

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141414.11 - phase: 0
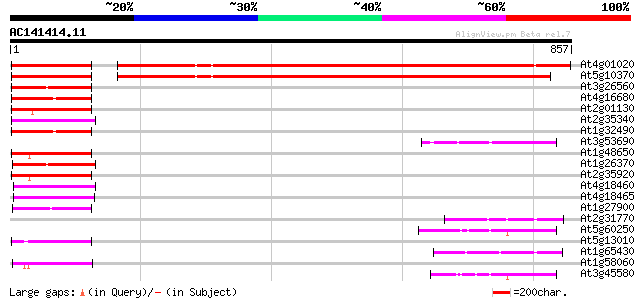
(857 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g01020 unknown protein 684 0.0
At5g10370 putative protein 629 e-180
At3g26560 ATP-dependent RNA helicase, putative 103 5e-22
At4g16680 RNA helicase 94 3e-19
At2g01130 ATP-dependent RNA like helicase A 94 4e-19
At2g35340 putative pre-mRNA splicing factor RNA helicase 92 9e-19
At1g32490 putative RNA helicase (At1g32490) 91 2e-18
At3g53690 unknown protein 90 5e-18
At1g48650 hypothetical protein 90 5e-18
At1g26370 hypothetical protein 90 5e-18
At2g35920 ATP-dependent RNA helicase A like protein 90 6e-18
At4g18460 RNA helicase - like protein 89 1e-17
At4g18465 RNA helicase - like protein 88 2e-17
At1g27900 ATP-dependent RNA helicase (At1g27900) 88 2e-17
At2g31770 putative ARI-like RING zinc finger protein 86 7e-17
At5g60250 unknown protein 85 2e-16
At5g13010 pre-mRNA splicing factor ATP-dependent RNA helicase -l... 84 3e-16
At1g65430 unknown protein 82 1e-15
At1g58060 hypothetical protein 82 1e-15
At3g45580 putative protein 82 2e-15
>At4g01020 unknown protein
Length = 1787
Score = 684 bits (1766), Expect = 0.0
Identities = 344/695 (49%), Positives = 466/695 (66%), Gaps = 6/695 (0%)
Query: 165 IDEMELLMFFEKNTSGCICDMQKFTGMVKDVEDKAKWGKITFMTSNAAKRAAELDGEEFC 224
+D+ ELL F EK GCIC + KF +D ++K KWG+ITF+T +A +A E+ +F
Sbjct: 1077 VDDRELLTFLEKKIDGCICSIYKFAANKQDCDEKEKWGRITFLTPESAMKATEIQKFDFK 1136
Query: 225 GSPLKIVHSQSAMGGDTTFS-FPAVEARISWLRRPIKAVGIIKCDKNDVDFIIRDFENLI 283
GS LK+ S S GG F +V A+I W R+ G +KC D+ I+ D +L
Sbjct: 1137 GSVLKVFPSLSTGGGIFKMPYFSSVTAKIRWPRKESSGRGCLKCPSGDIHSILGDITSLE 1196
Query: 284 VDGRRYVRCAPSDKYLDNILITGLDKEVPETKILDILRSATSRRILDFF-FKRGDAVENP 342
+ G YV D+ILI+GL ++ E ++LD+L T RR L+FF F++ +V+ P
Sbjct: 1197 I-GTNYVHIQRDQLSNDSILISGLG-DLSEAEVLDVLEFRTQRRDLNFFIFRKKYSVQCP 1254
Query: 343 PCSMIAETILKEISPLMPKKKPHISSCRVQVFPPKPKDYSMNALIHFDGRLHLEAAKALE 402
+ E + K I M K P + +VQVF PK +Y M ALI FDGRLHLEAAKAL+
Sbjct: 1255 SPTACEEELHKRIFARMSAKNPEPNCVQVQVFEPKEDNYFMRALIKFDGRLHLEAAKALQ 1314
Query: 403 KIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLEWKLDIT 462
+++G+VLPG WQKIK ++LF S++I S +Y+ +K QL +LARF +G E L+ T
Sbjct: 1315 ELNGEVLPGCLPWQKIKCEQLFQSSIICSASIYNTVKRQLNVLLARFERQKGGECCLEPT 1374
Query: 463 PNGSHRVKITANATKTVAEGRRLLEELWRGKVIVHDNLTPATLQPILSKDGSSLTSSIQK 522
NG++RVKITA AT+ VAE RR LEEL RGK I H TP +Q ++S+DG +L IQ+
Sbjct: 1375 HNGAYRVKITAYATRPVAEMRRELEELLRGKPINHPGFTPRVVQHLMSRDGINLMRKIQQ 1434
Query: 523 ATSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQ 582
T TYI DR N+ +RI G+ +KIA AE++L+QSL+ H+ KQ I L G ++ D MK+
Sbjct: 1435 ETETYILLDRHNLTVRICGTSEKIAKAEQELVQSLMDYHESKQLEIHLRGPEIRPDLMKE 1494
Query: 583 VVKNFGPDLHGLKEKVPGADLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERF 642
VVK FGP+L G+KEKV G DL+LNTR I HG+ E++ V+++ E+AR + E+
Sbjct: 1495 VVKRFGPELQGIKEKVHGVDLKLNTRYHVIQVHGSKEMRQEVQKMVNELAREKSALGEKP 1554
Query: 643 D-TGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHIL 701
D CPICL EV+DGY LEGC HLFC++C++EQ E++++N +FPI C+H CG I+
Sbjct: 1555 DEIELECPICLSEVDDGYSLEGCSHLFCKACLLEQFEASMRNFDAFPILCSHIDCGAPIV 1614
Query: 702 LVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGA 761
+ D R LLS +KL+EL ASL AFV SS G RFC +PDCPSIYRVA P + PF+CGA
Sbjct: 1615 VADMRALLSQEKLDELISASLSAFVTSSDGKLRFCSTPDCPSIYRVAGPQESGEPFICGA 1674
Query: 762 CYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCN 821
C+SETCTRCH+EYHP ++CERY++FK++PD SL+DW KGK+ VK CP C IEK DGCN
Sbjct: 1675 CHSETCTRCHLEYHPLITCERYKKFKENPDLSLKDWAKGKD-VKECPICKSTIEKTDGCN 1733
Query: 822 HIECKCGKHICWVCLEFFTTSGECYSHMDTIHLGV 856
H++C+CGKHICW CL+ FT + CY+H+ TIH G+
Sbjct: 1734 HLQCRCGKHICWTCLDVFTQAEPCYAHLRTIHGGI 1768
Score = 184 bits (467), Expect = 2e-46
Identities = 85/121 (70%), Positives = 107/121 (88%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI VLIGETGSGKSTQLVQFLADSGV A+ESIVCTQPR+IAA +L +RVREES GCYE++
Sbjct: 320 QIMVLIGETGSGKSTQLVQFLADSGVAASESIVCTQPRKIAAMTLTDRVREESSGCYEEN 379
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
++ C +FSS + S++++MTD+CLLQHYM D++ +GISC+I+DEAHERS+NTDLLLAL
Sbjct: 380 TVSCTPTFSSTEEISSKVVYMTDNCLLQHYMKDRSLSGISCVIIDEAHERSLNTDLLLAL 439
Query: 124 V 124
+
Sbjct: 440 L 440
>At5g10370 putative protein
Length = 1751
Score = 629 bits (1621), Expect = e-180
Identities = 323/665 (48%), Positives = 440/665 (65%), Gaps = 6/665 (0%)
Query: 165 IDEMELLMFFEKNTSGCICDMQKFTGMVKDVEDKAKWGKITFMTSNAAKRAAELDGEEFC 224
+D+ ELL F EK G IC + KF +D ++K KWG+ITF+T +A +A E+ F
Sbjct: 1080 VDDRELLTFLEKKIDGSICSIYKFAANKQDCDEKEKWGRITFLTPESAMKATEIQKFYFK 1139
Query: 225 GSPLKIVHSQSAMGGDTTFS-FPAVEARISWLRRPIKAVGIIKCDKNDVDFIIRDFENLI 283
GS LK+ S S GG F +V A+I W RR G +KC D+ I+ D +L
Sbjct: 1140 GSVLKLFPSLSTGGGIFKMPYFSSVTAKIRWPRRESSGRGCLKCPSGDIHRILGDISSLE 1199
Query: 284 VDGRRYVRCAPSDKYLDNILITGLDKEVPETKILDILRSATSRRILDFF-FKRGDAVENP 342
+ G YV + D+ILI+GL ++ E ++LD+L T RR L+FF F++ +V+ P
Sbjct: 1200 I-GTNYVHIQRDQQSNDSILISGLG-DLSEAEVLDVLEFRTQRRDLNFFIFRKKYSVQCP 1257
Query: 343 PCSMIAETILKEISPLMPKKKPHISSCRVQVFPPKPKDYSMNALIHFDGRLHLEAAKALE 402
+ E + K I M K P + +VQVF PK +Y M ALI FDGRLH EAAKAL+
Sbjct: 1258 SPTACEEELHKRIFARMSAKNPEPNCVQVQVFEPKEDNYFMRALIKFDGRLHFEAAKALQ 1317
Query: 403 KIDGKVLPGFHSWQKIKTQRLFHSTLIFSPPVYHVIKGQLEKVLARFNNLEGLEWKLDIT 462
+++G+VLPG WQKIK ++LF S++I S +Y+ +K QL +LARF +G E L+ T
Sbjct: 1318 ELNGEVLPGCLPWQKIKCEQLFQSSIICSASIYNTVKRQLNVLLARFERQKGGECCLEPT 1377
Query: 463 PNGSHRVKITANATKTVAEGRRLLEELWRGKVIVHDNLTPATLQPILSKDGSSLTSSIQK 522
NG++RVKITA AT+ VAE RR LEEL RG+ I H T LQ ++S+DG +L IQ+
Sbjct: 1378 HNGAYRVKITAYATRPVAEMRRELEELLRGRPINHPGFTRRVLQHLMSRDGINLMRKIQQ 1437
Query: 523 ATSTYIQFDRRNMKLRIFGSPDKIALAEKKLIQSLLSLHDEKQSVICLSGRDLPSDFMKQ 582
T TYI DR N+ +RI G+ +KIA AE++LIQ+L+ H+ KQ I L G ++ D MK+
Sbjct: 1438 ETETYILLDRHNLTVRICGTSEKIAKAEQELIQALMDYHESKQLEIHLRGPEIRPDLMKE 1497
Query: 583 VVKNFGPDLHGLKEKVPGADLRLNTRNRTILCHGNSELKSRVEEITFEIARLSNPSSERF 642
VVK FGP+L G+KEKV G DL+LNTR I HG+ E++ V+++ E+AR + E+
Sbjct: 1498 VVKRFGPELQGIKEKVHGVDLKLNTRYHVIQVHGSKEMRQEVQKMVNELAREKSALGEKP 1557
Query: 643 D-TGPSCPICLCEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNHIL 701
D CPICL EV+DGY LEGC HLFC++C++EQ E++++N +FPI C+H CG I+
Sbjct: 1558 DEIEVECPICLSEVDDGYSLEGCSHLFCKACLLEQFEASMRNFDAFPILCSHIDCGAPIV 1617
Query: 702 LVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGA 761
L D R LLS +KL+ELF ASL +FV SS G +RFC +PDCPS+YRVA P + PF+CGA
Sbjct: 1618 LADMRALLSQEKLDELFSASLSSFVTSSDGKFRFCSTPDCPSVYRVAGPQESGEPFICGA 1677
Query: 762 CYSETCTRCHIEYHPYVSCERYRQFKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCN 821
C+SE CTRCH+EYHP ++CERY++FK++PD SL+DW KGK VK CP C IEK DGCN
Sbjct: 1678 CHSEICTRCHLEYHPLITCERYKKFKENPDLSLKDWAKGK-NVKECPICKSTIEKTDGCN 1736
Query: 822 HIECK 826
H++C+
Sbjct: 1737 HMKCR 1741
Score = 186 bits (471), Expect = 6e-47
Identities = 86/121 (71%), Positives = 108/121 (89%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
QI VLIGETGSGKSTQLVQFLADSGV A+ESIVCTQPR+IAA +LA+RVREES GCYE++
Sbjct: 323 QIMVLIGETGSGKSTQLVQFLADSGVAASESIVCTQPRKIAAMTLADRVREESSGCYEEN 382
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
++ C +FSS + S++++MTD+CLLQHYM D++ +GISC+I+DEAHERS+NTDLLLAL
Sbjct: 383 TVSCTPTFSSTEEISSKVVYMTDNCLLQHYMKDRSLSGISCVIIDEAHERSLNTDLLLAL 442
Query: 124 V 124
+
Sbjct: 443 L 443
>At3g26560 ATP-dependent RNA helicase, putative
Length = 1168
Score = 103 bits (256), Expect = 5e-22
Identities = 56/121 (46%), Positives = 79/121 (65%), Gaps = 1/121 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ V+IGETGSGK+TQ+ Q+LA++G I CTQPRR+AA S+A+RV EE GC
Sbjct: 532 QVLVVIGETGSGKTTQVTQYLAEAGYTTKGKIGCTQPRRVAAMSVAKRVAEEF-GCRLGE 590
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F D+ I +MTD LL+ + D+N + S I++DEAHER+I+TD+L L
Sbjct: 591 EVGYAIRFEDCTGPDTVIKYMTDGMLLREILIDENLSQYSVIMLDEAHERTIHTDVLFGL 650
Query: 124 V 124
+
Sbjct: 651 L 651
>At4g16680 RNA helicase
Length = 883
Score = 94.0 bits (232), Expect = 3e-19
Identities = 54/124 (43%), Positives = 76/124 (60%), Gaps = 7/124 (5%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESG---GCY 60
Q+ V++GETGSGK+TQ+ Q+L ++G I CTQPRR+AA S+A RV +E G G
Sbjct: 239 QVLVIVGETGSGKTTQIPQYLQEAGYTKRGKIGCTQPRRVAAMSVASRVAQEVGVKLGHE 298
Query: 61 EDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLL 120
SI+ F + I +MTD LL+ + + S II+DEAHER+++TD+L
Sbjct: 299 VGYSIR----FEDCTSEKTVIKYMTDGMLLRELLIEPKLDSYSVIIIDEAHERTLSTDIL 354
Query: 121 LALV 124
ALV
Sbjct: 355 FALV 358
>At2g01130 ATP-dependent RNA like helicase A
Length = 899
Score = 93.6 bits (231), Expect = 4e-19
Identities = 52/125 (41%), Positives = 77/125 (61%), Gaps = 5/125 (4%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANE----SIVCTQPRRIAAKSLAERVREESGGC 59
Q+ V+ GETG GK+TQ+ QF+ +S + AN SI+CTQPRRI+A S++ERV E G
Sbjct: 25 QVIVISGETGCGKTTQIPQFILESEIEANRGAFSSIICTQPRRISAMSVSERVAYERGEQ 84
Query: 60 YEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
+S + D+R++F T LL+ + D+N G++ +IVDE HER +N D
Sbjct: 85 LGES-VGYKVRLEGVKGRDTRLLFCTTGILLRRLLVDRNLRGVTHVIVDEIHERGMNEDF 143
Query: 120 LLALV 124
LL ++
Sbjct: 144 LLIIL 148
>At2g35340 putative pre-mRNA splicing factor RNA helicase
Length = 1087
Score = 92.4 bits (228), Expect = 9e-19
Identities = 50/127 (39%), Positives = 75/127 (58%), Gaps = 1/127 (0%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ +++GETGSGK+TQ+ Q+L ++G + CTQPRR+AA S+A RV +E GG
Sbjct: 464 QVLIIVGETGSGKTTQIPQYLHEAGYTKLGKVGCTQPRRVAAMSVAARVAQEMGGKL-GH 522
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
+ F + + +MTD LL+ + + + S IIVDEAHER++ TD+L L
Sbjct: 523 EVGYSIRFEDCTSEKTILKYMTDGMLLRELLGEPDLGSYSVIIVDEAHERTLRTDILFGL 582
Query: 124 VPSSNRS 130
V R+
Sbjct: 583 VKDIARA 589
>At1g32490 putative RNA helicase (At1g32490)
Length = 1044
Score = 91.3 bits (225), Expect = 2e-18
Identities = 50/124 (40%), Positives = 76/124 (60%), Gaps = 7/124 (5%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESG---GCY 60
Q+ V++G+TGSGK+TQ+ Q+L ++G + CTQPRR+AA S+A RV +E G G
Sbjct: 421 QVLVIVGDTGSGKTTQIPQYLHEAGYTKRGKVGCTQPRRVAAMSVAARVAQEMGVKLGHE 480
Query: 61 EDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLL 120
SI+ F + + +MTD LL+ + + + S +IVDEAHER+++TD+L
Sbjct: 481 VGYSIR----FEDCTSDKTVLKYMTDGMLLRELLGEPDLASYSVVIVDEAHERTLSTDIL 536
Query: 121 LALV 124
LV
Sbjct: 537 FGLV 540
>At3g53690 unknown protein
Length = 320
Score = 90.1 bits (222), Expect = 5e-18
Identities = 51/211 (24%), Positives = 94/211 (44%), Gaps = 12/211 (5%)
Query: 630 EIARLSNPSSERFDTGPSCPICLCE--VEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSF 687
E + S+ + FD C IC+ + + +++ GC H +C C+ + + +++
Sbjct: 101 EKGQSSSSKTATFD----CEICVDSKSIIESFRIGGCSHFYCNDCVSKYIAAKLQDN-IL 155
Query: 688 PIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRV 747
I C GC + R +L + + A A V S Y CP DC ++ +
Sbjct: 156 SIECPVSGCSGRLEPDQCRQILPKEVFDRWGDALCEAVVMRSKKFY--CPYKDCSALVFL 213
Query: 748 ADPDTASAPFVCGACYSETCTRCHIEYHPYVSCERYRQFKDDP---DSSLRDWCKGKEQV 804
+ + C C+ C C ++HP ++CE +++ + D L +++
Sbjct: 214 EESEVKMKDSECPHCHRMVCVECGTQWHPEMTCEEFQKLAANERGRDDILLATMAKQKKW 273
Query: 805 KNCPACGHVIEKVDGCNHIECKCGKHICWVC 835
K CP+C IEK GC +++C+CG C+ C
Sbjct: 274 KRCPSCKFYIEKSQGCLYMKCRCGLAFCYNC 304
>At1g48650 hypothetical protein
Length = 1167
Score = 90.1 bits (222), Expect = 5e-18
Identities = 50/125 (40%), Positives = 77/125 (61%), Gaps = 5/125 (4%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGV----GANESIVCTQPRRIAAKSLAERVREESGGC 59
Q+ V+ GETG GK+TQL Q++ +S + GA SI+CTQPRRI+A S++ERV E G
Sbjct: 286 QVVVVSGETGCGKTTQLPQYILESEIEAARGATCSIICTQPRRISAISVSERVAAERGEQ 345
Query: 60 YEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
+S + D+R++F T LL+ + D++ G++ ++VDE HER +N D
Sbjct: 346 IGES-VGYKVRLEGMRGRDTRLLFCTTGVLLRRLLVDRSLKGVTHVVVDEIHERGMNEDF 404
Query: 120 LLALV 124
LL ++
Sbjct: 405 LLIVL 409
>At1g26370 hypothetical protein
Length = 710
Score = 90.1 bits (222), Expect = 5e-18
Identities = 56/129 (43%), Positives = 80/129 (61%), Gaps = 6/129 (4%)
Query: 5 ITVLIGETGSGKSTQLVQFLADSGVGANESIV-CTQPRRIAAKSLAERVREESGGCYEDS 63
I +++GETGSGK+TQL QFL ++G ++ TQPRRIAA ++A+RV EE C
Sbjct: 56 ILIIVGETGSGKTTQLPQFLYNAGFCREGKMIGITQPRRIAAVTVAKRVAEE---CEVQL 112
Query: 64 SIKCYSS--FSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLL 121
K S F +R+ +MTD LL+ + D + + S IIVDEAH+RS++TD+LL
Sbjct: 113 GQKVGYSIRFDDTTSGSTRLKYMTDGLLLREALLDPHLSRYSVIIVDEAHDRSVHTDVLL 172
Query: 122 ALVPSSNRS 130
AL+ R+
Sbjct: 173 ALLKKIQRT 181
>At2g35920 ATP-dependent RNA helicase A like protein
Length = 995
Score = 89.7 bits (221), Expect = 6e-18
Identities = 50/125 (40%), Positives = 75/125 (60%), Gaps = 5/125 (4%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGV----GANESIVCTQPRRIAAKSLAERVREESGGC 59
Q+ V+ GETG GK+TQL QF+ + + GA+ +I+CTQPRRI+A S+A R+ E G
Sbjct: 245 QVLVVSGETGCGKTTQLPQFILEEEISSLRGADCNIICTQPRRISAISVASRISAERGES 304
Query: 60 YEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDL 119
+S + S +R++F T LL+ + D N T +S ++VDE HER +N D
Sbjct: 305 IGES-VGYQIRLESKRSDQTRLLFCTTGVLLRRLIEDPNLTNVSHLLVDEIHERGMNEDF 363
Query: 120 LLALV 124
LL ++
Sbjct: 364 LLIIL 368
>At4g18460 RNA helicase - like protein
Length = 982
Score = 88.6 bits (218), Expect = 1e-17
Identities = 49/126 (38%), Positives = 75/126 (58%), Gaps = 1/126 (0%)
Query: 6 TVLIGETGSGKSTQLVQFLADSG-VGANESIVCTQPRRIAAKSLAERVREESGGCYEDSS 64
T+++GETGSGK+TQ+ Q+L ++G I CTQPRR+A ++++ RV EE G +
Sbjct: 468 TIIVGETGSGKTTQIPQYLKEAGWAEGGRVIACTQPRRLAVQAVSARVAEEMGVNLGEEV 527
Query: 65 IKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
+ + F+TD L++ M D T S I++DEAHERSI+TD+LL L+
Sbjct: 528 GYTIRFEDHTTSGVTSVKFLTDGVLIREMMEDPLLTKYSVIMIDEAHERSISTDILLGLL 587
Query: 125 PSSNRS 130
++S
Sbjct: 588 KKVSQS 593
>At4g18465 RNA helicase - like protein
Length = 720
Score = 88.2 bits (217), Expect = 2e-17
Identities = 49/125 (39%), Positives = 73/125 (58%), Gaps = 1/125 (0%)
Query: 6 TVLIGETGSGKSTQLVQFLADSG-VGANESIVCTQPRRIAAKSLAERVREESGGCYEDSS 64
T+++GETGSGK+TQ+ Q+L ++G I CTQPRR+A ++++ RV EE G +
Sbjct: 67 TIIVGETGSGKTTQIPQYLKEAGWAEGGRVIACTQPRRLAVQAVSARVAEEMGVNLGEEV 126
Query: 65 IKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
+ + F+TD L++ M D T S I++DEAHERSI+TD+LL L+
Sbjct: 127 GYTIRFEDHTTSGVTSVKFLTDGVLIREMMEDPLLTKYSVIMIDEAHERSISTDILLGLL 186
Query: 125 PSSNR 129
R
Sbjct: 187 KKIQR 191
>At1g27900 ATP-dependent RNA helicase (At1g27900)
Length = 700
Score = 88.2 bits (217), Expect = 2e-17
Identities = 54/120 (45%), Positives = 71/120 (59%), Gaps = 1/120 (0%)
Query: 5 ITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDSS 64
+ V+IGETGSGKSTQL Q L G + I TQPRR+AA S+A RV +E +
Sbjct: 22 VVVIIGETGSGKSTQLSQILHRHGYTKSGVIAITQPRRVAAVSVARRVAQELDVPLGE-D 80
Query: 65 IKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLALV 124
+ F +RI ++TD LL+ +S+ S II+DEAHERS+NTD+LL LV
Sbjct: 81 VGYAIRFEDRTTSKTRIKYLTDGVLLRESLSNPMLDDYSVIILDEAHERSLNTDILLGLV 140
>At2g31770 putative ARI-like RING zinc finger protein
Length = 543
Score = 86.3 bits (212), Expect = 7e-17
Identities = 52/188 (27%), Positives = 81/188 (42%), Gaps = 10/188 (5%)
Query: 664 CGHLFCQSCMVEQCESAIKN-QGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASL 722
CGH +C++C + I++ G ++C C + + E+ R L
Sbjct: 145 CGHPYCKTCWAGYITTKIEDGPGCLRVKCPEPSCSAAVGKDMIEDVTETKVNEKYSRYIL 204
Query: 723 GAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGACYSETCTRCHIEYHPYVSCER 782
++V ++CPSP C +++S C C C C + H V C+
Sbjct: 205 RSYVEDGK-KIKWCPSPGCGYAVEFGGSESSSYDVSC-LCSYRFCWNCSEDAHSPVDCDT 262
Query: 783 YRQ--FKDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIECK--CGKHICWVCLEF 838
+ FK+ +S ++W K CP C IEK DGCNH+ C CG CW+CL+
Sbjct: 263 VSKWIFKNQDESENKNWMLANS--KPCPECKRPIEKNDGCNHMTCSAPCGHEFCWICLKA 320
Query: 839 FTT-SGEC 845
+ SG C
Sbjct: 321 YRRHSGAC 328
>At5g60250 unknown protein
Length = 655
Score = 85.1 bits (209), Expect = 2e-16
Identities = 57/226 (25%), Positives = 94/226 (41%), Gaps = 20/226 (8%)
Query: 625 EEITFEIARLSNPSSERFDTGPSCPICLCEV--EDGYQLEGCGHLFCQSCMVEQCESAIK 682
E I + +P + C IC ++ E + ++ C H FC C+ + E +
Sbjct: 278 ESILSMVTPHEDPRQAKAVLKEECAICFNDIVAEGMFSVDKCRHRFCFQCVKQHVEVKLL 337
Query: 683 NQGSFPIRCAHQGCGNHILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCP 742
+ G P +C H GC + L++D L KL +L++ L + +CP P C
Sbjct: 338 H-GMAP-KCPHDGCKSE-LVIDACGKLLTPKLSKLWQQRLQENAIPVTERV-YCPYPRCS 393
Query: 743 SIYRVADPDTASAPFV----------CGACYSETCTRCHIEYHPYVSCERYRQFKDDP-- 790
++ ++ + C C C C + +H +SC Y++ +P
Sbjct: 394 ALMSKTKISESAKSLLSLYPKSGVRRCVECRGLFCVDCKVPWHGNLSCTEYKKLHPEPPA 453
Query: 791 -DSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHIECKCGKHICWVC 835
D L+ K + C C H+IE GCNHI C+CG C+ C
Sbjct: 454 DDVKLKSLANNK-MWRQCGKCQHMIELSQGCNHITCRCGHEFCYNC 498
>At5g13010 pre-mRNA splicing factor ATP-dependent RNA helicase -like
protein
Length = 1226
Score = 84.0 bits (206), Expect = 3e-16
Identities = 50/121 (41%), Positives = 71/121 (58%), Gaps = 5/121 (4%)
Query: 4 QITVLIGETGSGKSTQLVQFLADSGVGANESIVCTQPRRIAAKSLAERVREESGGCYEDS 63
Q+ V++GETGSGK+TQL Q G N + CTQPRR+AA S+A+RV EE D
Sbjct: 550 QVIVVVGETGSGKTTQLTQ----DGYTINGIVGCTQPRRVAAMSVAKRVSEEMETELGDK 605
Query: 64 SIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGISCIIVDEAHERSINTDLLLAL 123
I F ++ I +MTD LL+ + D + +++DEAHERS+NTD+L +
Sbjct: 606 -IGYAIRFEDVTGPNTVIKYMTDGVLLRETLKDSDLDKYRVVVMDEAHERSLNTDVLFGI 664
Query: 124 V 124
+
Sbjct: 665 L 665
>At1g65430 unknown protein
Length = 567
Score = 82.4 bits (202), Expect = 1e-15
Identities = 59/203 (29%), Positives = 83/203 (40%), Gaps = 11/203 (5%)
Query: 648 CPICL-CEVEDGYQLEGCGHLFCQSCMVEQCESAIKN-QGSFPIRCAHQGCGNHILLVDF 705
C IC + D CGH FC SC +AI + G +RC C + D
Sbjct: 128 CGICFETFLSDKLHAAACGHPFCDSCWEGYITTAINDGPGCLTLRCPDPSC-RAAVGQDM 186
Query: 706 RTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFVCGACYSE 765
LL+ DK ++ + + + ++CP+P C + + C CYS
Sbjct: 187 INLLAPDKDKQKYTSYFVRSYVEDNRKTKWCPAPGCDYAVNFV-VGSGNYDVNCRCCYS- 244
Query: 766 TCTRCHIEYHPYVSCERYRQF--KDDPDSSLRDWCKGKEQVKNCPACGHVIEKVDGCNHI 823
C C E H V C+ ++ K+ +S +W K CP C IEK GC HI
Sbjct: 245 FCWNCAEEAHRPVDCDTVSKWVLKNSAESENMNWILANS--KPCPKCKRPIEKNQGCMHI 302
Query: 824 EC--KCGKHICWVCLEFFTTSGE 844
C C CW+CL +T GE
Sbjct: 303 TCTPPCKFEFCWLCLGAWTEHGE 325
>At1g58060 hypothetical protein
Length = 1389
Score = 82.4 bits (202), Expect = 1e-15
Identities = 55/143 (38%), Positives = 77/143 (53%), Gaps = 21/143 (14%)
Query: 5 ITVLIGETGSGKSTQLV--------------QFLAD----SGVGANESIVCTQPRRIAAK 46
+ V+ GETGSGK+TQL+ QF+ D SG G +I+CTQPRRIAA
Sbjct: 632 VLVVCGETGSGKTTQLIALSYFNSATLFQVPQFILDDMIDSGHGGYCNIICTQPRRIAAI 691
Query: 47 SLAERVREE---SGGCYEDSSIKCYSSFSSWNKFDSRIIFMTDHCLLQHYMSDKNFTGIS 103
S+A+RV +E S +DS + S +R++F T LL+ D+ ++
Sbjct: 692 SVAQRVADERCESSPGLDDSLVGYQVRLESARSDKTRLLFCTTGILLRKLAGDRTLNDVT 751
Query: 104 CIIVDEAHERSINTDLLLALVPS 126
IIVDE HERS+ D LL ++ S
Sbjct: 752 HIIVDEVHERSLLGDFLLIILKS 774
>At3g45580 putative protein
Length = 408
Score = 81.6 bits (200), Expect = 2e-15
Identities = 57/203 (28%), Positives = 87/203 (42%), Gaps = 15/203 (7%)
Query: 643 DTGPSCPICL---CEVEDGYQLEGCGHLFCQSCMVEQCESAIKNQGSFPIRCAHQGCGNH 699
D +C IC E E + + CGH FC C+ E + G P RC H C +
Sbjct: 168 DHDKTCSICSDDNFEPELMFSVALCGHEFCVECVKRHIEVRLL-AGGVP-RCLHYQCESK 225
Query: 700 ILLVDFRTLLSNDKLEELFRASLGAFVASSSGTYRFCPSPDCPSIYRVADPDTASAPFV- 758
+ L + LL++ KL+ ++ + +CP+P C S+ V ++ V
Sbjct: 226 LTLANCANLLTS-KLKAMWELRIEEESIPVEERV-YCPNPRCSSLMSVTKLSNSTREDVT 283
Query: 759 ---CGACYSETCTRCHIEYHPYVSCERYRQFKDDP---DSSLRDWCKGKEQVKNCPACGH 812
C C C C + +H +SC Y+ +P D L+ K + C C +
Sbjct: 284 MRSCVKCGEPFCINCKLPWHSNLSCNDYKSLGPNPTADDIKLKALANQK-MWRQCENCKN 342
Query: 813 VIEKVDGCNHIECKCGKHICWVC 835
VIE +GC HI C+CG C+ C
Sbjct: 343 VIELSEGCMHITCRCGHQFCYKC 365
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,573,303
Number of Sequences: 26719
Number of extensions: 864207
Number of successful extensions: 2607
Number of sequences better than 10.0: 103
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 2403
Number of HSP's gapped (non-prelim): 141
length of query: 857
length of database: 11,318,596
effective HSP length: 108
effective length of query: 749
effective length of database: 8,432,944
effective search space: 6316275056
effective search space used: 6316275056
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC141414.11


