

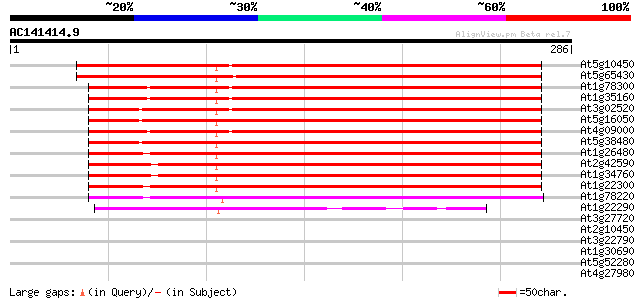
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.9 + phase: 0
(286 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g10450 14-3-3-like protein AFT1 292 1e-79
At5g65430 GF14 Kappa isoform 285 2e-77
At1g78300 putative tyrosine activation protein 258 3e-69
At1g35160 endonuclease like protein (32KD) 254 3e-68
At3g02520 putative 14-3-3 protein 252 1e-67
At5g16050 14-3-3-LIKE PROTEIN GF14 UPSILON 251 4e-67
At4g09000 14-3-3-like protein 251 4e-67
At5g38480 14-3-3 protein homolog RCI1 (pir||S47969) 250 7e-67
At1g26480 14-3-3 protein epsilon like 225 2e-59
At2g42590 14-3-3 regulatory protein (GF14 mu) 212 2e-55
At1g34760 putative protein 212 2e-55
At1g22300 unknown protein 211 4e-55
At1g78220 14-3-3 GF14 Pi protein (GRF13) 145 2e-35
At1g22290 hypothetical protein 93 2e-19
At3g27720 hypothetical protein 40 0.001
At2g10450 14-3-3 regulatory protein 39 0.002
At3g22790 unknown protein 36 0.027
At1g30690 putative phosphatidyl-inositol-transfer protein 35 0.061
At5g52280 hyaluronan mediated motility receptor-like protein 33 0.13
At4g27980 putative protein 32 0.30
>At5g10450 14-3-3-like protein AFT1
Length = 248
Score = 292 bits (748), Expect = 1e-79
Identities = 156/241 (64%), Positives = 191/241 (78%), Gaps = 5/241 (2%)
Query: 35 VAERLRREEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATE 94
+A L R++YV+ AKLA+QAERYEEMV FM+++V G TPA EL++EE NLLSVAYKN
Sbjct: 1 MAATLGRDQYVYMAKLAEQAERYEEMVQFMEQLVTGATPAEELTVEERNLLSVAYKNVIG 60
Query: 95 PLRAALRILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSAS 151
LRAA RI+S ++EE RKN D+H VK Y+SKVESEL +VC IL+LLDS LIPSA
Sbjct: 61 SLRAAWRIVSSIEQKEESRKN-DEHVSLVKDYRSKVESELSSVCSGILKLLDSHLIPSAG 119
Query: 152 SSEIRVVYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGL 211
+SE +V Y +MKGDY RYMAEFK GD++K+A ED +L+YKAAQDIAAAD+ +HPIRLGL
Sbjct: 120 ASESKVFYLKMKGDYHRYMAEFKSGDERKTAAEDTMLAYKAAQDIAAADMAPTHPIRLGL 179
Query: 212 ALNFSVFYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTF 270
ALNFSVFYYEILN D+ +MA+QA +EA EL LG+E YKDST+ MQLLR+N+ LWT
Sbjct: 180 ALNFSVFYYEILNSSDKACNMAKQAFEEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTS 239
Query: 271 D 271
D
Sbjct: 240 D 240
>At5g65430 GF14 Kappa isoform
Length = 248
Score = 285 bits (729), Expect = 2e-77
Identities = 150/241 (62%), Positives = 189/241 (78%), Gaps = 5/241 (2%)
Query: 35 VAERLRREEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATE 94
+A L R++YV+ AKLA+QAERYEEMV FM+++V G TPA EL++EE NLLSVAYKN
Sbjct: 1 MATTLSRDQYVYMAKLAEQAERYEEMVQFMEQLVSGATPAGELTVEERNLLSVAYKNVIG 60
Query: 95 PLRAALRILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSAS 151
LRAA RI+S ++EE RKNE+ H VK Y+SKVE+EL ++C IL LLDS LIPSA+
Sbjct: 61 SLRAAWRIVSSIEQKEESRKNEE-HVSLVKDYRSKVETELSSICSGILRLLDSHLIPSAT 119
Query: 152 SSEIRVVYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGL 211
+SE +V Y +MKGDY RY+AEFK GD++K+A ED +++YKAAQD+A ADL +HPIRLGL
Sbjct: 120 ASESKVFYLKMKGDYHRYLAEFKSGDERKTAAEDTMIAYKAAQDVAVADLAPTHPIRLGL 179
Query: 212 ALNFSVFYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTF 270
ALNFSVFYYEILN ++ MA+QA +EA EL LG+E YKDST+ MQLLR+N+ LWT
Sbjct: 180 ALNFSVFYYEILNSSEKACSMAKQAFEEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTS 239
Query: 271 D 271
D
Sbjct: 240 D 240
>At1g78300 putative tyrosine activation protein
Length = 259
Score = 258 bits (658), Expect = 3e-69
Identities = 136/235 (57%), Positives = 176/235 (74%), Gaps = 6/235 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE+V+ AKLA+QAERYEEMV FM+K+ EL++EE NLLSVAYKN RA+
Sbjct: 5 REEFVYMAKLAEQAERYEEMVEFMEKVSAA-VDGDELTVEERNLLSVAYKNVIGARRASW 63
Query: 101 RILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRV 157
RI+S ++EE R N DDH +++Y+SK+E+EL +C IL+LLDS+LIP+A+S + +V
Sbjct: 64 RIISSIEQKEESRGN-DDHVTAIREYRSKIETELSGICDGILKLLDSRLIPAAASGDSKV 122
Query: 158 VYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSV 217
Y +MKGDY RY+AEFK G ++K A E + +YK+AQDIA A+L +HPIRLGLALNFSV
Sbjct: 123 FYLKMKGDYHRYLAEFKTGQERKDAAEHTLAAYKSAQDIANAELAPTHPIRLGLALNFSV 182
Query: 218 FYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
FYYEILN D ++A+QA DEA EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 183 FYYEILNSPDRACNLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTSD 237
>At1g35160 endonuclease like protein (32KD)
Length = 267
Score = 254 bits (650), Expect = 3e-68
Identities = 134/235 (57%), Positives = 175/235 (74%), Gaps = 6/235 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE+V+ AKLA+QAERYEEMV FM+K+ EL++EE NLLSVAYKN RA+
Sbjct: 11 REEFVYLAKLAEQAERYEEMVEFMEKVAEA-VDKDELTVEERNLLSVAYKNVIGARRASW 69
Query: 101 RILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRV 157
RI+S ++EE R N DDH ++ Y+SK+ESEL +C IL+LLD++L+P++++ + +V
Sbjct: 70 RIISSIEQKEESRGN-DDHVTTIRDYRSKIESELSKICDGILKLLDTRLVPASANGDSKV 128
Query: 158 VYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSV 217
Y +MKGDY RY+AEFK G ++K A E + +YKAAQDIA A+L +HPIRLGLALNFSV
Sbjct: 129 FYLKMKGDYHRYLAEFKTGQERKDAAEHTLTAYKAAQDIANAELAPTHPIRLGLALNFSV 188
Query: 218 FYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
FYYEILN D ++A+QA DEA EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 189 FYYEILNSPDRACNLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTSD 243
>At3g02520 putative 14-3-3 protein
Length = 265
Score = 252 bits (644), Expect = 1e-67
Identities = 137/235 (58%), Positives = 171/235 (72%), Gaps = 6/235 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE V+ AKLA+QAERYEEMV FM+K V EL++EE NLLSVAYKN RA+
Sbjct: 5 REENVYLAKLAEQAERYEEMVEFMEK-VAKTVDTDELTVEERNLLSVAYKNVIGARRASW 63
Query: 101 RILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRV 157
RI+S ++EE R N DDH +K Y+ K+E+EL +C IL LLDS L+P+AS +E +V
Sbjct: 64 RIISSIEQKEESRGN-DDHVSIIKDYRGKIETELSKICDGILNLLDSHLVPTASLAESKV 122
Query: 158 VYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSV 217
Y +MKGDY RY+AEFK G ++K A E +++YK+AQDIA ADL +HPIRLGLALNFSV
Sbjct: 123 FYLKMKGDYHRYLAEFKTGAERKEAAESTLVAYKSAQDIALADLAPTHPIRLGLALNFSV 182
Query: 218 FYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
FYYEILN D +A+QA DEA +EL LG+E YKDST+ MQLLR+N+ LW D
Sbjct: 183 FYYEILNSPDRACSLAKQAFDEAISELDTLGEESYKDSTLIMQLLRDNLTLWNSD 237
>At5g16050 14-3-3-LIKE PROTEIN GF14 UPSILON
Length = 268
Score = 251 bits (640), Expect = 4e-67
Identities = 133/234 (56%), Positives = 170/234 (71%), Gaps = 4/234 (1%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE V+ AKLA+QAERYEEMV FM+K V EL++EE NLLSVAYKN RA+
Sbjct: 7 REENVYLAKLAEQAERYEEMVEFMEK-VAKTVETEELTVEERNLLSVAYKNVIGARRASW 65
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RI+S +++E + DH +K Y+ K+E+EL +C IL LL++ LIP+AS +E +V
Sbjct: 66 RIISSIEQKEDSRGNSDHVSIIKDYRGKIETELSKICDGILNLLEAHLIPAASLAESKVF 125
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVF 218
Y +MKGDY RY+AEFK G ++K A E +++YK+AQDIA ADL +HPIRLGLALNFSVF
Sbjct: 126 YLKMKGDYHRYLAEFKTGAERKEAAESTLVAYKSAQDIALADLAPTHPIRLGLALNFSVF 185
Query: 219 YYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
YYEILN D +A+QA DEA +EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 186 YYEILNSSDRACSLAKQAFDEAISELDTLGEESYKDSTLIMQLLRDNLTLWTSD 239
>At4g09000 14-3-3-like protein
Length = 267
Score = 251 bits (640), Expect = 4e-67
Identities = 133/235 (56%), Positives = 175/235 (73%), Gaps = 6/235 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
R+E+V+ AKLA+QAERYEEMV FM+K+ EL++EE NLLSVAYKN RA+
Sbjct: 10 RDEFVYMAKLAEQAERYEEMVEFMEKVAKA-VDKDELTVEERNLLSVAYKNVIGARRASW 68
Query: 101 RILS---KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRV 157
RI+S ++EE R N DDH ++ Y+SK+E+EL ++C IL+LLD+ L+P+A+S + +V
Sbjct: 69 RIISSIEQKEESRGN-DDHVSLIRDYRSKIETELSDICDGILKLLDTILVPAAASGDSKV 127
Query: 158 VYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSV 217
Y +MKGDY RY+AEFK G ++K A E + +YKAAQDIA ++L +HPIRLGLALNFSV
Sbjct: 128 FYLKMKGDYHRYLAEFKSGQERKDAAEHTLTAYKAAQDIANSELAPTHPIRLGLALNFSV 187
Query: 218 FYYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
FYYEILN D ++A+QA DEA EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 188 FYYEILNSPDRACNLAKQAFDEAIAELDTLGEESYKDSTLIMQLLRDNLTLWTSD 242
>At5g38480 14-3-3 protein homolog RCI1 (pir||S47969)
Length = 255
Score = 250 bits (638), Expect = 7e-67
Identities = 133/234 (56%), Positives = 169/234 (71%), Gaps = 4/234 (1%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
REE V+ AKLA+QAERYEEMV FM+K V ELS+EE NLLSVAYKN RA+
Sbjct: 4 REENVYMAKLAEQAERYEEMVEFMEK-VAKTVDVEELSVEERNLLSVAYKNVIGARRASW 62
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RI+S +++E K +DH +K Y+ K+ESEL +C IL +L++ LIPSAS +E +V
Sbjct: 63 RIISSIEQKEESKGNEDHVAIIKDYRGKIESELSKICDGILNVLEAHLIPSASPAESKVF 122
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVF 218
Y +MKGDY RY+AEFK G ++K A E +++YK+A DIA A+L +HPIRLGLALNFSVF
Sbjct: 123 YLKMKGDYHRYLAEFKAGAERKEAAESTLVAYKSASDIATAELAPTHPIRLGLALNFSVF 182
Query: 219 YYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
YYEILN D +A+QA D+A EL LG+E YKDST+ MQLLR+N+ LWT D
Sbjct: 183 YYEILNSPDRACSLAKQAFDDAIAELDTLGEESYKDSTLIMQLLRDNLTLWTSD 236
>At1g26480 14-3-3 protein epsilon like
Length = 268
Score = 225 bits (573), Expect = 2e-59
Identities = 121/234 (51%), Positives = 160/234 (67%), Gaps = 6/234 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
RE +V+ AKL++QAERY+EMV M+K+ SEL++EE NLLSV YKN RA+
Sbjct: 10 RETFVYMAKLSEQAERYDEMVETMKKVA---RVNSELTVEERNLLSVGYKNVIGARRASW 66
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RI+S +++E K + + +K Y+ KVE EL N+C IL ++D LIP A+S E V
Sbjct: 67 RIMSSIEQKEESKGNESNVKQIKGYRQKVEDELANICQDILTIIDQHLIPHATSGEATVF 126
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVF 218
YY+MKGDY RY+AEFK ++K A E + Y+AA A+ +L S+HPIRLGLALNFSVF
Sbjct: 127 YYKMKGDYYRYLAEFKTEQERKEAAEQSLKGYEAATQAASTELPSTHPIRLGLALNFSVF 186
Query: 219 YYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
YYEI+N + +A+QA DEA EL L +E YKDST+ MQLLR+N+ LWT D
Sbjct: 187 YYEIMNSPERACHLAKQAFDEAIAELDTLSEESYKDSTLIMQLLRDNLTLWTSD 240
>At2g42590 14-3-3 regulatory protein (GF14 mu)
Length = 263
Score = 212 bits (540), Expect = 2e-55
Identities = 117/234 (50%), Positives = 157/234 (67%), Gaps = 6/234 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
R+ +V+ AKL++QAERYEEMV M+ + +L++EE NLLSV YKN RA+
Sbjct: 7 RDTFVYLAKLSEQAERYEEMVESMKSVAKLNV---DLTVEERNLLSVGYKNVIGSRRASW 63
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RI S +++E K D + +K+Y KVE EL N+C I+ +LD LIPSAS E V
Sbjct: 64 RIFSSIEQKEAVKGNDVNVKRIKEYMEKVELELSNICIDIMSVLDEHLIPSASEGESTVF 123
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVF 218
+ +MKGDY RY+AEFK G+++K A + + +Y+ A A A L +HPIRLGLALNFSVF
Sbjct: 124 FNKMKGDYYRYLAEFKSGNERKEAADQSLKAYEIATTAAEAKLPPTHPIRLGLALNFSVF 183
Query: 219 YYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
YYEI+N + +A+QA DEA +EL L +E YKDST+ MQLLR+N+ LWT D
Sbjct: 184 YYEIMNAPERACHLAKQAFDEAISELDTLNEESYKDSTLIMQLLRDNLTLWTSD 237
>At1g34760 putative protein
Length = 241
Score = 212 bits (539), Expect = 2e-55
Identities = 116/234 (49%), Positives = 159/234 (67%), Gaps = 6/234 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
R + V+ AKL +QAERY+EMV M+K+ EL++EE NLLSV YKN RA+
Sbjct: 5 RAKQVYLAKLNEQAERYDEMVEAMKKVAALDV---ELTIEERNLLSVGYKNVIGARRASW 61
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RILS +++E K + + +K Y++KVE EL +C IL ++D L+P A+S E V
Sbjct: 62 RILSSIEQKEESKGNEQNAKRIKDYRTKVEEELSKICYDILAVIDKHLVPFATSGESTVF 121
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVF 218
YY+MKGDY RY+AEFK G D++ A + + +Y+AA A+ +L ++HPIRLGLALNFSVF
Sbjct: 122 YYKMKGDYFRYLAEFKSGADREEAADLSLKAYEAATSSASTELSTTHPIRLGLALNFSVF 181
Query: 219 YYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
YYEILN + +A++A DEA EL L ++ YKDST+ MQLLR+N+ LWT D
Sbjct: 182 YYEILNSPERACHLAKRAFDEAIAELDSLNEDSYKDSTLIMQLLRDNLTLWTSD 235
>At1g22300 unknown protein
Length = 254
Score = 211 bits (537), Expect = 4e-55
Identities = 115/234 (49%), Positives = 157/234 (66%), Gaps = 6/234 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
RE+ V+ AKL++Q ERY+EMV M+K+ EL++EE NL+SV YKN RA+
Sbjct: 5 REKQVYLAKLSEQTERYDEMVEAMKKVA---QLDVELTVEERNLVSVGYKNVIGARRASW 61
Query: 101 RILS--KEEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVV 158
RILS +++E K D++ +K Y+ +VE EL VC IL ++D LIPS+++ E V
Sbjct: 62 RILSSIEQKEESKGNDENVKRLKNYRKRVEDELAKVCNDILSVIDKHLIPSSNAVESTVF 121
Query: 159 YYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSVF 218
+Y+MKGDY RY+AEF G ++K A + + +YKAA A L +HP+RLGLALNFSVF
Sbjct: 122 FYKMKGDYYRYLAEFSSGAERKEAADQSLEAYKAAVAAAENGLAPTHPVRLGLALNFSVF 181
Query: 219 YYEILNRFDEGLDMARQALDEARNEL-KLGDEYYKDSTVRMQLLRNNIILWTFD 271
YYEILN + +A+QA D+A EL L +E YKDST+ MQLLR+N+ LWT D
Sbjct: 182 YYEILNSPESACQLAKQAFDDAIAELDSLNEESYKDSTLIMQLLRDNLTLWTSD 235
>At1g78220 14-3-3 GF14 Pi protein (GRF13)
Length = 245
Score = 145 bits (367), Expect = 2e-35
Identities = 89/236 (37%), Positives = 138/236 (57%), Gaps = 7/236 (2%)
Query: 41 REEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASELSLEEMNLLSVAYKNATEPLRAAL 100
RE+ ++ AKL QA RY++++ M+K+ ELS EE +LL+ YKN E R +L
Sbjct: 5 REKLIYLAKLGCQAGRYDDVMKSMRKVC---ELDIELSEEERDLLTTGYKNVMEAKRVSL 61
Query: 101 RILSKEE--EGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSS-EIRV 157
R++S E E K D + +K + V+ E NVC IL L+DS LIPS +++ E V
Sbjct: 62 RVISSIEKMEDSKGNDQNVKLIKGQQEMVKYEFFNVCNDILSLIDSHLIPSTTTNVESIV 121
Query: 158 VYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNFSV 217
++ ++KGDY RYMAEF ++K ++ + +YK A ++A L ++ +RLGLALNFS+
Sbjct: 122 LFNRVKGDYFRYMAEFGSDAERKENADNSLDAYKVAMEMAENSLAPTNMVRLGLALNFSI 181
Query: 218 FYYEILNRFDEGLDMARQALDEARNELK-LGDEYYKDSTVRMQLLRNNIILWTFDD 272
F YEI + + ++A DEA EL L ++S +++L+ N+ WT D
Sbjct: 182 FNYEIHKSIESACKLVKKAYDEAITELDGLDKNICEESMYIIEMLKYNLSTWTSGD 237
>At1g22290 hypothetical protein
Length = 196
Score = 92.8 bits (229), Expect = 2e-19
Identities = 66/204 (32%), Positives = 108/204 (52%), Gaps = 23/204 (11%)
Query: 44 YVFHAKLAQQAERYEEMVSFMQKIVVGYTPA-SELSLEEMNLLSVAYKNATEPLRAALRI 102
+V A L+ +ERY E ++K + +ELS +E NL+SV YKN RA+L I
Sbjct: 8 HVHFASLSSSSERYNETFEEIKKAMKKSVQLKAELSAKERNLVSVGYKNVISARRASLEI 67
Query: 103 LSK--EEEGRKNEDDHFVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVVYY 160
LS ++E K +++ +K Y++KVE EL +C IL +++ +LIPS+++ + V++Y
Sbjct: 68 LSSIVQKEESKGNEENVKKLKNYRNKVEDELAKICNDILSVINKQLIPSSTTVDSSVLFY 127
Query: 161 QMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDIAAADLRSSHPIRLGLALNF-SVFY 219
M +A+F + K A + + +YK L +L + LN+ SVF
Sbjct: 128 NM-------LADFSSNAESKEATDQSLDAYKR--------LVWYQQFQLYMTLNWTSVF- 171
Query: 220 YEILNRFDEGLDMARQALDEARNE 243
LN + +A+QA D+A NE
Sbjct: 172 ---LNSPESAYQLAKQAFDDAINE 192
>At3g27720 hypothetical protein
Length = 493
Score = 40.0 bits (92), Expect = 0.001
Identities = 43/177 (24%), Positives = 83/177 (46%), Gaps = 12/177 (6%)
Query: 4 YRYRDSHNHTLTLDRKTKQSNKREKRIMGFTVAERLRREEYVFHAKLAQQAERYEEMVSF 63
YRY+ +H +L L+ K K+S + K ++ ++ +EY + + R ++S+
Sbjct: 313 YRYK-AHIDSLKLEDKLKKSILK-KAVLNSETKDQKVFKEYSWIIDAVNRLFRSRRILSY 370
Query: 64 MQKIVVGYTPASEL-----SLEEMNLLSVAYKNATEPLRAALRILSKEEEGRKNEDDH-- 116
V Y EL S EE N+ +++ + L + LSK E +E DH
Sbjct: 371 SYPFVF-YMFGKELFKDDMSDEERNIKKNLFEDQQQQLEGNVERLSKILEEPFDEYDHEK 429
Query: 117 FVHVKKYKSKVESELENVCGSILELLDSKLIPSASSSEIRVVYYQMKGDYQRYMAEF 173
V + ++ + + + ++N+C + E ++++L+ S + Y+ KG Q AEF
Sbjct: 430 VVEMMRHLTNLTAVVDNLCKEMYECIENELLGPLISGIHNIAPYRSKGIEQ--AAEF 484
>At2g10450 14-3-3 regulatory protein
Length = 82
Score = 39.3 bits (90), Expect = 0.002
Identities = 16/26 (61%), Positives = 21/26 (80%)
Query: 246 LGDEYYKDSTVRMQLLRNNIILWTFD 271
LGDE YKDST+ M++LR+N+ WT D
Sbjct: 24 LGDELYKDSTLIMKILRDNLTFWTSD 49
>At3g22790 unknown protein
Length = 1694
Score = 35.8 bits (81), Expect = 0.027
Identities = 57/226 (25%), Positives = 91/226 (40%), Gaps = 31/226 (13%)
Query: 14 LTLDRKTKQSNKREKRIMGFTVAERLRREEYVFHAKLAQQAERYEEMVSFMQKIVVGYTP 73
L L+ TK REK E LR E+Y F A+ A + + M MQK+
Sbjct: 626 LLLESNTKLDGSREKTKDLQERCESLRGEKYEFIAERANLLSQLQIMTENMQKL------ 679
Query: 74 ASELSLEEMNLLSVAYKNATEPLRAALRILSKEEEGRKNEDDHFVHVKKYKSKVESELEN 133
LE+ +LL + A L+ +E K ++ F +K K+++ E E+
Sbjct: 680 -----LEKNSLLETSLSGANIELQCV-------KEKSKCFEEFFQLLKNDKAELIKERES 727
Query: 134 VCGSILELLDSKLIPSASSSEIRVVYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAA 193
+ S L + KL + + +++G Y E + K VE++ +S
Sbjct: 728 LI-SQLNAVKEKL------GVLEKKFTELEGKYADLQREKQF---KNLQVEELRVSLATE 777
Query: 194 QDIAAADLRSSHPIRLGLALNFSVFYYEILNR---FDEGLDMARQA 236
+ A+ RS+ L N S E +R F+E LD A A
Sbjct: 778 KQERASYERSTDTRLADLQNNVSFLREECRSRKKEFEEELDRAVNA 823
>At1g30690 putative phosphatidyl-inositol-transfer protein
Length = 540
Score = 34.7 bits (78), Expect = 0.061
Identities = 52/207 (25%), Positives = 89/207 (42%), Gaps = 20/207 (9%)
Query: 11 NHTLTLDRKTKQSNKREKRIMGFTVAERLRREEYVFHAKLAQQAERYEEMVSFMQKIVVG 70
++TL +K + S +EK+ E + + E K + AE E + +V
Sbjct: 85 DNTLLKTKKKESSPMKEKK-------EEVVKPEAEVEKKKEEAAEEKVEEEKKSEAVVTE 137
Query: 71 YTPASELSLEEMNLLSVAYKNATEPLRAALRILSKEEEGRKNEDDHFVHVKKYKSKVESE 130
P +E ++E + + K + + +KEEE +K ED VK +VE E
Sbjct: 138 EAPKAE-TVEAVVTEEIIPKEEVTTVVEKVEEETKEEE-KKTEDVVTEEVKAETIEVEDE 195
Query: 131 LENVCGSILELLDSKLIPS--ASSSEIRVVYYQMKGDY---QRYMAEFKIGDDKKSAVED 185
E+V I EL L+PS A S+++ ++ + D+ + + K +K D
Sbjct: 196 DESVDKDI-ELWGVPLLPSKGAESTDVILLKFLRARDFKVNEAFEMLKKTLKWRKQNKID 254
Query: 186 IILSYKAAQDIAAADL-----RSSHPI 207
IL + +D+A A R SHP+
Sbjct: 255 SILGEEFGEDLATAAYMNGVDRESHPV 281
>At5g52280 hyaluronan mediated motility receptor-like protein
Length = 853
Score = 33.5 bits (75), Expect = 0.13
Identities = 40/243 (16%), Positives = 95/243 (38%), Gaps = 7/243 (2%)
Query: 17 DRKTKQSNKREKRIMGFTVAERLRREEYVFHAKLAQQAERYEEMVSFMQKIVVGYTPASE 76
D T K E+ E LR+ + + E+ + + M+ + + ++
Sbjct: 547 DIDTMMREKTEQEQRAIKAEENLRKTRWNNAITAERLQEKCKRLSLEMESKLSEHENLTK 606
Query: 77 LSLEEMNLLSVAYKNATEPLRAALRILSKEEEGRKNEDDHFVHVKKYKSKVESELENVCG 136
+L E N L + K E +++E+E RK HV++ + +++ +
Sbjct: 607 KTLAEANNLRLQNKTLEEMQEKTHTEITQEKEQRK-------HVEEKNKALSMKVQMLES 659
Query: 137 SILELLDSKLIPSASSSEIRVVYYQMKGDYQRYMAEFKIGDDKKSAVEDIILSYKAAQDI 196
+L+L + SA+++E + + + + + + + + + + K++ D
Sbjct: 660 EVLKLTKLRDESSAAATETEKIIQEWRKERDEFERKLSLAKEVAKTAQKELTLTKSSNDD 719
Query: 197 AAADLRSSHPIRLGLALNFSVFYYEILNRFDEGLDMARQALDEARNELKLGDEYYKDSTV 256
LR+ GL+L +S + E ++ +Q + + + +E K
Sbjct: 720 KETRLRNLKTEVEGLSLQYSELQNSFVQEKMENDELRKQVSNLKVDIRRKEEEMTKILDA 779
Query: 257 RMQ 259
RM+
Sbjct: 780 RME 782
>At4g27980 putative protein
Length = 565
Score = 32.3 bits (72), Expect = 0.30
Identities = 23/101 (22%), Positives = 45/101 (43%), Gaps = 10/101 (9%)
Query: 17 DRKTKQSNKREKRIMGFTVAER-----LRREEYVFHAKLAQQAERYEEMVSFMQKIVVGY 71
+R+ ++S REK + A + L+R+E K+ ++AE+ E M+K +
Sbjct: 170 ERRNEESEAREKDLRALEEAVKEKTAELKRKEETLELKMKEEAEKLREETELMRKGLEIK 229
Query: 72 TPASELSLEEMNLLSVAYKNATEPLRAALRILSKEEEGRKN 112
E L+E+ L + + + P +++ E R N
Sbjct: 230 EKTLEKRLKELELKQMELEETSRP-----QLVEAESRKRSN 265
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.134 0.371
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,894,284
Number of Sequences: 26719
Number of extensions: 233335
Number of successful extensions: 949
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 886
Number of HSP's gapped (non-prelim): 53
length of query: 286
length of database: 11,318,596
effective HSP length: 98
effective length of query: 188
effective length of database: 8,700,134
effective search space: 1635625192
effective search space used: 1635625192
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141414.9


