I + D++S+ C RC K C +H C CH C PC+ L++ SC C M
Sbjct: 527 IRSSSLDKRSESCEKCDLRCQKERTPRC-QHPCPRRCHPEDCPPCKTLVKRSCHCGAMVH 585
Query: 403 VLFC---GEMAMKGEFEAEGGVFSCGSNCGNVLGCSNHICREVCHPGSCGECEFLPSRVK 459
C M+ K + +A SC C L H+C E+CHPG C E +V
Sbjct: 586 AFECIYYNTMSEKDQMKAR----SCRGPCHRKLPNCTHLCPEICHPGQCPLPEKCGKKVS 641
Query: 460 AC 461
C
Sbjct: 642 IC 643
>At2g21440 unknown protein (At2g21440)
Length = 1003
Score = 41.6 bits (96), Expect = 0.003
Identities = 25/68 (36%), Positives = 35/68 (50%)
Query: 100 VEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVEREHVAHRVEREHV 159
VER+ V +R V VER++ +E + V ER+ V VER+ V VER+ V
Sbjct: 147 VERKKVEKPIERKQVEKPVERKKAEKPIELKQVEKPFERKQVEKPVERKQVEKPVERKQV 206
Query: 160 AHRVERGR 167
+ER R
Sbjct: 207 EKPIERKR 214
>At3g52100 putative protein
Length = 763
Score = 39.3 bits (90), Expect = 0.013
Identities = 27/85 (31%), Positives = 41/85 (47%), Gaps = 12/85 (14%)
Query: 200 TVECMICYDM-VRRS--APIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNWRCP 256
++ C +CY + V +S A + SC C +H NC+K WA+ + S +W CP
Sbjct: 141 SITCHMCYLVEVGKSERAKMLSCKCCGKKYHRNCVKSWAQHRDLFNWS-------SWACP 193
Query: 257 GCQSVQHTSS-KDIKYACFCGKRVD 280
C+ + + D K FC KR D
Sbjct: 194 SCRICEGCGTLGDPKKFMFC-KRCD 217
>At5g62240 putative protein
Length = 366
Score = 38.9 bits (89), Expect = 0.017
Identities = 29/127 (22%), Positives = 48/127 (36%), Gaps = 1/127 (0%)
Query: 28 KGAGASSSASTTSTTTTASTTTAAAATTTVDQPTSLHSHEKNTNSDGGSSNQGAVVAPTF 87
K S S T T+ + T A+ ++P ++S + T + G VAP
Sbjct: 113 KDVSKKKSKSKTKTSNSTLTRPTASLLARQNKPLDIYSVQLLTRCQRSLAKFGENVAPVL 172
Query: 88 ARHRSNHHVAHRVEREHVAHRTDRDHVAHRVERE-RVAHRVEREHVAHRMEREHVAHRVE 146
N + + VAH + R + E R A R ER +RE +A
Sbjct: 173 VSKLQNQDTNRQKQEAKVAHVSSRAKLTVPKEPNLRTAERSERHRSKVNTQREQIATSNS 232
Query: 147 REHVAHR 153
+ H+ ++
Sbjct: 233 KRHIRNK 239
>At5g55670 unknown protein
Length = 710
Score = 38.9 bits (89), Expect = 0.017
Identities = 32/96 (33%), Positives = 37/96 (38%), Gaps = 13/96 (13%)
Query: 89 RHRSNHHVAHRVEREHVAHR-TDRDHVAHRVERERVA--HRVEREHVAHRMEREHVAHRV 145
R R +HH REHV R +R+ HR ERER HR HR E EH
Sbjct: 619 REREHHHKDRERSREHVRDRERERERDRHREERERYGGDHRTR-----HRDEPEH----- 668
Query: 146 EREHVAHRVEREHVAHRVERGRGRSGNMAGRQYGSR 181
+ E R R H R+ R YG R
Sbjct: 669 DEEWNRGRSSRGHNKSRLSREDNHRSKSRDTDYGKR 704
>At2g40040 unknown protein
Length = 839
Score = 37.7 bits (86), Expect = 0.039
Identities = 34/139 (24%), Positives = 54/139 (38%), Gaps = 27/139 (19%)
Query: 1059 SNPWKKAAVLDPGWKEDSWGDEQWTTAGDS--ANIQPSALKKEAPIPASLNPWNVLNHES 1116
S+ W +V D W + +WG E A S A + S+ KK + + W + +
Sbjct: 275 SDVWGHKSVSDKSWDKKNWGTESAPAAWGSTDAAVWGSSDKKNSETESDAAAWGSRDKNN 334
Query: 1117 SSSSSPATVI--------------RSVASGKQTESG----------NVSTKVEPSAGGAD 1152
S S A V+ + S +T+SG N+ T EP+A G+
Sbjct: 335 SDVGSGAGVLGPWNKKSSETESNGATWGSSDKTKSGAAAWNSWDKKNIETDSEPAAWGSQ 394
Query: 1153 GGNSDATEAAEVV-DDWEK 1170
G + TE+ W+K
Sbjct: 395 GKKNSETESGPAAWGAWDK 413
>At1g32190 unknown protein
Length = 422
Score = 37.0 bits (84), Expect = 0.066
Identities = 16/33 (48%), Positives = 18/33 (54%), Gaps = 5/33 (15%)
Query: 735 SGCRHMCMALCHPGCPCPDARCEFPVTITCSCG 767
+GC C LC P C CP RC P +CSCG
Sbjct: 293 TGC--CCSGLCRPSCSCPKPRCPKP---SCSCG 320
>At3g08020 unknown protein
Length = 764
Score = 36.2 bits (82), Expect = 0.11
Identities = 32/121 (26%), Positives = 42/121 (34%), Gaps = 42/121 (34%)
Query: 201 VECMICY-------DMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLGFNW 253
+ C +C+ D RR + SC C +H NC+K WA+ S +W
Sbjct: 146 IMCRMCFLGEGEGSDRARR---MLSCKDCGKKYHKNCLKSWAQHRDLFHWS-------SW 195
Query: 254 RCPGCQ------------SVQHTSSKDIKYACFCGKRVDPP-----SDLYLTP-----HS 291
CP C+ D Y C+C PP S YL P HS
Sbjct: 196 SCPSCRVCEVCRRTGDPNKFMFCKRCDAAYHCYC---QHPPHKNVSSGPYLCPKHTRCHS 252
Query: 292 C 292
C
Sbjct: 253 C 253
>At5g58410 unknown protein
Length = 1873
Score = 35.8 bits (81), Expect = 0.15
Identities = 22/70 (31%), Positives = 32/70 (45%), Gaps = 15/70 (21%)
Query: 194 EKLTKGTVECMICYDMVR---RSAPIWSCSSCYSIFHLNCIKKWARAPTSVDLSAEKNLG 250
+K +G +C ICY ++ S P +C +C FH C+ KW ++ K L
Sbjct: 1814 DKEFEGVEDCPICYSVIHIGNHSLPRRACVTCKYKFHKACLDKWF-------YTSNKKL- 1865
Query: 251 FNWRCPGCQS 260
CP CQS
Sbjct: 1866 ----CPLCQS 1871
>At2g18670 putative RING zinc finger protein
Length = 181
Score = 35.8 bits (81), Expect = 0.15
Identities = 28/112 (25%), Positives = 42/112 (37%), Gaps = 12/112 (10%)
Query: 154 VEREHVAHRVERGRGRSGNMAGRQYGSRDSSLPQLVQEIQEKLTKGTVECMICYDMVRRS 213
+ R H R R + S ++ R LPQ T+ +C++C+D R+
Sbjct: 65 LHRHHRRRRRNRRQESSDGLSSRFV----KKLPQFKFSEPSTYTRYESDCVVCFDGFRQG 120
Query: 214 APIWSCSSCYSIFHLNCIKKW-ARAPTSVDLSAEKNL-------GFNWRCPG 257
+ C +FH C+ W +A T A L G WRC G
Sbjct: 121 QWCRNLPGCGHVFHRKCVDTWLLKASTCPICRARVRLWEEDPQEGELWRCFG 172
>At5g22920 PGPD14 protein
Length = 291
Score = 35.4 bits (80), Expect = 0.19
Identities = 49/199 (24%), Positives = 68/199 (33%), Gaps = 35/199 (17%)
Query: 416 EAEGGVFSCGSNCGNVLG---CS------NHICREVCHPGSCGEC----EFLPSRVKACC 462
E E V SNCG +G CS + + ++ H CG C E K C
Sbjct: 82 ETEQDVQQNCSNCGVCMGKYFCSKCKFFDDDLSKKQYHCDECGICRTGGEENFFHCKRCR 141
Query: 463 CGKTKLEDERKSCVDPI-----PTCSKVCSKTLR-CGVHACKETCHVG-----------E 505
C +K+ +++ CV+ P C + + R V C T H+
Sbjct: 142 CCYSKIMEDKHQCVEGAMHHNCPVCFEYLFDSTRDITVLRCGHTMHLECTKDMGLHNRYT 201
Query: 506 CPPC-KVLISQKCRCGSTSRTVECY---KTTENQK-FTCQKPCGAKKNCGRHRCSEKCCP 560
CP C K + V Y K EN+ + CG+ N H + KC
Sbjct: 202 CPVCSKSICDMSNLWKKLDEEVAAYPMPKMYENKMVWILCNDCGSNTNVRFHLIAHKCSS 261
Query: 561 LSGPNNGLTTPDWDPHFCS 579
N T D H CS
Sbjct: 262 CGSYNTRQTQRGSDSHSCS 280
>At3g05200 RING-H2 zinc finger protein ATL6 (ATL6)
Length = 398
Score = 35.0 bits (79), Expect = 0.25
Identities = 15/44 (34%), Positives = 21/44 (47%)
Query: 194 EKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARA 237
+KL KG +EC IC + + C +FH +CI W A
Sbjct: 119 QKLGKGELECAICLNEFEDDETLRLLPKCDHVFHPHCIDAWLEA 162
>At5g66070 putative protein
Length = 221
Score = 34.7 bits (78), Expect = 0.33
Identities = 15/43 (34%), Positives = 20/43 (45%)
Query: 201 VECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPTSVDL 243
V C +C + + S C+ +FHL CI KW R S L
Sbjct: 174 VSCSVCLQDFQVGETVRSLPHCHHMFHLPCIDKWLRRHASCPL 216
>At5g47610 unknown protein
Length = 166
Score = 34.7 bits (78), Expect = 0.33
Identities = 18/61 (29%), Positives = 23/61 (37%)
Query: 180 SRDSSLPQLVQEIQEKLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKWARAPT 239
S + P LV +L EC IC + I C FH+ CI KW +
Sbjct: 84 SSTPTTPTLVYSSDLELAGAEAECAICLSEFEQGESIQVLEKCQHGFHVKCIHKWLSTRS 143
Query: 240 S 240
S
Sbjct: 144 S 144
>At3g15400 anther development protein, ATA20
Length = 416
Score = 34.7 bits (78), Expect = 0.33
Identities = 23/95 (24%), Positives = 41/95 (42%), Gaps = 2/95 (2%)
Query: 103 EHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVEREHVAHRVEREHVAHR 162
EH H+ DH + E H+ + +H M EH H+ + +H + EH H+
Sbjct: 98 EHPEHKEKPDHKLQSMLGEHPEHKEKPDHKLQSMLGEHPEHKEKPDHKLQSMLGEHPEHK 157
Query: 163 VERGRGRSGNMAGRQYGSRDSSLP-QLVQEIQEKL 196
E+ + +M+G P Q++Q + +L
Sbjct: 158 -EKPDHKLQSMSGESKHPEHKVKPDQIMQTMASEL 191
>At5g67320 unknown protein
Length = 613
Score = 34.3 bits (77), Expect = 0.43
Identities = 22/98 (22%), Positives = 40/98 (40%), Gaps = 10/98 (10%)
Query: 88 ARHRSNHHVAHRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVER 147
A+ + H ERE +++ ++ERE+ R + E E++ + ER
Sbjct: 133 AKEKDRHEKQKEREREREKLEREKEREREKIEREKEREREKMEREIFEREKDRLKLEKER 192
Query: 148 EHVAHRVEREHVAHRVERGRGRSGNMAGRQYGSRDSSL 185
E +ERE ++ER + +Q G D +
Sbjct: 193 E-----IEREREREKIEREKSHE-----KQLGDADREM 220
>At5g44180 unknown protein
Length = 1694
Score = 34.3 bits (77), Expect = 0.43
Identities = 22/79 (27%), Positives = 34/79 (42%), Gaps = 5/79 (6%)
Query: 89 RHRSNHHVAHRVEREHVAHRTDRDHVAHRVERERVAHRVEREHVAHRMEREHVAHRVERE 148
RHR N R+ RE AH + +E++ + R E + MER+ R E E
Sbjct: 326 RHRKNEEA--RIAREVEAHEK---RIRRELEKQDMLRRKREEQIRKEMERQDRERRKEEE 380
Query: 149 HVAHRVEREHVAHRVERGR 167
+ +RE + E+ R
Sbjct: 381 RLLREKQREEERYLKEQMR 399
>At4g09130 putative protein
Length = 357
Score = 34.3 bits (77), Expect = 0.43
Identities = 15/40 (37%), Positives = 18/40 (44%)
Query: 195 KLTKGTVECMICYDMVRRSAPIWSCSSCYSIFHLNCIKKW 234
K+ G VEC IC P+ C FH NCI +W
Sbjct: 112 KIGNGGVECAICLCEFEDEEPLRWMPPCSHTFHANCIDEW 151
>At1g74760
Length = 255
Score = 34.3 bits (77), Expect = 0.43
Identities = 24/73 (32%), Positives = 27/73 (36%), Gaps = 18/73 (24%)
Query: 590 QHVCETLCHSGHCPPCLETIFTDLACACGMTSIPPPLPCGTMPPLCQLPCSVPQPCGHSG 649
+HVC C +CP C E IFT +S LPCG L C C H
Sbjct: 137 EHVCREKCLEDNCPICHEYIFTS-------SSPVKALPCG---HLMHSTCFQEYTCSHY- 185
Query: 650 SHSCHFGDCPPCS 662
CP CS
Sbjct: 186 -------TCPVCS 191
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.134 0.445
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,538,947
Number of Sequences: 26719
Number of extensions: 1390292
Number of successful extensions: 5360
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 75
Number of HSP's that attempted gapping in prelim test: 4991
Number of HSP's gapped (non-prelim): 229
length of query: 1173
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1063
effective length of database: 8,379,506
effective search space: 8907414878
effective search space used: 8907414878
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC141436.6


