

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141323.10 + phase: 0 /pseudo
(337 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
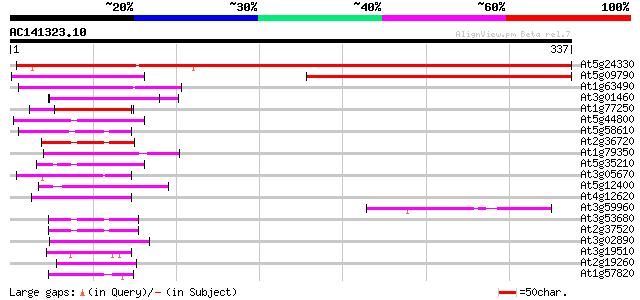
Score E
Sequences producing significant alignments: (bits) Value
At5g24330 putative protein 369 e-102
At5g09790 unknown protein 250 9e-67
At1g63490 RB-binding protein -like 64 9e-11
At3g01460 unknown protein 62 4e-10
At1g77250 unknown protein 60 1e-09
At5g44800 helicase-like protein 54 2e-07
At5g58610 putative protein 51 8e-07
At2g36720 putative PHD-type zinc finger protein 49 5e-06
At1g79350 hypothetical protein 48 7e-06
At5g35210 putative protein 48 9e-06
At3g05670 unknown protein 47 1e-05
At5g12400 putative protein 47 2e-05
At4g12620 origin recognition complex subunit 1 -like protein 47 2e-05
At3g59960 putative protein 45 4e-05
At3g53680 putative protein 45 6e-05
At2g37520 unknown protein 45 7e-05
At3g02890 unknown protein 44 2e-04
At3g19510 putative homeobox protein, HAT3.1 43 2e-04
At2g19260 unknown protein 43 2e-04
At1g57820 putative transcription factor protein (At1g57820) 43 2e-04
>At5g24330 putative protein
Length = 349
Score = 369 bits (947), Expect = e-102
Identities = 194/346 (56%), Positives = 246/346 (71%), Gaps = 14/346 (4%)
Query: 5 LRRRRIPA-------PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLT 57
+RRRR A P+ S +++D D VC++C+SGK P KLLLCD CD G+HLFCL
Sbjct: 4 VRRRRTQASNPRSEPPQHMSDHDSDSDWDTVCEECSSGKQPAKLLLCDKCDKGFHLFCLR 63
Query: 58 PILPSVPKSSWFCPSCSHNPKIPK-FPLVQTKIIDFFKIQRTSDASQILNHEE----EKA 112
PIL SVPK SWFCPSCS + +IPK FPL+QTKIIDFF+I+R+ D+SQI + + ++
Sbjct: 64 PILVSVPKGSWFCPSCSKH-QIPKSFPLIQTKIIDFFRIKRSPDSSQISSSSDSIGKKRK 122
Query: 113 E**LGCFKKEKKVVGICAK**FEEEIRTDGVVGDRVDSY*NRVQ**AYVYAWNGAKGC*F 172
+ L KK+++++ + + + + + + + +
Sbjct: 123 KTSLVMSKKKRRLLPYNPSNDPQRRLEQMASLATALRASNTKFSNELTYVSGKAPRSANQ 182
Query: 173 SCS*TWGNAVLSKEDTETLNLCRSMMERGECPPLMVVYDPVEGFTIEADKSIKDLTIITE 232
+ G VLSKE ETL LC+ MM+ GECPPLMVV+DP EGFT+EAD+ IKD TIITE
Sbjct: 183 AAFEKGGMQVLSKEGVETLALCKKMMDLGECPPLMVVFDPYEGFTVEADRFIKDWTIITE 242
Query: 233 YVGDVDFLKNREHD-DGDSIMTLLSASNPSQSLVICPDKRSNIARFINGINNHTPEGKKK 291
YVGDVD+L NRE D DGDS+MTLL AS+PSQ LVICPD+RSNIARFI+GINNH+PEG+KK
Sbjct: 243 YVGDVDYLSNREDDYDGDSMMTLLHASDPSQCLVICPDRRSNIARFISGINNHSPEGRKK 302
Query: 292 QNLKCVRYNVDGECRVLLIANRDIAKGERLYYDYNGLEHEYPTEHF 337
QNLKCVR+N++GE RVLL+ANRDI+KGERLYYDYNG EHEYPTEHF
Sbjct: 303 QNLKCVRFNINGEARVLLVANRDISKGERLYYDYNGYEHEYPTEHF 348
>At5g09790 unknown protein
Length = 352
Score = 250 bits (638), Expect = 9e-67
Identities = 116/159 (72%), Positives = 133/159 (82%)
Query: 179 GNAVLSKEDTETLNLCRSMMERGECPPLMVVYDPVEGFTIEADKSIKDLTIITEYVGDVD 238
G VL KED ETL C+SM RGECPPL+VV+DP+EG+T+EAD IKDLT I EY GDVD
Sbjct: 193 GMQVLCKEDLETLEQCQSMYRRGECPPLVVVFDPLEGYTVEADGPIKDLTFIAEYTGDVD 252
Query: 239 FLKNREHDDGDSIMTLLSASNPSQSLVICPDKRSNIARFINGINNHTPEGKKKQNLKCVR 298
+LKNRE DD DSIMTLL + +PS++LVICPDK NI+RFINGINNH P KKKQN KCVR
Sbjct: 253 YLKNREKDDCDSIMTLLLSEDPSKTLVICPDKFGNISRFINGINNHNPVAKKKQNCKCVR 312
Query: 299 YNVDGECRVLLIANRDIAKGERLYYDYNGLEHEYPTEHF 337
Y+++GECRVLL+A RDI+KGERLYYDYNG EHEYPT HF
Sbjct: 313 YSINGECRVLLVATRDISKGERLYYDYNGYEHEYPTHHF 351
Score = 69.7 bits (169), Expect = 2e-12
Identities = 27/80 (33%), Positives = 46/80 (56%)
Query: 2 VSPLRRRRIPAPKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILP 61
++ + + +P +++ ++ +V C+KC SG+ +LLLCD CD G+H+ CL PI+
Sbjct: 40 MAEIMAKSVPVVEQEEEEDEDSYSNVTCEKCGSGEGDDELLLCDKCDRGFHMKCLRPIVV 99
Query: 62 SVPKSSWFCPSCSHNPKIPK 81
VP +W C CS + K
Sbjct: 100 RVPIGTWLCVDCSDQRPVRK 119
>At1g63490 RB-binding protein -like
Length = 1458
Score = 64.3 bits (155), Expect = 9e-11
Identities = 31/98 (31%), Positives = 50/98 (50%), Gaps = 1/98 (1%)
Query: 6 RRRRIPAPKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPK 65
+RR+ A K N D C++C S K +LLCD+C+ G+H++CL+P L +P
Sbjct: 215 KRRKRNADVKNPKVENEEGVDQACEQCKSDKHGEVMLLCDSCNKGWHIYCLSPPLKHIPL 274
Query: 66 SSWFCPSCSHNPKIPKFPLVQTKIIDFFKIQRTSDASQ 103
+W+C C N F V K + +R +D ++
Sbjct: 275 GNWYCLECL-NTDEETFGFVPGKCLLLEDFKRIADRAK 311
>At3g01460 unknown protein
Length = 2176
Score = 62.0 bits (149), Expect = 4e-10
Identities = 25/66 (37%), Positives = 37/66 (55%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNPKIPKFPL 84
D+ VC+ C K +LLCD CD YH +CL P L +P +W+CPSC ++ + L
Sbjct: 1286 DEGVCKVCGVDKDDDSVLLCDTCDAEYHTYCLNPPLIRIPDGNWYCPSCVIAKRMAQEAL 1345
Query: 85 VQTKII 90
K++
Sbjct: 1346 ESYKLV 1351
Score = 47.0 bits (110), Expect = 1e-05
Identities = 24/80 (30%), Positives = 40/80 (50%), Gaps = 2/80 (2%)
Query: 24 DDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNPKIPK-F 82
D D C C +S +++CD C+ G+H+ C+ + + P + W C C + K +
Sbjct: 81 DRDASCGACGRPESIELVVVCDACERGFHMSCVNDGVEAAPSADWMCSDCRTGGERSKLW 140
Query: 83 PL-VQTKIIDFFKIQRTSDA 101
PL V++K+I SDA
Sbjct: 141 PLGVKSKLILDMNASPPSDA 160
>At1g77250 unknown protein
Length = 522
Score = 60.5 bits (145), Expect = 1e-09
Identities = 20/46 (43%), Positives = 30/46 (64%)
Query: 28 VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSC 73
+C+ C + K K++LCD CD+ YH++C+ P SVP WFC +C
Sbjct: 404 LCRNCLTDKDDDKIVLCDGCDDAYHIYCMRPPCESVPNGEWFCTAC 449
Score = 44.7 bits (104), Expect = 7e-05
Identities = 17/62 (27%), Positives = 28/62 (44%)
Query: 13 PKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPS 72
P + N+ +C+ C L CD+C++ YH+ C P +P SW+C
Sbjct: 227 PCPNAENIRNDSVSDICKLCGEKAEARDCLACDHCEDMYHVSCAQPGGKGMPTHSWYCLD 286
Query: 73 CS 74
C+
Sbjct: 287 CT 288
>At5g44800 helicase-like protein
Length = 2228
Score = 53.5 bits (127), Expect = 2e-07
Identities = 28/80 (35%), Positives = 37/80 (46%), Gaps = 4/80 (5%)
Query: 3 SPLRRRRIPAPKKQSTTFNNNDDDVV-CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILP 61
S R + P++ S+ ND + C C+ G LL CD+C YH CL P L
Sbjct: 37 SKQRLKTDSTPERNSSKRKGNDGNYFECVICDLGGD---LLCCDSCPRTYHTACLNPPLK 93
Query: 62 SVPKSSWFCPSCSHNPKIPK 81
+P W CP CS N + K
Sbjct: 94 RIPNGKWICPKCSPNSEALK 113
>At5g58610 putative protein
Length = 1065
Score = 51.2 bits (121), Expect = 8e-07
Identities = 24/68 (35%), Positives = 33/68 (48%), Gaps = 5/68 (7%)
Query: 6 RRRRIPAPKKQSTTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPK 65
+R++ P ++DV C C+ G KL+LCD C + +H CL L VP
Sbjct: 674 KRKKAQPPDMLKMKLRQGENDVFCSVCHYGG---KLILCDGCPSAFHANCLG--LEDVPD 728
Query: 66 SSWFCPSC 73
WFC SC
Sbjct: 729 GDWFCQSC 736
>At2g36720 putative PHD-type zinc finger protein
Length = 958
Score = 48.5 bits (114), Expect = 5e-06
Identities = 20/56 (35%), Positives = 35/56 (61%), Gaps = 5/56 (8%)
Query: 20 FNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSH 75
++ ND++ +C C G + LLLCD+C +H+ C++ LPS+P+ +W C C +
Sbjct: 571 YSANDNNDLCVICADGGN---LLLCDSCPRAFHIECVS--LPSIPRGNWHCKYCEN 621
Score = 40.8 bits (94), Expect = 0.001
Identities = 16/41 (39%), Positives = 24/41 (58%), Gaps = 4/41 (9%)
Query: 34 SGKSPTKLLLCDNCDNGYHLFCLTP----ILPSVPKSSWFC 70
SG P +++CD C+ YH+ CL+ L +PK +WFC
Sbjct: 668 SGFGPRTIIICDQCEKEYHIGCLSSQNIVDLKELPKGNWFC 708
>At1g79350 hypothetical protein
Length = 1318
Score = 48.1 bits (113), Expect = 7e-06
Identities = 23/83 (27%), Positives = 42/83 (49%), Gaps = 5/83 (6%)
Query: 21 NNNDDDV-VCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNPKI 79
N++DD+ +CQ C+ KLL C CD +H C+ P + +P +W C SC +
Sbjct: 677 NDSDDEFQICQICSGEDERKKLLHCSECDKLFHPDCVVPPVIDLPSEAWICFSCKEKTE- 735
Query: 80 PKFPLVQTKIIDFFKIQRTSDAS 102
+Q + + ++Q+ +A+
Sbjct: 736 ---EYIQARRLYIAELQKRYEAA 755
>At5g35210 putative protein
Length = 1606
Score = 47.8 bits (112), Expect = 9e-06
Identities = 23/65 (35%), Positives = 32/65 (48%), Gaps = 5/65 (7%)
Query: 17 STTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHN 76
S+ + N D+ C+ C + LL CD C YH C+ + +P WFCP C+ N
Sbjct: 404 SSDLDGNSDE--CRICGMDGT---LLCCDGCPLAYHSRCIGVVKMYIPDGPWFCPECTIN 458
Query: 77 PKIPK 81
K PK
Sbjct: 459 KKGPK 463
>At3g05670 unknown protein
Length = 883
Score = 47.4 bits (111), Expect = 1e-05
Identities = 21/78 (26%), Positives = 40/78 (50%), Gaps = 10/78 (12%)
Query: 5 LRRRRIPAPKKQST---------TFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFC 55
LR IP P++ ++ + ++++C +C+ G +LLCD CD+ H +C
Sbjct: 473 LREVVIPVPERDQVYQPTEEELRSYLDPYENIICTECHQGDDDGLMLLCDLCDSSAHTYC 532
Query: 56 LTPILPSVPKSSWFCPSC 73
+ + VP+ +W+C C
Sbjct: 533 V-GLGREVPEGNWYCEGC 549
>At5g12400 putative protein
Length = 1595
Score = 46.6 bits (109), Expect = 2e-05
Identities = 25/79 (31%), Positives = 37/79 (46%), Gaps = 6/79 (7%)
Query: 18 TTFNNNDDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNP 77
T+F+ N DD C K LL CD C YH C+ +P+ W+CP C+ +
Sbjct: 602 TSFDRNSDD-----CCFCKMDGSLLCCDGCPAAYHSKCVGLASHLLPEGDWYCPECAFDR 656
Query: 78 KIPKF-PLVQTKIIDFFKI 95
+ P P Q + +F +I
Sbjct: 657 RAPGLKPDKQIRGAEFIEI 675
>At4g12620 origin recognition complex subunit 1 -like protein
Length = 813
Score = 46.6 bits (109), Expect = 2e-05
Identities = 21/62 (33%), Positives = 35/62 (55%), Gaps = 2/62 (3%)
Query: 14 KKQSTTFNNNDDDVVCQKCN-SGKSPTKLLL-CDNCDNGYHLFCLTPILPSVPKSSWFCP 71
K++ + ++ ++D + C KS T +++ CD+C G+HL CL P L VP+ W C
Sbjct: 151 KRREDSNSDEEEDPEIEDCQICFKSDTNIMIECDDCLGGFHLKCLKPPLKEVPEGDWICQ 210
Query: 72 SC 73
C
Sbjct: 211 FC 212
>At3g59960 putative protein
Length = 352
Score = 45.4 bits (106), Expect = 4e-05
Identities = 35/114 (30%), Positives = 56/114 (48%), Gaps = 11/114 (9%)
Query: 215 GFTIEADKSIKDLTIITEYVGDV---DFLKNREHDDGDSIMTLLSASNPSQSLVICPDKR 271
G+ I AD+ I I EYVG+V + R + T + ++VI +
Sbjct: 122 GYGIVADEDINSGEFIIEYVGEVIDDKICEERLWKLNHKVETNFYLCQINWNMVIDATHK 181
Query: 272 SNIARFINGINNHTPEGKKKQNLKCVRYNVDGECRVLLIANRDIAKGERLYYDY 325
N +R+IN ++ +P N + ++ +DGE R+ + A R I KGE+L YDY
Sbjct: 182 GNKSRYIN--HSCSP------NTEMQKWIIDGETRIGIFATRFINKGEQLTYDY 227
>At3g53680 putative protein
Length = 839
Score = 45.1 bits (105), Expect = 6e-05
Identities = 20/54 (37%), Positives = 28/54 (51%), Gaps = 5/54 (9%)
Query: 24 DDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNP 77
D D +C C +G LLLC C +H CL S+P+ +W+C SC+ P
Sbjct: 484 DSDDMCSICGNGGD---LLLCAGCPQAFHTACLK--FQSMPEGTWYCSSCNDGP 532
Score = 35.0 bits (79), Expect = 0.058
Identities = 16/39 (41%), Positives = 22/39 (56%), Gaps = 5/39 (12%)
Query: 41 LLLCDNCDNGYHLFCLTP----ILPSVPKSSWFCPS-CS 74
++LCD C+ YH+ CL L +P+ WFC S CS
Sbjct: 582 VILCDQCEKEYHVGCLRENELCDLKGIPQDKWFCCSDCS 620
>At2g37520 unknown protein
Length = 825
Score = 44.7 bits (104), Expect = 7e-05
Identities = 20/54 (37%), Positives = 27/54 (49%), Gaps = 5/54 (9%)
Query: 24 DDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKSSWFCPSCSHNP 77
D D +C C G LLLC C +H CL S+P+ +W+C SC+ P
Sbjct: 462 DSDDMCSICGDGGD---LLLCAGCPQAFHTACLK--FQSMPEGTWYCSSCNDGP 510
Score = 35.4 bits (80), Expect = 0.044
Identities = 16/43 (37%), Positives = 24/43 (55%), Gaps = 6/43 (13%)
Query: 41 LLLCDNCDNGYHLFCLTP----ILPSVPKSSWFCPSCSHNPKI 79
++LCD C+ YH+ CL L +P+ WFC CS+ +I
Sbjct: 564 VILCDQCEKEYHVGCLRENGFCDLKEIPQEKWFC--CSNCSRI 604
>At3g02890 unknown protein
Length = 963
Score = 43.5 bits (101), Expect = 2e-04
Identities = 20/61 (32%), Positives = 25/61 (40%), Gaps = 1/61 (1%)
Query: 25 DDVVCQKCNSGKSPTKLLLCDNCDNGY-HLFCLTPILPSVPKSSWFCPSCSHNPKIPKFP 83
D VC C L +C C +G H +C+ +L VPK W C C K K
Sbjct: 201 DVKVCDTCGDAGREDLLAICSRCSDGAEHTYCMRVMLKKVPKGYWLCEECKFAEKAEKHK 260
Query: 84 L 84
L
Sbjct: 261 L 261
>At3g19510 putative homeobox protein, HAT3.1
Length = 723
Score = 43.1 bits (100), Expect = 2e-04
Identities = 21/58 (36%), Positives = 29/58 (49%), Gaps = 7/58 (12%)
Query: 23 NDDDVVCQKCNSG--KSPTKLLLCDN-CDNGYHLFCLTPIL--PSVP--KSSWFCPSC 73
+ +D+ C KC S ++LCD CD G+H +CL P L +P W CP C
Sbjct: 262 SSEDIFCAKCGSKDLSVDNDIILCDGFCDRGFHQYCLEPPLRKEDIPPDDEGWLCPGC 319
>At2g19260 unknown protein
Length = 631
Score = 43.1 bits (100), Expect = 2e-04
Identities = 17/49 (34%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 29 CQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPK-SSWFCPSCSHN 76
C+ C+ + K+L+CD C+ YH C + V + W CPSC N
Sbjct: 412 CKHCDKPGTVEKMLICDECEEAYHTRCCGVQMKDVAEIDEWLCPSCLKN 460
>At1g57820 putative transcription factor protein (At1g57820)
Length = 645
Score = 43.1 bits (100), Expect = 2e-04
Identities = 21/56 (37%), Positives = 27/56 (47%), Gaps = 9/56 (16%)
Query: 24 DDDVVCQKCNSGKSPTKLLLCDNCDNGYHLFCLTPILPSVPKS-----SWFCPSCS 74
D D VC +C S P + L C C +H+ CL+ S PK+ W CP CS
Sbjct: 10 DGDGVCMRCKSNPPPEESLTCGTCVTPWHVSCLS----SPPKTLASTLQWHCPDCS 61
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.333 0.146 0.480
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,125,397
Number of Sequences: 26719
Number of extensions: 352951
Number of successful extensions: 1191
Number of sequences better than 10.0: 76
Number of HSP's better than 10.0 without gapping: 44
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 1091
Number of HSP's gapped (non-prelim): 105
length of query: 337
length of database: 11,318,596
effective HSP length: 100
effective length of query: 237
effective length of database: 8,646,696
effective search space: 2049266952
effective search space used: 2049266952
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC141323.10


