

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.3 + phase: 0 /pseudo
(223 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
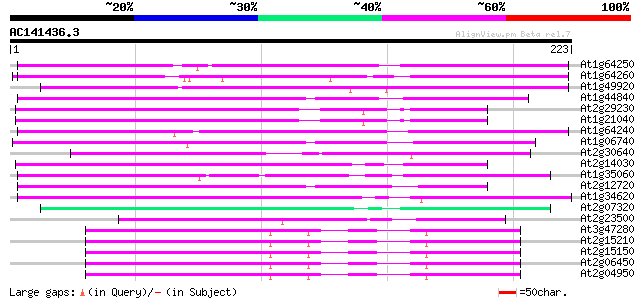
Score E
Sequences producing significant alignments: (bits) Value
At1g64250 hypothetical protein 92 3e-19
At1g64260 hypothetical protein 81 5e-16
At1g49920 hypothetical protein 78 3e-15
At1g44840 hypothetical protein 75 3e-14
At2g29230 Mutator-like transposase 74 5e-14
At1g21040 hypothetical protein 74 8e-14
At1g64240 hypothetical protein 73 1e-13
At1g06740 mudrA-like protein 70 9e-13
At2g30640 Mutator-like transposase 70 1e-12
At2g14030 Mutator-like transposase 69 2e-12
At1g35060 hypothetical protein 67 6e-12
At2g12720 putative protein 62 3e-10
At1g34620 Mutator-like protein 61 5e-10
At2g07320 Mutator-like transposase 60 9e-10
At2g23500 Mutator-like transposase 55 4e-08
At3g47280 putative protein 54 9e-08
At2g15210 putaive transposase of transposon FARE2.7 (cds1) 54 9e-08
At2g15150 putative transposase of FARE2.2 (CDS1) 54 9e-08
At2g06450 putative transposase of FARE2.11 54 9e-08
At2g04950 putative transposase of transposon FARE2.9 (cds1) 54 9e-08
>At1g64250 hypothetical protein
Length = 790
Score = 91.7 bits (226), Expect = 3e-19
Identities = 60/224 (26%), Positives = 104/224 (45%), Gaps = 17/224 (7%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
F+D LK V G F+ Y +EI+ + +A W+D P+ +W Q+HD GRR+
Sbjct: 510 FQDDYLKNLVYEAGSTSEKEEFDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDSGRRYRV 569
Query: 64 MTSNLVESQN-----NVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSE 118
MT ++++N ++ + GLP+TA + + F +++ RVN G+ +++
Sbjct: 570 MT---IDAENLFAFCESFQSL-GLPVTATALLLFDEMRFFFYSGLCDSSGRVNRGDMYTK 625
Query: 119 NSMKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFR 178
M L + S H V ++ + + E L + E+ V L C CG F+
Sbjct: 626 PVMDQLEKLMTDSIPHVVMPLEKGLFQVT--------EPLQEDEWIVQLSEWSCTCGEFQ 677
Query: 179 ALHLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
PC HV+A C + +VD+ ++ + +Y Y F V
Sbjct: 678 LKKFPCLHVLAVCEKLKINPLQYVDDCYSLDRLYKTYAATFSPV 721
>At1g64260 hypothetical protein
Length = 1490
Score = 80.9 bits (198), Expect = 5e-16
Identities = 62/225 (27%), Positives = 99/225 (43%), Gaps = 19/225 (8%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
F+D L+ VE G F+ Y ++I + +A W+D IP+ KW +HD G R+G
Sbjct: 1253 FRDYNLESLVEQAGSTNQKEEFDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGLRYG- 1311
Query: 64 MTSNLVE-SQNNVYKGIRGLP-----ITAIVKASYYRLAALFAKRGHEAAARVNSGEPFS 117
++E + ++ RG P +T V + L + F K + +N G ++
Sbjct: 1312 ----IIEIDREALFAVCRGFPYCTVAMTGGVMLMFDELRSSFDKSLSSIYSSLNRGVVYT 1367
Query: 118 ENSMKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRF 177
E M L + S + +TQ +RD +F V E+ E K E+ V L C C +F
Sbjct: 1368 EPFMDKLEEFMTDSIPYVITQLERD--SFKVSESSE------KEEWIVQLNVSTCTCRKF 1419
Query: 178 RALHLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
++ PC H +A + +VD +T E Y F V
Sbjct: 1420 QSYKFPCLHALAVFEKLKINPLQYVDECYTVEQYCKTYAATFSPV 1464
Score = 73.9 bits (180), Expect = 6e-14
Identities = 58/225 (25%), Positives = 96/225 (41%), Gaps = 13/225 (5%)
Query: 2 RSFKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRW 61
R F L ++ G F Y ++I + +A W+D P+ +W +HD+GRR+
Sbjct: 487 RVFPSFCLGARIRRAGSTSQKDEFVSYMNDIKEKNPEARKWLDQFPQNRWALAHDNGRRY 546
Query: 62 GHMTSNL--VESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSEN 119
G M N + + N ++ G +T V + L + F K + + +N G+ ++E
Sbjct: 547 GIMEINTKALFAVCNAFEQA-GHVVTGSVLLLFDELRSKFDKSFSCSRSSLNCGDVYTEP 605
Query: 120 SMKYLRN--EVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRF 177
M L + S+ VT D + + + L KGE V L + C CG F
Sbjct: 606 VMDKLEEFRTTFVTYSYIVTPLDNNAFQVAT--------ALDKGECIVQLSDCSCTCGDF 657
Query: 178 RALHLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
+ PC H +A C + +VD+ +T E + Y F V
Sbjct: 658 QRYKFPCLHALAVCKKLKFNPLQYVDDCYTLERLKRTYATIFSHV 702
>At1g49920 hypothetical protein
Length = 785
Score = 78.2 bits (191), Expect = 3e-15
Identities = 56/215 (26%), Positives = 96/215 (44%), Gaps = 6/215 (2%)
Query: 13 VEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQ 72
V+ G + F+ Y EI + +A W+D P +W +HDDGRR+G M + E+
Sbjct: 496 VDEAGSSSQKEEFDSYMKEIKERNPEAWKWLDQFPPHQWALAHDDGRRYGIMRID-TEAL 554
Query: 73 NNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSN 132
V K R + + V + +L FA+ + + G+ ++E+ M+ L S+
Sbjct: 555 FAVCKRFRKVAMAGGVMLLFGQLKDAFAESFKLSRGSLKHGDVYTEHVMEKLEEFETDSD 614
Query: 133 SH--HVTQFDRDRYTFSV---RETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHV 187
+ +T +RD Y S+ ++T + V L + C CG F+ PC H
Sbjct: 615 TWVITITPLERDAYQVSMAPKKKTRLMGQSNDSTSGIVQLNDTTCTCGEFQKNKFPCLHA 674
Query: 188 IAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
+A C + +VD+ +T E + Y+ F V
Sbjct: 675 LAVCDELKINPLQYVDDCYTVERYHKTYSAKFSPV 709
>At1g44840 hypothetical protein
Length = 926
Score = 75.1 bits (183), Expect = 3e-14
Identities = 50/203 (24%), Positives = 88/203 (42%), Gaps = 12/203 (5%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
+K+ L + V+ GYA F+ + +I+ ++ ++ +I W + + G+R+
Sbjct: 673 YKNKALTQLVKNAGYAFTGTKFKEFYGQIETTNQNCGKYLHDIGMANWTRHYFRGQRFNL 732
Query: 64 MTSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKY 123
MTSN+ E+ N R I +++ L F R ++ P + K
Sbjct: 733 MTSNIAETLNKALNKGRSSHIVELIRFIRSMLTRWFNARRKKSLKHKGPVPPEVD---KQ 789
Query: 124 LRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLP 183
+ ++ +N V + Y + G+ G VDL+ K C C R+ L +P
Sbjct: 790 ITKTMLTTNGSKVGRITNWSYEIN---------GMLGGRNVVDLEKKQCTCKRYDKLKIP 840
Query: 184 CSHVIAACSSFSHDYKTFVDNKF 206
CSH + A +SF YK VD+ F
Sbjct: 841 CSHALVAANSFKISYKALVDDCF 863
>At2g29230 Mutator-like transposase
Length = 915
Score = 74.3 bits (181), Expect = 5e-14
Identities = 58/191 (30%), Positives = 83/191 (43%), Gaps = 18/191 (9%)
Query: 3 SFKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWG 62
S+K L V A I F Y +E+ D +++++ W +++ G+R+
Sbjct: 640 SYKKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTRAYFLGKRYN 699
Query: 63 HMTSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMK 122
MTSN+ ES N V K R LPI ++++ L + FA R A + P
Sbjct: 700 VMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAARTEASPLPP------- 752
Query: 123 YLRNEVIKSNSHHVTQF---DRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRA 179
EV+ N +F DRY + +RE EG Y V L + C C F
Sbjct: 753 -KMREVVHRNFEKSVRFAVHRLDRYDYEIRE-----EG--ASVYHVKLMERTCSCRAFDL 804
Query: 180 LHLPCSHVIAA 190
LHLPC H IAA
Sbjct: 805 LHLPCPHAIAA 815
>At1g21040 hypothetical protein
Length = 904
Score = 73.6 bits (179), Expect = 8e-14
Identities = 58/191 (30%), Positives = 82/191 (42%), Gaps = 18/191 (9%)
Query: 3 SFKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWG 62
S+K L V A I F Y +E+ D +++++ W +++ G R+
Sbjct: 640 SYKKKHLLFHVSRAARAFRICEFHTYFNEVIRLDPACARYLESVGFCHWTRAYFLGERYN 699
Query: 63 HMTSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMK 122
MTSN+ ES N V K R LPI ++++ L + FA R A + P
Sbjct: 700 VMTSNVAESLNAVLKEARELPIISLLEFIRTTLISWFAMRREAARTEASPLPP------- 752
Query: 123 YLRNEVIKSNSHHVTQF---DRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRA 179
EV+ N +F DRY + +RE EG Y V L + C C F
Sbjct: 753 -KMREVVHRNFEKSVRFAVHRLDRYDYEIRE-----EG--ASVYHVKLMERTCSCRAFDL 804
Query: 180 LHLPCSHVIAA 190
LHLPC H IAA
Sbjct: 805 LHLPCPHAIAA 815
>At1g64240 hypothetical protein
Length = 938
Score = 72.8 bits (177), Expect = 1e-13
Identities = 54/222 (24%), Positives = 93/222 (41%), Gaps = 13/222 (5%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
F+D L+ V+ G F+ Y +I+ + +A W+D P+ +W +HD+GRR+G
Sbjct: 679 FRDYYLEDLVKRAGSTSQKEEFDSYMKDIEKKNSEARKWLDQFPQNQWALAHDNGRRYGI 738
Query: 64 M---TSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENS 120
M T+ L E N + +T V + L F + + G ++E
Sbjct: 739 MEIETTTLFEDFN--VSHLDNHVLTGYVLRLFDELRHSFDEFFCFSRGSRKCGNVYTEPV 796
Query: 121 MKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRAL 180
+ L S ++ V D + + + + E+ V L + C CG F++
Sbjct: 797 TEKLAESRKDSVTYDVMPLDNNAFQVTAPQ--------ENDEWTVQLSDCSCTCGEFQSC 848
Query: 181 HLPCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVV 222
PC H +A C + +VD+ +T E +Y Y F V
Sbjct: 849 KFPCLHALAVCKLLKINPLEYVDDCYTLERLYKTYTATFSPV 890
>At1g06740 mudrA-like protein
Length = 726
Score = 70.1 bits (170), Expect = 9e-13
Identities = 50/209 (23%), Positives = 93/209 (43%), Gaps = 14/209 (6%)
Query: 2 RSFKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRW 61
R F+ L + + + F+ ++I+ +A W+ N +W S+ +G R+
Sbjct: 453 REFQSSVLVDLFWEAAHCLTVLEFKSKINKIEQISPEASLWIQNKSPARWASSYFEGTRF 512
Query: 62 GHMTSNLV-ESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENS 120
G +T+N++ ES +N + GLPI ++ + L + +R + N P +E
Sbjct: 513 GQLTANVITESLSNWVEDTSGLPIIQTMECIHRHLINMLKERRETSLHWSNVLVPSAE-- 570
Query: 121 MKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRAL 180
K + + +S +H V + + + E G V+++N C CGR++
Sbjct: 571 -KQMLAAIEQSRAHRVYRANEAEFEVMTCE----------GNVVVNIENCSCLCGRWQVY 619
Query: 181 HLPCSHVIAACSSFSHDYKTFVDNKFTNE 209
LPCSH + A S D + ++ FT E
Sbjct: 620 GLPCSHAVGALLSCEEDVYRYTESCFTVE 648
>At2g30640 Mutator-like transposase
Length = 754
Score = 69.7 bits (169), Expect = 1e-12
Identities = 49/185 (26%), Positives = 88/185 (47%), Gaps = 17/185 (9%)
Query: 25 FEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQNNVYKGIRGLPI 84
FE EI +A +W+ NI +W +G R+GH+T+N+ ES N+ + GLPI
Sbjct: 498 FEGKMGEIAQISPEAASWIRNIQHSQWATYCFEGTRFGHLTANVSESLNSWVQDASGLPI 557
Query: 85 TAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRY 144
++++ +L LF +R E SM++ ++ S HV + +
Sbjct: 558 IQMLESIRRQLMTLFNER--------------RETSMQW-SGMLVPSAERHVLEAIEECR 602
Query: 145 TFSVRETIEHKEGL--PKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYKTFV 202
+ V + E + + +G++ VD++ + C C + LPCSH +AA + + F
Sbjct: 603 LYPVHKANEAQFEVMTSEGKWIVDIRCRTCYCRGWELYGLPCSHAVAALLACRQNVYRFT 662
Query: 203 DNKFT 207
++ FT
Sbjct: 663 ESYFT 667
>At2g14030 Mutator-like transposase
Length = 874
Score = 68.9 bits (167), Expect = 2e-12
Identities = 54/188 (28%), Positives = 82/188 (42%), Gaps = 12/188 (6%)
Query: 3 SFKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWG 62
S+K L V A I F+ Y +E D +++ + W +++ G R+
Sbjct: 625 SYKKKHLLFHVSRAARAYRICEFQTYFNEGIRLDPACARYLELVGFCHWTRAYFLGERYN 684
Query: 63 HMTSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMK 122
MTSN+VES N V K R LPI ++++ L + FA R A ++ ++ P +
Sbjct: 685 VMTSNVVESLNPVLKEARELPIFSLLEFIRTTLISWFAMRRKAARSKTSALLPKMREVVH 744
Query: 123 YLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHL 182
+ + H + DRY + VR G + V L + C C F LHL
Sbjct: 745 QNFEKSVTFAVHRI-----DRYDYKVR-------GEGSSVFHVKLMERTCSCRAFDLLHL 792
Query: 183 PCSHVIAA 190
PC H IAA
Sbjct: 793 PCPHAIAA 800
>At1g35060 hypothetical protein
Length = 873
Score = 67.4 bits (163), Expect = 6e-12
Identities = 55/213 (25%), Positives = 92/213 (42%), Gaps = 14/213 (6%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
FK+ L + V+ GY F+ ++I+ + + ++ ++ W + + G+R+
Sbjct: 624 FKNRGLTQLVKNAGYEFTSGKFKTLYNQINAINPLCIKYLHDVGMAHWTRLYFPGQRFNL 683
Query: 64 MTSNLVESQNN-VYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMK 122
MTSN+ E+ N ++KG R I +++ L F H ++ +SG E +
Sbjct: 684 MTSNIAETLNKALFKG-RSSHIVELLRFIRSMLTRWF--NAHRKKSQAHSGPVPPEVDKQ 740
Query: 123 YLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHL 182
+N S S R T E + G G VDL+ K C C R+ L +
Sbjct: 741 ISKNLTTSSGSKV------GRVTSWSYEVV----GKLGGSNVVDLEKKQCTCKRYDKLKI 790
Query: 183 PCSHVIAACSSFSHDYKTFVDNKFTNECVYAVY 215
PC H + A +S + YK VD+ F A Y
Sbjct: 791 PCGHALVAANSINLSYKALVDDYFKPHSWVASY 823
>At2g12720 putative protein
Length = 819
Score = 61.6 bits (148), Expect = 3e-10
Identities = 48/187 (25%), Positives = 77/187 (40%), Gaps = 13/187 (6%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
FK +L V+ + F +EI+ D A++ + W ++H G R+
Sbjct: 548 FKRKDLFPLVKKAARCYRLNDFTNAFNEIEELDPLLHAYLQRAGVEMWARAHFPGDRYNL 607
Query: 64 MTSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKY 123
MT+N+ ES N + LPI +++A + FA+R +A+ + P E K
Sbjct: 608 MTTNIAESMNRALSQAKNLPIVRMLEAIRQMMTRWFAERRDDASKQHTQLTPGVE---KL 664
Query: 124 LRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLP 183
L+ V S V D R + ++ + V++ K C C F LP
Sbjct: 665 LQTRVTSSRLLDVQTIDASRVQVAYEASL----------HVVNVDEKQCTCRLFNKEKLP 714
Query: 184 CSHVIAA 190
C H IAA
Sbjct: 715 CIHAIAA 721
>At1g34620 Mutator-like protein
Length = 1034
Score = 60.8 bits (146), Expect = 5e-10
Identities = 51/221 (23%), Positives = 89/221 (40%), Gaps = 14/221 (6%)
Query: 4 FKDGELKKKVEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGH 63
+K L + + A ++ F +I + A++ ++ +W + H GRR+
Sbjct: 662 YKPKSLCRLMSEAAQAFHVTDFNRIFLKIQKLNPGCAAYLVDLGFSEWTRVHSKGRRFNI 721
Query: 64 MTSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKY 123
M SN+ ES NNV + R P+ +++ L FA R +A + P + ++
Sbjct: 722 MDSNICESWNNVIREAREYPLICMLEYIRTTLMDWFATRRAQAEDCPTTLAPRVQERVEE 781
Query: 124 LRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKGE-YKVDLQNKWCDCGRFRALHL 182
+ + + F+ F V+E GE + V L C C F+ L +
Sbjct: 782 NYQSAMSMSVKPICNFE-----FQVQER--------TGECFIVKLDESTCSCLEFQGLGI 828
Query: 183 PCSHVIAACSSFSHDYKTFVDNKFTNECVYAVYNIHFDVVH 223
PC+H IAA + + N + NE V Y +H
Sbjct: 829 PCAHAIAAAARIGVPTDSLAANGYFNELVKLSYEEKIYPIH 869
>At2g07320 Mutator-like transposase
Length = 534
Score = 60.1 bits (144), Expect = 9e-10
Identities = 46/203 (22%), Positives = 81/203 (39%), Gaps = 12/203 (5%)
Query: 13 VEGMGYAMNIPTFEYYRSEIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQ 72
V +G + F EI + + +++ I +W++ + G R+ MTSN+ E+
Sbjct: 2 VLNVGCVYKMSVFRNLYKEIKSRNYECDKYLEKIGPARWSRVYFQGERYNLMTSNIAETL 61
Query: 73 NNVYKGIRGLPITAIVKASYYRLAALFAKRGHEAAARVNSGEPFSENSMKYLRNEVIKSN 132
NN + R PI +VK L + R ++ +P+ + M ++ S
Sbjct: 62 NNAIRLGRCAPILELVKFIRAMLTRWCSARRKKSIKHTGMVQPYVDKVMAQTMMKMEGSK 121
Query: 133 SHHVTQFDRDRYTFSVRETIEHKEGLPKGEYKVDLQNKWCDCGRFRALHLPCSHVIAACS 192
+VT +T+ + G G + V + C C F L +PC H + A +
Sbjct: 122 IQNVT-----NWTYEI-------VGRFGGNHMVSFDDMKCSCKVFDKLKIPCGHAMLAAN 169
Query: 193 SFSHDYKTFVDNKFTNECVYAVY 215
S D+ T V F + Y
Sbjct: 170 SIGLDFGTLVGECFKTKTWVLTY 192
>At2g23500 Mutator-like transposase
Length = 784
Score = 54.7 bits (130), Expect = 4e-08
Identities = 46/156 (29%), Positives = 66/156 (41%), Gaps = 12/156 (7%)
Query: 44 DNIPKQKWNQSHDDGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKASYYRLAALFAKRG 103
D K+KW + G R+ TSN VES NNV+K R + +V A +++ F +
Sbjct: 539 DTTVKEKWARCCFPGERYNLDTSNCVESLNNVFKNARKYSLIPMVDAIIKKISGWFNEHR 598
Query: 104 HEAA--ARVNSGEPFSENSMKYLRNEVIKSNSHHVTQFDRDRYTFSVRETIEHKEGLPKG 161
EAA + N P EN + L K + F+R+ Y + + I
Sbjct: 599 MEAASGSLENKMVPLVENYLHDLWVFAEKLKVVELNSFERE-YVVTCDKGI--------- 648
Query: 162 EYKVDLQNKWCDCGRFRALHLPCSHVIAACSSFSHD 197
+Y V L K C C F PC H +AA + D
Sbjct: 649 DYTVSLLLKTCSCKVFDIQKYPCIHALAAFINIMDD 684
>At3g47280 putative protein
Length = 739
Score = 53.5 bits (127), Expect = 9e-08
Identities = 44/184 (23%), Positives = 79/184 (42%), Gaps = 34/184 (18%)
Query: 31 EIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKA 90
EI +D+K ++ + +KW++++ R+ MTSNL ES N + K R P+ + ++
Sbjct: 491 EITNSDKKLAQYLCEVDVRKWSRAYSPSNRYNIMTSNLAESVNALLKQNREYPVVCLFES 550
Query: 91 SYYRLAALFAKR-----GHEAAARVNSGEPFS---ENSMKYLRNEVIKSNSHHVTQFDRD 142
+ F +R H +A +N G+ + S ++L V Q +++
Sbjct: 551 IRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWL----------EVCQVNQE 600
Query: 143 RYTFSVRETIEHKEGLPKGEYK---VDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYK 199
+ KG+ K V+L + C C F PC+H IA+ + +
Sbjct: 601 EFEV-------------KGDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASAKHINLNEN 647
Query: 200 TFVD 203
FVD
Sbjct: 648 MFVD 651
>At2g15210 putaive transposase of transposon FARE2.7 (cds1)
Length = 580
Score = 53.5 bits (127), Expect = 9e-08
Identities = 45/184 (24%), Positives = 80/184 (43%), Gaps = 34/184 (18%)
Query: 31 EIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKA 90
EI +D+K ++ + +KW++++ R+ MTSNL ES N + K R PI + ++
Sbjct: 332 EITNSDKKLAQYLWEVHVRKWSRAYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFES 391
Query: 91 SYYRLAALFAKR-----GHEAAARVNSGEPFS---ENSMKYLRNEVIKSNSHHVTQFDRD 142
+ F +R H +A +N G+ + S ++L V Q +++
Sbjct: 392 IRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWL----------EVCQVNQE 441
Query: 143 RYTFSVRETIEHKEGLPKGEYK---VDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYK 199
+ KG+ K V+L + C C F PC+H IA+ + + +
Sbjct: 442 EFEV-------------KGDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASANHINLNEN 488
Query: 200 TFVD 203
FVD
Sbjct: 489 MFVD 492
>At2g15150 putative transposase of FARE2.2 (CDS1)
Length = 607
Score = 53.5 bits (127), Expect = 9e-08
Identities = 45/184 (24%), Positives = 80/184 (43%), Gaps = 34/184 (18%)
Query: 31 EIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKA 90
EI +D+K ++ + +KW++++ R+ MTSNL ES N + K R PI + ++
Sbjct: 368 EITNSDKKLAQYLWEVHVRKWSRAYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFES 427
Query: 91 SYYRLAALFAKR-----GHEAAARVNSGEPFS---ENSMKYLRNEVIKSNSHHVTQFDRD 142
+ F +R H +A +N G+ + S ++L V Q +++
Sbjct: 428 IRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWL----------EVCQVNQE 477
Query: 143 RYTFSVRETIEHKEGLPKGEYK---VDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYK 199
+ KG+ K V+L + C C F PC+H IA+ + + +
Sbjct: 478 EFEV-------------KGDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASANHINLNEN 524
Query: 200 TFVD 203
FVD
Sbjct: 525 MFVD 528
>At2g06450 putative transposase of FARE2.11
Length = 580
Score = 53.5 bits (127), Expect = 9e-08
Identities = 45/184 (24%), Positives = 79/184 (42%), Gaps = 34/184 (18%)
Query: 31 EIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKA 90
EI +D+K ++ + +KW++++ R+ MTSNL ES N + K R PI + ++
Sbjct: 332 EITNSDKKLAQYLWEVDVRKWSRTYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFES 391
Query: 91 SYYRLAALFAKR-----GHEAAARVNSGEPFS---ENSMKYLRNEVIKSNSHHVTQFDRD 142
+ F +R H +A +N G+ + S ++L V Q +++
Sbjct: 392 IRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWL----------EVCQVNQE 441
Query: 143 RYTFSVRETIEHKEGLPKGEYK---VDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYK 199
+ KG+ K V+L + C C F PC+H IA+ + +
Sbjct: 442 EFEV-------------KGDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASAKHINLNEN 488
Query: 200 TFVD 203
FVD
Sbjct: 489 MFVD 492
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 53.5 bits (127), Expect = 9e-08
Identities = 45/184 (24%), Positives = 79/184 (42%), Gaps = 34/184 (18%)
Query: 31 EIDVADRKALAWVDNIPKQKWNQSHDDGRRWGHMTSNLVESQNNVYKGIRGLPITAIVKA 90
EI +D+K ++ + +KW++++ R+ MTSNL ES N + K R PI + ++
Sbjct: 368 EITNSDKKLAQYLWEVDVRKWSRAYSPSNRYNIMTSNLAESVNALLKQNREYPIVCLFES 427
Query: 91 SYYRLAALFAKR-----GHEAAARVNSGEPFS---ENSMKYLRNEVIKSNSHHVTQFDRD 142
+ F +R H +A +N G+ + S ++L V Q +++
Sbjct: 428 IRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWL----------EVCQVNQE 477
Query: 143 RYTFSVRETIEHKEGLPKGEYK---VDLQNKWCDCGRFRALHLPCSHVIAACSSFSHDYK 199
+ KG+ K V+L + C C F PC+H IA+ + +
Sbjct: 478 EFEV-------------KGDTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASAKHINLNEN 524
Query: 200 TFVD 203
FVD
Sbjct: 525 MFVD 528
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,233,737
Number of Sequences: 26719
Number of extensions: 218443
Number of successful extensions: 538
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 423
Number of HSP's gapped (non-prelim): 78
length of query: 223
length of database: 11,318,596
effective HSP length: 96
effective length of query: 127
effective length of database: 8,753,572
effective search space: 1111703644
effective search space used: 1111703644
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC141436.3


