

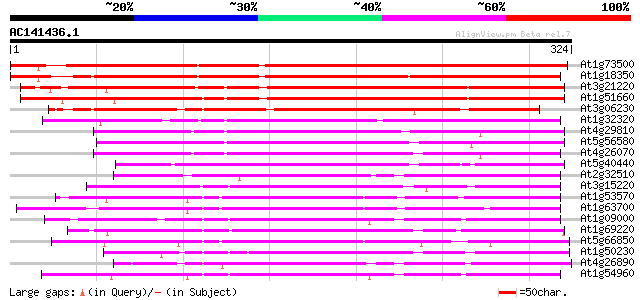
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141436.1 + phase: 0
(324 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g73500 unknown protein 361 e-100
At1g18350 MAP kinase kinase 5, putative 323 8e-89
At3g21220 MAP kinase kinase 5 285 2e-77
At1g51660 unknown protein 277 6e-75
At3g06230 putative MAP kinase 244 5e-65
At1g32320 MAP kinase, putative 225 2e-59
At4g29810 MAP kinase kinase 2 199 1e-51
At5g56580 protein kinase MEK1 homolog 194 5e-50
At4g26070 MEK1 191 5e-49
At5g40440 MAP kinase kinase 3 (ATMKK3) 175 3e-44
At2g32510 putative protein kinase 146 2e-35
At3g15220 putative MAP kinase 145 4e-35
At1g53570 MEK kinase MAP3Ka, putative 143 1e-34
At1g63700 putative protein kinase 132 3e-31
At1g09000 NPK1-related protein kinase 1S (ANP1) 130 1e-30
At1g69220 serine/threonine kinase (SIK1) 129 3e-30
At5g66850 MAP protein kinase like protein 127 1e-29
At1g50230 hypothetical protein 127 1e-29
At4g26890 putative NPK1-related protein kinase 126 1e-29
At1g54960 NPK1-related protein kinase 2 (ANP2) 126 1e-29
>At1g73500 unknown protein
Length = 310
Score = 361 bits (927), Expect = e-100
Identities = 192/325 (59%), Positives = 231/325 (71%), Gaps = 18/325 (5%)
Query: 1 MALVHRRNSPKLRLP--EISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEK 58
MALV R LRLP ISD R + S+SATT +VAG + ISA D EK
Sbjct: 1 MALVRERRQLNLRLPLPPISDRR-----------FSTSSSSATTTTVAGCNGISACDLEK 49
Query: 59 LSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKY 118
L+VLG GNGG VYKVRHK TS IYALK + D DP R+ + E+ ILRR TD VVK
Sbjct: 50 LNVLGCGNGGIVYKVRHKTTSEIYALKTVNGDMDPIFTRQLMREMEILRR-TDSPYVVKC 108
Query: 119 HGSFEKPT-GDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAH 177
HG FEKP G+V ILMEYMD G+LE+ G +E KL+ A+ IL GL+YLHA I H
Sbjct: 109 HGIFEKPVVGEVSILMEYMDGGTLESL---RGGVTEQKLAGFAKQILKGLSYLHALKIVH 165
Query: 178 RDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNG 237
RDIKP+N+L+N KNEVKIADFGVSK + R+L++CNSYVGTCAYMSPERFD E GG+ +
Sbjct: 166 RDIKPANLLLNSKNEVKIADFGVSKILVRSLDSCNSYVGTCAYMSPERFDSESSGGSSDI 225
Query: 238 FSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVEC 297
++ DIWS GL + EL VG+FP L GQRPDWA+LMCA+CF +PP PE S EFR+FVEC
Sbjct: 226 YAGDIWSFGLMMLELLVGHFPLLPPGQRPDWATLMCAVCFGEPPRAPEGCSEEFRSFVEC 285
Query: 298 CLKKESGERWSAAQLLTHPFLCKDM 322
CL+K+S +RW+A QLL HPFL +D+
Sbjct: 286 CLRKDSSKRWTAPQLLAHPFLREDL 310
>At1g18350 MAP kinase kinase 5, putative
Length = 307
Score = 323 bits (828), Expect = 8e-89
Identities = 175/321 (54%), Positives = 217/321 (67%), Gaps = 21/321 (6%)
Query: 1 MALVHRRNSPKLRLP--EISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEK 58
MALV +R LRLP +S H P F S +++TA V + ISA D EK
Sbjct: 1 MALVRKRRQINLRLPVPPLSVHLPWF------------SFASSTAPVIN-NGISASDVEK 47
Query: 59 LSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKY 118
L VLG G+ G VYKV HK T IYALK + D P R+ E+ ILRR TD VV+
Sbjct: 48 LHVLGRGSSGIVYKVHHKTTGEIYALKSVNGDMSPAFTRQLAREMEILRR-TDSPYVVRC 106
Query: 119 HGSFEKP-TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAH 177
G FEKP G+V ILMEYMD G+LE+ G +E +L+ +R IL GL+YLH+ I H
Sbjct: 107 QGIFEKPIVGEVSILMEYMDGGNLESL---RGAVTEKQLAGFSRQILKGLSYLHSLKIVH 163
Query: 178 RDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNG 237
RDIKP+N+L+N +NEVKIADFGVSK + R+L+ CNSYVGTCAYMSPERFD G N +
Sbjct: 164 RDIKPANLLLNSRNEVKIADFGVSKIITRSLDYCNSYVGTCAYMSPERFDSAA-GENSDV 222
Query: 238 FSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVEC 297
++ DIWS G+ + EL+VG+FP L GQRPDWA+LMC +CF +PP PE S EFR+FV+C
Sbjct: 223 YAGDIWSFGVMILELFVGHFPLLPQGQRPDWATLMCVVCFGEPPRAPEGCSDEFRSFVDC 282
Query: 298 CLKKESGERWSAAQLLTHPFL 318
CL+KES ERW+A+QLL HPFL
Sbjct: 283 CLRKESSERWTASQLLGHPFL 303
>At3g21220 MAP kinase kinase 5
Length = 348
Score = 285 bits (729), Expect = 2e-77
Identities = 157/321 (48%), Positives = 209/321 (64%), Gaps = 19/321 (5%)
Query: 7 RNSPKLRLPEISDHRP---RFPVPLPANIYKQPSTSATTASVAGGDNISAG----DFEKL 59
R P L LP HR P+PLP S+SA +S A NISA + E++
Sbjct: 19 RKRPDLSLP--LPHRDVALAVPLPLPP---PSSSSSAPASSSAISTNISAAKSLSELERV 73
Query: 60 SVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYH 119
+ +G G GGTVYKV H TS +ALK+ + + + T RR+ E+ ILR + D NVVK H
Sbjct: 74 NRIGSGAGGTVYKVIHTPTSRPFALKVIYGNHEDTVRRQICREIEILR-SVDHPNVVKCH 132
Query: 120 GSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRD 179
F+ G++ +L+E+MD GSLE A + E +L+ ++R IL+GL YLH R+I HRD
Sbjct: 133 DMFDH-NGEIQVLLEFMDQGSLEGA----HIWQEQELADLSRQILSGLAYLHRRHIVHRD 187
Query: 180 IKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFS 239
IKPSN+L+N VKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y+G++
Sbjct: 188 IKPSNLLINSAKNVKIADFGVSRILAQTMDPCNSSVGTIAYMSPERINTDLNHGRYDGYA 247
Query: 240 ADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCL 299
D+WSLG+++ E Y+G FPF S Q DWASLMCAIC S PP P TAS EFR+FV CCL
Sbjct: 248 GDVWSLGVSILEFYLGRFPFAVSRQ-GDWASLMCAICMSQPPEAPATASQEFRHFVSCCL 306
Query: 300 KKESGERWSAAQLLTHPFLCK 320
+ + +RWSA QLL HPF+ K
Sbjct: 307 QSDPPKRWSAQQLLQHPFILK 327
>At1g51660 unknown protein
Length = 366
Score = 277 bits (708), Expect = 6e-75
Identities = 151/325 (46%), Positives = 209/325 (63%), Gaps = 18/325 (5%)
Query: 7 RNSPKLRLP-EISDHRPRFPVPLP-------ANIYKQPSTSATTASVAGGDNISAGDFEK 58
R P L LP D P+PLP + PS+ + +S +I A ++
Sbjct: 19 RRRPDLTLPLPQRDVSLAVPLPLPPTSGGSGGSSGSAPSSGGSASSTNTNSSIEAKNYSD 78
Query: 59 L---SVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNV 115
L + +G G GGTVYKV H+ +S +YALK+ + + + T RR+ E+ ILR NV
Sbjct: 79 LVRGNRIGSGAGGTVYKVIHRPSSRLYALKVIYGNHEETVRRQICREIEILRDVNH-PNV 137
Query: 116 VKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNI 175
VK H F++ G++ +L+E+MD GSLE A + E +L+ ++R IL+GL YLH+R+I
Sbjct: 138 VKCHEMFDQ-NGEIQVLLEFMDKGSLEGA----HVWKEQQLADLSRQILSGLAYLHSRHI 192
Query: 176 AHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNY 235
HRDIKPSN+L+N VKIADFGVS+ + +T++ CNS VGT AYMSPER + ++ G Y
Sbjct: 193 VHRDIKPSNLLINSAKNVKIADFGVSRILAQTMDPCNSSVGTIAYMSPERINTDLNQGKY 252
Query: 236 NGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFV 295
+G++ DIWSLG+++ E Y+G FPF S Q DWASLMCAIC S PP P TAS EFR+F+
Sbjct: 253 DGYAGDIWSLGVSILEFYLGRFPFPVSRQ-GDWASLMCAICMSQPPEAPATASPEFRHFI 311
Query: 296 ECCLKKESGERWSAAQLLTHPFLCK 320
CCL++E G+R SA QLL HPF+ +
Sbjct: 312 SCCLQREPGKRRSAMQLLQHPFILR 336
>At3g06230 putative MAP kinase
Length = 293
Score = 244 bits (623), Expect = 5e-65
Identities = 136/288 (47%), Positives = 183/288 (63%), Gaps = 25/288 (8%)
Query: 23 RFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIY 82
RFP+ +PA SAT +S A + S + +++SVLG GNGGTV+KV+ K TS IY
Sbjct: 27 RFPI-IPAT-----KVSATVSSCAS-NTFSVANLDRISVLGSGNGGTVFKVKDKTTSEIY 79
Query: 83 ALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLE 142
ALK + D T+ R E+ ILR V K H F+ P+G+V ILM+YMD GSLE
Sbjct: 80 ALKKVKENWDSTSLR----EIEILRMVNS-PYVAKCHDIFQNPSGEVSILMDYMDLGSLE 134
Query: 143 TALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSK 202
+ T E +L+ ++R +L G YLH I HRDIKP+N+L + K EVKIADFGVSK
Sbjct: 135 SLRGVT----EKQLALMSRQVLEGKNYLHEHKIVHRDIKPANLLRSSKEEVKIADFGVSK 190
Query: 203 FMGRTLEACNSYVGTCAYMSPERFDPEVYG----GNYNGFSADIWSLGLTLFELYVGYFP 258
+ R+L CNS+VGT AYMSPER D E G N ++ DIWS GLT+ E+ VGY+P
Sbjct: 191 IVVRSLNKCNSFVGTFAYMSPERLDSEADGVTEEDKSNVYAGDIWSFGLTMLEILVGYYP 250
Query: 259 FLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGER 306
L PD A+++CA+CF +PP PE S + ++F++CCL+K++ ER
Sbjct: 251 ML-----PDQAAIVCAVCFGEPPKAPEECSDDLKSFMDCCLRKKASER 293
>At1g32320 MAP kinase, putative
Length = 305
Score = 225 bits (574), Expect = 2e-59
Identities = 127/303 (41%), Positives = 178/303 (57%), Gaps = 13/303 (4%)
Query: 20 HRPRFPVPLPANIYKQPSTSATTASVAGGDNI---SAGDFEKLSVLGHGNGGTVYKVRHK 76
H+ + +P IY + S ++S + + + D EKLSVLG G+GGTVYK RH+
Sbjct: 9 HQEPLTLSIPPLIYHGTAFSVASSSSSSPETSPIQTLNDLEKLSVLGQGSGGTVYKTRHR 68
Query: 77 LTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYM 136
T +YALK+ P E +IL+R + + ++K + F D+C +ME M
Sbjct: 69 RTKTLYALKVLR----PNLNTTVTVEADILKRI-ESSFIIKCYAVFVS-LYDLCFVMELM 122
Query: 137 DSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIA 196
+ GSL AL FSE +S++A IL GL YL I H DIKPSN+L+N K EVKIA
Sbjct: 123 EKGSLHDALLAQQVFSEPMVSSLANRILQGLRYLQKMGIVHGDIKPSNLLINKKGEVKIA 182
Query: 197 DFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYG-GNYNGFSADIWSLGLTLFELYVG 255
DFG S+ + N GTCAYMSPER D E +G G GF+ D+WSLG+ + E Y+G
Sbjct: 183 DFGASRIVAGGDYGSN---GTCAYMSPERVDLEKWGFGGEVGFAGDVWSLGVVVLECYIG 239
Query: 256 YFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTH 315
+P + G +PDWA+L CAIC ++ +P + S EFR+FV CL+K+ +R + +LL H
Sbjct: 240 RYPLTKVGDKPDWATLFCAICCNEKVDIPVSCSLEFRDFVGRCLEKDWRKRDTVEELLRH 299
Query: 316 PFL 318
F+
Sbjct: 300 SFV 302
>At4g29810 MAP kinase kinase 2
Length = 363
Score = 199 bits (507), Expect = 1e-51
Identities = 116/277 (41%), Positives = 157/277 (55%), Gaps = 12/277 (4%)
Query: 49 DNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRR 108
D +S D + + V+G G+ G V V+HK T +ALK+ + D R+ E+ I +
Sbjct: 63 DQLSLSDLDMVKVIGKGSSGVVQLVQHKWTGQFFALKVIQLNIDEAIRKAIAQELKI-NQ 121
Query: 109 ATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLT 168
++ C N+V + SF G + +++EYMD GSL LK+ +S LS + R +L GL
Sbjct: 122 SSQCPNLVTSYQSFYD-NGAISLILEYMDGGSLADFLKSVKAIPDSYLSAIFRQVLQGLI 180
Query: 169 YLHA-RNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFD 227
YLH R+I HRD+KPSN+L+N + EVKI DFGVS M T N++VGT YMSPER
Sbjct: 181 YLHHDRHIIHRDLKPSNLLINHRGEVKITDFGVSTVMTNTAGLANTFVGTYNYMSPERI- 239
Query: 228 PEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWAS---LMCAICFSDPPSLP 284
GN G +DIWSLGL + E G FP+ Q W S LM AI PP+LP
Sbjct: 240 ----VGNKYGNKSDIWSLGLVVLECATGKFPYAPPNQEETWTSVFELMEAIVDQPPPALP 295
Query: 285 E-TASSEFRNFVECCLKKESGERWSAAQLLTHPFLCK 320
S E +F+ CL+K+ R SA +L+ HPFL K
Sbjct: 296 SGNFSPELSSFISTCLQKDPNSRSSAKELMEHPFLNK 332
>At5g56580 protein kinase MEK1 homolog
Length = 356
Score = 194 bits (493), Expect = 5e-50
Identities = 109/275 (39%), Positives = 165/275 (59%), Gaps = 11/275 (4%)
Query: 51 ISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRAT 110
I+A D E + V+G G+GG V VRHK +A+K+ + R++ + E+ I + ++
Sbjct: 65 ITAEDLETVKVIGKGSGGVVQLVRHKWVGKFFAMKVIQMNIQEEIRKQIVQELKINQASS 124
Query: 111 DCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYL 170
C +VV + SF G +++EYMD GSL ++ T E L+ V + +L GL YL
Sbjct: 125 QCPHVVVCYHSFYH-NGAFSLVLEYMDRGSLADVIRQVKTILEPYLAVVCKQVLLGLVYL 183
Query: 171 H-ARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPE 229
H R++ HRDIKPSN+LVN K EVKI+DFGVS + ++ +++VGT YMSPER
Sbjct: 184 HNERHVIHRDIKPSNLLVNHKGEVKISDFGVSASLASSMGQRDTFVGTYNYMSPERISGS 243
Query: 230 VYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQR---PDWASLMCAICFSDPPSLP-E 285
Y +S+DIWSLG+++ E +G FP+L+S + P + L+ AI + PP+ P +
Sbjct: 244 TY-----DYSSDIWSLGMSVLECAIGRFPYLESEDQQNPPSFYELLAAIVENPPPTAPSD 298
Query: 286 TASSEFRNFVECCLKKESGERWSAAQLLTHPFLCK 320
S EF +FV C++K+ R S+ LL+HPF+ K
Sbjct: 299 QFSPEFCSFVSACIQKDPPARASSLDLLSHPFIKK 333
>At4g26070 MEK1
Length = 354
Score = 191 bits (485), Expect = 5e-49
Identities = 111/275 (40%), Positives = 157/275 (56%), Gaps = 12/275 (4%)
Query: 49 DNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRR 108
+ +S D E + V+G G+ G V V+HKLT +ALK+ +++ +T R E+ I
Sbjct: 61 NQLSLADLEVIKVIGKGSSGNVQLVKHKLTQQFFALKVIQLNTEESTCRAISQELRI-NL 119
Query: 109 ATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLT 168
++ C +V + SF G V I++E+MD GSL LK G E+ LS + + +L GL
Sbjct: 120 SSQCPYLVSCYQSFYH-NGLVSIILEFMDGGSLADLLKKVGKVPENMLSAICKRVLRGLC 178
Query: 169 YLH-ARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFD 227
Y+H R I HRD+KPSN+L+N + EVKI DFGVSK + T NS+VGT YMSPER
Sbjct: 179 YIHHERRIIHRDLKPSNLLINHRGEVKITDFGVSKILTSTSSLANSFVGTYPYMSPERIS 238
Query: 228 PEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWAS---LMCAICFSDPPSLP 284
+Y +DIWSLGL L E G FP+ + W+S L+ AI + PP P
Sbjct: 239 GSLYSN-----KSDIWSLGLVLLECATGKFPYTPPEHKKGWSSVYELVDAIVENPPPCAP 293
Query: 285 ETA-SSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
S EF +F+ C++K+ +R SA +LL H F+
Sbjct: 294 SNLFSPEFCSFISQCVQKDPRDRKSAKELLEHKFV 328
>At5g40440 MAP kinase kinase 3 (ATMKK3)
Length = 520
Score = 175 bits (443), Expect = 3e-44
Identities = 106/263 (40%), Positives = 154/263 (58%), Gaps = 14/263 (5%)
Query: 62 LGHGNGGTVYKVRHKLTSIIYALK-INHYDSDPTTRRRALTEVNILRRATDCTNVVKYHG 120
+G G V + H I ALK IN ++ + R++ LTE+ L A +V +HG
Sbjct: 89 IGSGASSVVQRAIHIPNHRILALKKINIFEREK--RQQLLTEIRTLCEAPCHEGLVDFHG 146
Query: 121 SFEKP-TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHA-RNIAHR 178
+F P +G + I +EYM+ GSL LK T E LS++ +L GL+YLH R++ HR
Sbjct: 147 AFYSPDSGQISIALEYMNGGSLADILKVTKKIPEPVLSSLFHKLLQGLSYLHGVRHLVHR 206
Query: 179 DIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGF 238
DIKP+N+L+N+K E KI DFG+S + ++ C ++VGT YMSPER + Y +
Sbjct: 207 DIKPANLLINLKGEPKITDFGISAGLENSMAMCATFVGTVTYMSPERIRNDSY-----SY 261
Query: 239 SADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLP-ETASSEFRNFVEC 297
ADIWSLGL LFE G FP++ + + P +LM I P+ P + S EF +F++
Sbjct: 262 PADIWSLGLALFECGTGEFPYI-ANEGP--VNLMLQILDDPSPTPPKQEFSPEFCSFIDA 318
Query: 298 CLKKESGERWSAAQLLTHPFLCK 320
CL+K+ R +A QLL+HPF+ K
Sbjct: 319 CLQKDPDARPTADQLLSHPFITK 341
>At2g32510 putative protein kinase
Length = 372
Score = 146 bits (368), Expect = 2e-35
Identities = 92/264 (34%), Positives = 142/264 (52%), Gaps = 18/264 (6%)
Query: 61 VLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHG 120
+LG G+ TVY + I A+K + +R A + + V+ Y G
Sbjct: 8 ILGRGSTATVYAAAGHNSDEILAVKSSEVHRSEFLQREAK-----ILSSLSSPYVIGYRG 62
Query: 121 SFEKPTGDVCI----LMEYMDSGSL-ETALKTTGTFSESKLSTVARDILNGLTYLHARNI 175
S K + + LMEY G+L + A K G E+++ RDIL GL Y+H++ I
Sbjct: 63 SETKRESNGVVMYNLLMEYAPYGTLTDAAAKDGGRVDETRVVKYTRDILKGLEYIHSKGI 122
Query: 176 AHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNY 235
H D+K SN++++ K E KIADFG +K + E + +GT A+M+ PEV G
Sbjct: 123 VHCDVKGSNVVISEKGEAKIADFGCAKRVDPVFE--SPVMGTPAFMA-----PEVARGEK 175
Query: 236 NGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICF-SDPPSLPETASSEFRNF 294
G +DIW++G T+ E+ G P+ ++ R D S++ + + S+ P LP + E ++F
Sbjct: 176 QGKESDIWAVGCTMIEMVTGSPPWTKADSREDPVSVLYRVGYSSETPELPCLLAEEAKDF 235
Query: 295 VECCLKKESGERWSAAQLLTHPFL 318
+E CLK+E+ ERW+A QLL HPFL
Sbjct: 236 LEKCLKREANERWTATQLLNHPFL 259
>At3g15220 putative MAP kinase
Length = 690
Score = 145 bits (365), Expect = 4e-35
Identities = 91/276 (32%), Positives = 144/276 (51%), Gaps = 14/276 (5%)
Query: 45 VAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVN 104
VAG + F ++ ++G G+ G VYK K + A+K+ + E++
Sbjct: 4 VAGLQEAAGARFSQIELIGRGSFGDVYKAFDKDLNKEVAIKVIDLEESEDEIEDIQKEIS 63
Query: 105 ILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDIL 164
+L + C + +Y+GS+ T + I+MEYM GS+ L++ E+ ++ + RD+L
Sbjct: 64 VLSQCR-CPYITEYYGSYLHQT-KLWIIMEYMAGGSVADLLQSNNPLDETSIACITRDLL 121
Query: 165 NGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPE 224
+ + YLH HRDIK +NIL++ +VK+ADFGVS + RT+ ++VGT +M+PE
Sbjct: 122 HAVEYLHNEGKIHRDIKAANILLSENGDVKVADFGVSAQLTRTISRRKTFVGTPFWMAPE 181
Query: 225 RFDPEVYGGNYNGFS--ADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPS 282
N G++ ADIWSLG+T+ E+ G P ++ I PP
Sbjct: 182 VIQ------NSEGYNEKADIWSLGITVIEMAKGEPPLADLHP----MRVLFIIPRETPPQ 231
Query: 283 LPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
L E S + + FV CLKK ER SA +L+ H F+
Sbjct: 232 LDEHFSRQVKEFVSLCLKKAPAERPSAKELIKHRFI 267
>At1g53570 MEK kinase MAP3Ka, putative
Length = 609
Score = 143 bits (361), Expect = 1e-34
Identities = 103/305 (33%), Positives = 152/305 (49%), Gaps = 28/305 (9%)
Query: 27 PLPANIYKQPSTSATTASVAGGDNISAG---------DFEKLSVLGHGNGGTVYKVRHKL 77
PLP +P TS T+ S G I G ++K LG G G VY +
Sbjct: 181 PLP-----RPPTSPTSPSAVHGSRIGGGYETSPSGFSTWKKGKFLGSGTFGQVYLGFNSE 235
Query: 78 TSIIYALKINHYDSDPTTRRRALT----EVNILRRATDCTNVVKYHGSFEKPTGDVCILM 133
+ A+K SD T + L E+N+L + N+V+Y+GS E + + +
Sbjct: 236 KGKMCAIKEVKVISDDQTSKECLKQLNQEINLLNQLCH-PNIVQYYGS-ELSEETLSVYL 293
Query: 134 EYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEV 193
EY+ GS+ LK G+F+E + R IL GL YLH RN HRDIK +NILV+ E+
Sbjct: 294 EYVSGGSIHKLLKDYGSFTEPVIQNYTRQILAGLAYLHGRNTVHRDIKGANILVDPNGEI 353
Query: 194 KIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELY 253
K+ADFG++K + S+ G+ +M+PE V N + DIWSLG T+ E+
Sbjct: 354 KLADFGMAKHV-TAFSTMLSFKGSPYWMAPE----VVMSQNGYTHAVDIWSLGCTILEMA 408
Query: 254 VGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLL 313
P+ Q A++ D P +P+ S++ +NF+ CL++ R +A+QLL
Sbjct: 409 TSKPPW---SQFEGVAAIFKIGNSKDTPEIPDHLSNDAKNFIRLCLQRNPTVRPTASQLL 465
Query: 314 THPFL 318
HPFL
Sbjct: 466 EHPFL 470
>At1g63700 putative protein kinase
Length = 883
Score = 132 bits (331), Expect = 3e-31
Identities = 97/318 (30%), Positives = 157/318 (48%), Gaps = 21/318 (6%)
Query: 5 HRRNSPKLRLPEISDHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGH 64
HR P L + P + ++ + P+ + T S ++K +LG
Sbjct: 356 HRLPLPPLLISNTCPFSPTYSAATSPSVPRSPARAEATVS-------PGSRWKKGRLLGM 408
Query: 65 GNGGTVYKVRHKLTSIIYALK-INHYDSDPTTRRRALT---EVNILRRATDCTNVVKYHG 120
G+ G VY + + + A+K + DP +R A E+++L R N+V+Y+G
Sbjct: 409 GSFGHVYLGFNSESGEMCAMKEVTLCSDDPKSRESAQQLGQEISVLSRLRH-QNIVQYYG 467
Query: 121 SFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDI 180
S E + I +EY+ GS+ L+ G F E+ + + IL+GL YLHA+N HRDI
Sbjct: 468 S-ETVDDKLYIYLEYVSGGSIYKLLQEYGQFGENAIRNYTQQILSGLAYLHAKNTVHRDI 526
Query: 181 KPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSA 240
K +NILV+ VK+ADFG++K + S+ G+ +M+PE + N + +
Sbjct: 527 KGANILVDPHGRVKVADFGMAKHI-TAQSGPLSFKGSPYWMAPE----VIKNSNGSNLAV 581
Query: 241 DIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLK 300
DIWSLG T+ E+ P+ Q P + + + P +P+ S E ++FV CL+
Sbjct: 582 DIWSLGCTVLEMATTKPPWSQYEGVPAMFKIGNS---KELPDIPDHLSEEGKDFVRKCLQ 638
Query: 301 KESGERWSAAQLLTHPFL 318
+ R +AAQLL H F+
Sbjct: 639 RNPANRPTAAQLLDHAFV 656
>At1g09000 NPK1-related protein kinase 1S (ANP1)
Length = 666
Score = 130 bits (326), Expect = 1e-30
Identities = 98/300 (32%), Positives = 153/300 (50%), Gaps = 19/300 (6%)
Query: 21 RPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFEKLSVLGHGNGGTVYKVRHKLTSI 80
+P F P PAN A S G I G F + + + + G + V+ L +
Sbjct: 49 KPSFSPPPPANTVDM----APPISWRKGQLIGRGAFGTVYMGMNLDSGELLAVKQVLIAA 104
Query: 81 IYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGS 140
+A K + + EV +L+ + N+V+Y G+ + + IL+E++ GS
Sbjct: 105 NFASK----EKTQAHIQELEEEVKLLKNLSH-PNIVRYLGTVREDD-TLNILLEFVPGGS 158
Query: 141 LETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGV 200
+ + L+ G F ES + T R +L GL YLH I HRDIK +NILV+ K +K+ADFG
Sbjct: 159 ISSLLEKFGPFPESVVRTYTRQLLLGLEYLHNHAIMHRDIKGANILVDNKGCIKLADFGA 218
Query: 201 SKFMGR--TLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFP 258
SK + T+ S GT +M+ PEV + FSADIWS+G T+ E+ G P
Sbjct: 219 SKQVAELATMTGAKSMKGTPYWMA-----PEVILQTGHSFSADIWSVGCTVIEMVTGKAP 273
Query: 259 FLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL 318
+ Q Q + A++ P +P+T SS+ ++F+ CL++ R +A++LL HPF+
Sbjct: 274 WSQ--QYKEVAAIFFIGTTKSHPPIPDTLSSDAKDFLLKCLQEVPNLRPTASELLKHPFV 331
>At1g69220 serine/threonine kinase (SIK1)
Length = 836
Score = 129 bits (323), Expect = 3e-30
Identities = 97/294 (32%), Positives = 151/294 (50%), Gaps = 18/294 (6%)
Query: 34 KQPSTSATTASVAGGDNISAGD----FEKLSVLGHGNGGTVYKVRHKLTSIIYALKINHY 89
+Q ++ +T S+ D+I+ D +E L+ LG G+ G+VYK R TS I A+K+
Sbjct: 225 QQQNSKMSTTSLP--DSITREDPTTKYEFLNELGKGSYGSVYKARDLKTSEIVAVKVISL 282
Query: 90 DSDPTTRRRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTG 149
E+ +L++ NVV+Y GS++ + I+MEY GS+ + T
Sbjct: 283 TEGEEGYEEIRGEIEMLQQCNH-PNVVRYLGSYQGEDY-LWIVMEYCGGGSVADLMNVTE 340
Query: 150 -TFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTL 208
E +++ + R+ L GL YLH+ HRDIK NIL+ + EVK+ DFGV+ + RT+
Sbjct: 341 EALEEYQIAYICREALKGLAYLHSIYKVHRDIKGGNILLTEQGEVKLGDFGVAAQLTRTM 400
Query: 209 EACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDW 268
N+++GT +M+PE Y G D+W+LG++ E+ G P +S P
Sbjct: 401 SKRNTFIGTPHWMAPEVIQENRYDG-----KVDVWALGVSAIEMAEGLPP--RSSVHPMR 453
Query: 269 ASLMCAICFSDPPSLPETASSEFRNFVECCLKKESGERWSAAQLLTHPFL--CK 320
M +I + E S F +FV CL KE R +AA++L H F+ CK
Sbjct: 454 VLFMISIEPAPMLEDKEKWSLVFHDFVAKCLTKEPRLRPTAAEMLKHKFVERCK 507
>At5g66850 MAP protein kinase like protein
Length = 533
Score = 127 bits (318), Expect = 1e-29
Identities = 93/314 (29%), Positives = 154/314 (48%), Gaps = 27/314 (8%)
Query: 25 PVPLPANIYKQPSTSATTASVAGGDNISA----GDFEKLSVLGHGNGGTVYKVRHKLTSI 80
P+PLP S++A+ S + + ++K ++G G G+VY + T
Sbjct: 128 PLPLPPGATCSSSSAASVPSPQAPLKLDSFPMNSQWKKGKLIGRGTFGSVYVASNSETGA 187
Query: 81 IYALK-INHYDSDPTTR---RRALTEVNILRRATDCTNVVKYHGSFEKPTGDVCILMEYM 136
+ A+K + + DP + ++ E+ +L N+V+Y GS E I +EY+
Sbjct: 188 LCAMKEVELFPDDPKSAECIKQLEQEIKLLSNLQH-PNIVQYFGS-ETVEDRFFIYLEYV 245
Query: 137 DSGSLETALKT-TGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNILVNIKNEVKI 195
GS+ ++ GT +ES + R IL+GL YLH + HRDIK +N+LV+ VK+
Sbjct: 246 HPGSINKYIRDHCGTMTESVVRNFTRHILSGLAYLHNKKTVHRDIKGANLLVDASGVVKL 305
Query: 196 ADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYN--GFSADIWSLGLTLFELY 253
ADFG++K + A S G+ +M+PE + + F+ DIWSLG T+ E++
Sbjct: 306 ADFGMAKHL-TGQRADLSLKGSPYWMAPELMQAVMQKDSNPDLAFAVDIWSLGCTIIEMF 364
Query: 254 VGYFPFLQSGQRPDWASLMCAIC----FSDPPSLPETASSEFRNFVECCLKKESGERWSA 309
G +P W+ A D P +PE+ S E ++F+ C ++ ER +A
Sbjct: 365 TG---------KPPWSEFEGAAAMFKVMRDSPPIPESMSPEGKDFLRLCFQRNPAERPTA 415
Query: 310 AQLLTHPFLCKDME 323
+ LL H FL ++
Sbjct: 416 SMLLEHRFLKNSLQ 429
>At1g50230 hypothetical protein
Length = 269
Score = 127 bits (318), Expect = 1e-29
Identities = 85/273 (31%), Positives = 132/273 (48%), Gaps = 20/273 (7%)
Query: 55 DFEKLSVLGHGNGGTVYKVRHKLTSIIYALKI----NHYDSDPTTRRRALTEVNILRRAT 110
D+ + ++G G+ G VYK R K T A+K D D + R+ E+ ILR+
Sbjct: 5 DYHVIELVGEGSFGRVYKGRRKYTGQTVAMKFIMKQGKTDKDIHSLRQ---EIEILRKLK 61
Query: 111 DCTNVVKYHGSFEKPTGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYL 170
N+++ SFE + C++ E+ G L L+ E ++ +A+ ++ L YL
Sbjct: 62 H-ENIIEMLDSFENAR-EFCVVTEFAQ-GELFEILEDDKCLPEEQVQAIAKQLVKALDYL 118
Query: 171 HARNIAHRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEV 230
H+ I HRD+KP NIL+ + VK+ DFG ++ M S GT YM+PE +
Sbjct: 119 HSNRIIHRDMKPQNILIGAGSVVKLCDFGFARAMSTNTVVLRSIKGTPLYMAPELVKEQP 178
Query: 231 YGGNYNGFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSE 290
Y + D+WSLG+ L+ELYVG PF + + DP P+ S+
Sbjct: 179 YDR-----TVDLWSLGVILYELYVGQPPFYTNS-----VYALIRHIVKDPVKYPDEMSTY 228
Query: 291 FRNFVECCLKKESGERWSAAQLLTHPFLCKDME 323
F +F++ L KE R + L HPF+ + E
Sbjct: 229 FESFLKGLLNKEPHSRLTWPALREHPFVKETQE 261
>At4g26890 putative NPK1-related protein kinase
Length = 444
Score = 126 bits (317), Expect = 1e-29
Identities = 90/269 (33%), Positives = 143/269 (52%), Gaps = 24/269 (8%)
Query: 61 VLGHGNGGTVYKVRHKLTSIIYALKINHYDSDPTTRRRALTEVNILRRATDCTNVVKYHG 120
++G G+ TV + + ++A+K S ++ E +IL + ++VKY G
Sbjct: 10 IIGRGSTATV-SIAISSSGELFAVKSADLSSSSLLQK----EQSILSTLSS-PHMVKYIG 63
Query: 121 S--FEKPTGDVC-ILMEYMDSGSLETALKTTG-TFSESKLSTVARDILNGLTYLHARNIA 176
+ + G V ILMEY+ G+L +K +G E ++ + R ILNGL YLH R I
Sbjct: 64 TGLTRESNGLVYNILMEYVSGGNLHDLIKNSGGKLPEPEIRSYTRQILNGLVYLHERGIV 123
Query: 177 HRDIKPSNILVNIKNEVKIADFGVSKFMGRTLEACNSYVGTCAYMSPERFDPEVYGGNYN 236
H D+K N+LV +KIAD G +K + + + + GT A+M+ PEV G
Sbjct: 124 HCDLKSHNVLVEENGVLKIADMGCAKSVDK-----SEFSGTPAFMA-----PEVARGEEQ 173
Query: 237 GFSADIWSLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFS-DPPSLPETASSEFRNFV 295
F AD+W+LG T+ E+ G P+ + D + M I FS + P++P S + ++F+
Sbjct: 174 RFPADVWALGCTMIEMMTGSSPWPELN---DVVAAMYKIGFSGESPAIPAWISDKAKDFL 230
Query: 296 ECCLKKESGERWSAAQLLTHPFLCKDMES 324
+ CLK++ +RW+ +LL HPFL D ES
Sbjct: 231 KNCLKEDQKQRWTVEELLKHPFLDDDEES 259
>At1g54960 NPK1-related protein kinase 2 (ANP2)
Length = 651
Score = 126 bits (317), Expect = 1e-29
Identities = 99/315 (31%), Positives = 155/315 (48%), Gaps = 24/315 (7%)
Query: 19 DHRPRFPVPLPANIYKQPSTSATTASVAGGDNISAGDFE------KLSVLGHGNGGTVYK 72
+H P FP L I S A N S + K ++G G GTVY
Sbjct: 25 NHPPPFPSLLADKITSCIRKSMVFAKSQSPPNNSTVQIKPPIRWRKGQLIGRGAFGTVYM 84
Query: 73 VRHKLTSIIYALKINHYDSDPTTRRRALT-------EVNILRRATDCTNVVKYHGSFEKP 125
+ + + A+K S+ ++ + EV +L+ + N+V+Y G+ +
Sbjct: 85 GMNLDSGELLAVKQVLITSNCASKEKTQAHIQELEEEVKLLKNLSH-PNIVRYLGTVRED 143
Query: 126 TGDVCILMEYMDSGSLETALKTTGTFSESKLSTVARDILNGLTYLHARNIAHRDIKPSNI 185
+ IL+E++ GS+ + L+ G F ES + T +L GL YLH I HRDIK +NI
Sbjct: 144 E-TLNILLEFVPGGSISSLLEKFGAFPESVVRTYTNQLLLGLEYLHNHAIMHRDIKGANI 202
Query: 186 LVNIKNEVKIADFGVSKFMGR--TLEACNSYVGTCAYMSPERFDPEVYGGNYNGFSADIW 243
LV+ + +K+ADFG SK + T+ S GT +M+ PEV + FSADIW
Sbjct: 203 LVDNQGCIKLADFGASKQVAELATISGAKSMKGTPYWMA-----PEVILQTGHSFSADIW 257
Query: 244 SLGLTLFELYVGYFPFLQSGQRPDWASLMCAICFSDPPSLPETASSEFRNFVECCLKKES 303
S+G T+ E+ G P+ Q Q + A++ P +P+ SS+ +F+ CL++E
Sbjct: 258 SVGCTVIEMVTGKAPWSQ--QYKEIAAIFHIGTTKSHPPIPDNISSDANDFLLKCLQQEP 315
Query: 304 GERWSAAQLLTHPFL 318
R +A++LL HPF+
Sbjct: 316 NLRPTASELLKHPFV 330
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,835,414
Number of Sequences: 26719
Number of extensions: 347066
Number of successful extensions: 3007
Number of sequences better than 10.0: 977
Number of HSP's better than 10.0 without gapping: 891
Number of HSP's successfully gapped in prelim test: 86
Number of HSP's that attempted gapping in prelim test: 858
Number of HSP's gapped (non-prelim): 1087
length of query: 324
length of database: 11,318,596
effective HSP length: 99
effective length of query: 225
effective length of database: 8,673,415
effective search space: 1951518375
effective search space used: 1951518375
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 60 (27.7 bits)
Medicago: description of AC141436.1


