

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141114.7 + phase: 0
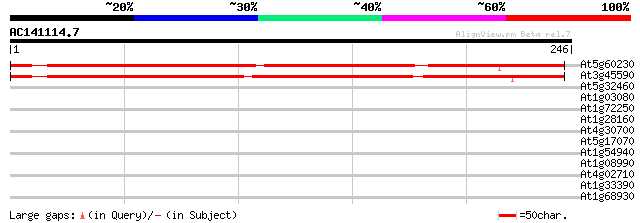
(246 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g60230 tRNA intron endonuclease - like protein 259 9e-70
At3g45590 putative protein 254 2e-68
At5g32460 meristem protein - like 32 0.41
At1g03080 unknown protein 30 1.2
At1g72250 kinesin, putative 30 1.5
At1g28160 AP2 domain transcription factor, putative 29 2.6
At4g30700 unknown protein 28 3.4
At5g17070 unknown protein 28 4.5
At1g54940 hypothetical protein 28 4.5
At1g08990 unknown protein 28 4.5
At4g02710 predicted protein of unknown function 28 5.9
At1g33390 RNA helicase, putative 28 5.9
At1g68930 hypothetical protein 27 7.7
>At5g60230 tRNA intron endonuclease - like protein
Length = 250
Score = 259 bits (662), Expect = 9e-70
Identities = 138/245 (56%), Positives = 166/245 (67%), Gaps = 16/245 (6%)
Query: 1 MAPRWKGKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCGFLSDNCVHLTVQAEQLD 60
MAPRWK K A EA+AL EP+SK +S+LQ SL ++ + G LS V L V+ EQ +
Sbjct: 1 MAPRWKWKGA------EAKALAEPISKSVSELQLSLAETESSGTLSSCNVLLAVEPEQAE 54
Query: 61 LLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYF 120
LLD+ CFGR V EK W QL+ EEAF+L Y+LKC+KI + N+ + W Y
Sbjct: 55 LLDRCCFGRLVLSAEKIKKWIQLSFEEAFFLHYNLKCIKISLQGRCLE---NEVDTWLYM 111
Query: 121 RSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGD 180
+SK+ FP F+KAYSHLR KNWV+RSG QYGVD + YRHHP+ VHSEY VLV S D D
Sbjct: 112 KSKRPNFPMFFKAYSHLRSKNWVLRSGLQYGVDFVAYRHHPSLVHSEYSVLVQSGDSD-- 169
Query: 181 LNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNKN--GNNDESLLCLTNYTVEERTISRWSP 238
RLRVWSD+HC RL G VAKTLL LYVN N G + L+CL N+TVEE+TISRWSP
Sbjct: 170 ---RLRVWSDIHCAVRLSGSVAKTLLTLYVNGNFKGEDVNLLVCLENFTVEEQTISRWSP 226
Query: 239 EQCRE 243
E RE
Sbjct: 227 ELSRE 231
>At3g45590 putative protein
Length = 237
Score = 254 bits (650), Expect = 2e-68
Identities = 136/245 (55%), Positives = 168/245 (68%), Gaps = 15/245 (6%)
Query: 1 MAPRWKGKDAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTCGFLSDNCVHLTVQAEQLD 60
MAPRWK K A EA+AL EP+SK +S+L+SSL Q+ GFLS V L+V++E+ +
Sbjct: 1 MAPRWKWKGA------EAKALAEPVSKTVSELRSSLTQTEALGFLSSCNVLLSVESEEAE 54
Query: 61 LLDKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYF 120
LLD+ CFGR V EKD W QL+ EEAF+L Y LKC+KI ++ N+ +LW
Sbjct: 55 LLDRCCFGRLVVGAEKDKRWIQLSFEEAFFLFYKLKCIKICLH---GRSLENEVDLWRSM 111
Query: 121 RSKKETFPYFYKAYSHLRMKNWVVRSGAQYGVDLIVYRHHPARVHSEYGVLVLSHDKDGD 180
S K+ F YKAYSHLR KNW+VRSG QYGVD +VYRHHP+ VHSEY VLV S
Sbjct: 112 SSFKQDFAILYKAYSHLRSKNWIVRSGLQYGVDFVVYRHHPSLVHSEYAVLVQSIGG--- 168
Query: 181 LNGRLRVWSDVHCTTRLLGGVAKTLLVLYVNKNGNNDES--LLCLTNYTVEERTISRWSP 238
N RL+VWSD+HC+ RL G VAK+LLVLYVN+ N ++ LCL +YTVEE+TI RWSP
Sbjct: 169 -NDRLKVWSDIHCSVRLTGSVAKSLLVLYVNRKVNTEKMNLPLCLEDYTVEEQTIRRWSP 227
Query: 239 EQCRE 243
E RE
Sbjct: 228 ELSRE 232
>At5g32460 meristem protein - like
Length = 517
Score = 31.6 bits (70), Expect = 0.41
Identities = 20/56 (35%), Positives = 31/56 (54%), Gaps = 2/56 (3%)
Query: 11 KSKKEAEAEALKEP--MSKIISQLQSSLVQSNTCGFLSDNCVHLTVQAEQLDLLDK 64
K K+E E E+ KE + + SS V S T LS++ +HL ++ + +LLDK
Sbjct: 392 KIKQEVEYESAKEEKNLESLSLSDNSSFVVSVTVSNLSEDILHLPIRLSRSNLLDK 447
>At1g03080 unknown protein
Length = 1744
Score = 30.0 bits (66), Expect = 1.2
Identities = 13/31 (41%), Positives = 21/31 (66%)
Query: 12 SKKEAEAEALKEPMSKIISQLQSSLVQSNTC 42
++ EAE E L+E +SK+ + +SSL+Q C
Sbjct: 265 TRAEAEVETLRESLSKVEVEKESSLLQYQQC 295
Score = 28.1 bits (61), Expect = 4.5
Identities = 13/27 (48%), Positives = 20/27 (73%)
Query: 12 SKKEAEAEALKEPMSKIISQLQSSLVQ 38
SK EAE ALK+ +SK+ ++ ++SL Q
Sbjct: 209 SKAEAEIVALKDALSKVQAEKEASLAQ 235
>At1g72250 kinesin, putative
Length = 1195
Score = 29.6 bits (65), Expect = 1.5
Identities = 14/42 (33%), Positives = 21/42 (49%)
Query: 192 HCTTRLLGGVAKTLLVLYVNKNGNNDESLLCLTNYTVEERTI 233
H LGG +KTL+ + ++ N N+ LC N+ R I
Sbjct: 776 HLLQDSLGGDSKTLMFVQISPNENDQSETLCSLNFASRVRGI 817
>At1g28160 AP2 domain transcription factor, putative
Length = 245
Score = 28.9 bits (63), Expect = 2.6
Identities = 18/48 (37%), Positives = 21/48 (43%), Gaps = 10/48 (20%)
Query: 38 QSNTCGFLSDNCVHLTVQAEQLD-----LLDKACFGRPVRIVEKDMYW 80
Q CG+ S +T AE D L CFG +RI E D YW
Sbjct: 162 QEAECGYQS-----ITSNAEHCDHELPPLPPSTCFGAELRIPETDSYW 204
>At4g30700 unknown protein
Length = 792
Score = 28.5 bits (62), Expect = 3.4
Identities = 23/103 (22%), Positives = 47/103 (45%), Gaps = 12/103 (11%)
Query: 118 HYFRSKKETFPYFYKAYSHL-RMKNWVVRSGAQYGVDLIVY------RHHPARVHSE--- 167
H F S ++ P + Y L +++ + +G Q +L ++ R +VHSE
Sbjct: 670 HVFTSGDQSHPQVKEIYEKLEKLEGKMREAGYQPETELALHDVEEEERELMVKVHSERLA 729
Query: 168 --YGVLVLSHDKDGDLNGRLRVWSDVHCTTRLLGGVAKTLLVL 208
+G++ + + LRV D H T+L+ + + ++V+
Sbjct: 730 IAFGLIATEPGTEIRIIKNLRVCLDCHTVTKLISKITERVIVV 772
>At5g17070 unknown protein
Length = 277
Score = 28.1 bits (61), Expect = 4.5
Identities = 12/30 (40%), Positives = 21/30 (70%)
Query: 9 DAKSKKEAEAEALKEPMSKIISQLQSSLVQ 38
+AK E + EAL EP S+++S+L +L++
Sbjct: 99 EAKMTDEQQKEALGEPYSELVSRLDEALLR 128
>At1g54940 hypothetical protein
Length = 557
Score = 28.1 bits (61), Expect = 4.5
Identities = 19/76 (25%), Positives = 34/76 (44%), Gaps = 9/76 (11%)
Query: 63 DKACFGRPVRIVEKDMYWFQLTVEEAFYLCYSLKCLKIKINVGADTGPLNDEELWHYFRS 122
DK F + ++E F+ +E++F KI+ G D G LN+ +W + S
Sbjct: 389 DKVLFNSGIMVLEPSACMFKDLMEKSF---------KIESYNGGDQGFLNEIFVWWHRLS 439
Query: 123 KKETFPYFYKAYSHLR 138
K+ ++ +H R
Sbjct: 440 KRVNTMKYFDEKNHRR 455
>At1g08990 unknown protein
Length = 550
Score = 28.1 bits (61), Expect = 4.5
Identities = 15/48 (31%), Positives = 21/48 (43%)
Query: 95 LKCLKIKINVGADTGPLNDEELWHYFRSKKETFPYFYKAYSHLRMKNW 142
LK KI G D G LN+ +W + K P + +L +K W
Sbjct: 419 LKSFKIGSYNGGDQGFLNEYFVWWHRHDKARNLPENLEGIHYLGLKPW 466
>At4g02710 predicted protein of unknown function
Length = 1111
Score = 27.7 bits (60), Expect = 5.9
Identities = 12/34 (35%), Positives = 21/34 (61%)
Query: 9 DAKSKKEAEAEALKEPMSKIISQLQSSLVQSNTC 42
D + EAE + L+E + K+ S+ +SS +Q + C
Sbjct: 261 DRAASAEAEIQTLRETLYKLESEKESSFLQYHKC 294
>At1g33390 RNA helicase, putative
Length = 1237
Score = 27.7 bits (60), Expect = 5.9
Identities = 19/60 (31%), Positives = 25/60 (41%), Gaps = 3/60 (5%)
Query: 8 KDAKSKKEAEAEALKEPMSKIIS---QLQSSLVQSNTCGFLSDNCVHLTVQAEQLDLLDK 64
KD K K +A + P S ++ L S V N GF N +HL E L D+
Sbjct: 897 KDRKEKIKAARDRFSNPSSDALTVAYALHSFEVSENGMGFCEANGLHLKTMDEMSKLKDQ 956
>At1g68930 hypothetical protein
Length = 743
Score = 27.3 bits (59), Expect = 7.7
Identities = 20/96 (20%), Positives = 41/96 (41%), Gaps = 12/96 (12%)
Query: 118 HYFRSKKETFPYFYKAYSHLR-MKNWVVRSG-----------AQYGVDLIVYRHHPARVH 165
H F + E+ PY + Y+ L + N ++ +G + V + + +H R+
Sbjct: 621 HSFSADDESSPYLDQIYAKLEELNNKIIDNGYKPDTSFVHHDVEEAVKVKMLNYHSERLA 680
Query: 166 SEYGVLVLSHDKDGDLNGRLRVWSDVHCTTRLLGGV 201
+G++ + + + LRV D H T+ + V
Sbjct: 681 IAFGLIFVPSGQPIRVGKNLRVCVDCHNATKHISSV 716
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,858,894
Number of Sequences: 26719
Number of extensions: 245949
Number of successful extensions: 648
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 623
Number of HSP's gapped (non-prelim): 20
length of query: 246
length of database: 11,318,596
effective HSP length: 97
effective length of query: 149
effective length of database: 8,726,853
effective search space: 1300301097
effective search space used: 1300301097
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141114.7


