

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
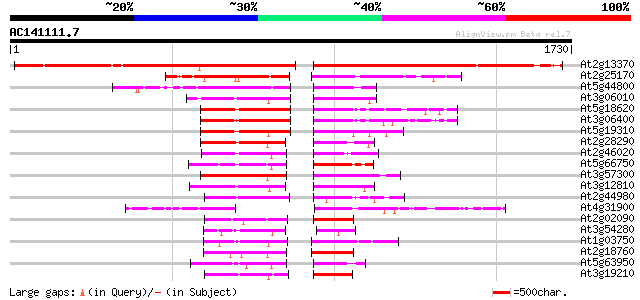
Query= AC141111.7 - phase: 0 /pseudo
(1730 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g13370 pseudogene 1058 0.0
At2g25170 putative chromodomain-helicase-DNA-binding protein 308 2e-83
At5g44800 helicase-like protein 280 5e-75
At3g06010 putative transcriptional regulator 233 1e-60
At5g18620 chromatin remodelling complex ATPase chain ISWI -like ... 231 2e-60
At3g06400 putative ATPase (ISW2-like) 231 2e-60
At5g19310 homeotic gene regulator - like protein 227 4e-59
At2g28290 putative SNF2 subfamily transcription regulator 223 6e-58
At2g46020 putative SNF2 subfamily transcriptional activator 213 1e-54
At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1) 206 7e-53
At3g57300 helicase-like protein 200 7e-51
At3g12810 Snf2-related CBP activator protein, putative 197 6e-50
At2g44980 putative transcription activator 170 6e-42
At4g31900 putative protein 166 8e-41
At2g02090 putative helicase 161 3e-39
At3g54280 TATA box binding protein (TBP) associated factor (TAF)... 134 6e-31
At1g03750 hypothetical protein 125 2e-28
At2g18760 putative SNF2/RAD54 family DNA repair and recombinatio... 115 2e-25
At5g63950 unknown protein 108 2e-23
At3g19210 DNA repair protein, putative 107 5e-23
>At2g13370 pseudogene
Length = 1738
Score = 1058 bits (2737), Expect = 0.0
Identities = 559/896 (62%), Positives = 673/896 (74%), Gaps = 42/896 (4%)
Query: 15 VGQNADRTRSEKEFYMNMEAQYESDAEPDDASGKQNEAAAVDRLSTRESNVETTSRNPSA 74
+ ++ D T SE+ F MNM+ QY+SD EP + + NE A + +S+ +++++
Sbjct: 34 LNEDVDGTYSERGFDMNMDVQYQSDPEPGCSIRQPNETAVDNVADPVDSHYQSSTKRLGV 93
Query: 75 SERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFEDNSSEGRGEKLGSEDEDGQK 134
+ RWGS++ KDCQPM + GS+ DS+SG Y+ EDN S R EKL SE+E+ +
Sbjct: 94 TGRWGSTFWKDCQPMGQREGSDPAKDSQSG--YKEAYHSEDNHSNDRSEKLDSENENDNE 151
Query: 135 DS------GKGQRGDSDVPAEEMLSDDSYGQDGEEQGESVHSRGFRPSTGSNSCLQPTS- 187
+ K Q G +DVPA+EMLSD+ Y QD + Q + VH +G+ T S S + S
Sbjct: 152 NEEEDNEMNKHQSGQADVPADEMLSDEYYEQDEDNQSDHVHYKGYSNPTNSRSLPKAGSA 211
Query: 188 ----TNVNRRVHRKSRILDDAEDDDDDADYE-EDEPDEDDPDDADFEP--ATSGRGANKY 240
+ +R +H+ D D + DAD + E+E DEDDP+DADFEP A GA+K
Sbjct: 212 VHSNSRTSRAIHKNIHYSDSNHDHNGDADMDYEEEEDEDDPEDADFEPYDAADDGGASKK 271
Query: 241 KDWEGEDSDEVDDSDEDIDVSDNDDLYFDKKAKGRQRGKFGPSVRSTRDCKAFTASSRQR 300
+ SDE +SDE+ID+SD +D Y KK K RQ+ K + + K+F SSRQ+
Sbjct: 272 HGQGWDVSDEDPESDEEIDLSDYEDDYGTKKPKVRQQSKGFRKSSAGLERKSFHVSSRQK 331
Query: 301 RVKSSFEDEDENSTAEDSDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSS 360
R K+S++D+D EDS++++DE F+SL +RG +R+NNGRS T+ S+EVRSS+
Sbjct: 332 R-KTSYQDDDSE---EDSENDNDEGFRSLARRGTTLRQNNGRS---TNTIGQSSEVRSST 384
Query: 361 RTIRKVSYVESDESEGADEG-TKKSQKEEIEVDDGDSVEKVLWHQPKGMAAEAQRNNQSM 419
R++RKVSYVES++SE D+G +K+QK++IE +D D +EKVLWHQ KGM + Q NN+S
Sbjct: 385 RSVRKVSYVESEDSEDIDDGKNRKNQKDDIEEEDADVIEKVLWHQLKGMGEDVQTNNKST 444
Query: 420 EPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRVTEEI 479
PVL+S LFD+EPDWN MEFLIKWKGQSHLHCQWK+ DLQNLSGFKKVLNYTK+VTEEI
Sbjct: 445 VPVLVSQLFDTEPDWNEMEFLIKWKGQSHLHCQWKTLSDLQNLSGFKKVLNYTKKVTEEI 504
Query: 480 RNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLSY 539
R R +SREEIEVNDVSKEMD+DIIKQNSQVERIIADRIS D G+V PEYLVKWQGLSY
Sbjct: 505 RYRTALSREEIEVNDVSKEMDLDIIKQNSQVERIIADRISKDGLGDVVPEYLVKWQGLSY 564
Query: 540 AEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKG---------------- 583
AE TWEKD+DIAFAQ IDEYK RE +++VQGKMV+ QR + KG
Sbjct: 565 AEATWEKDVDIAFAQVAIDEYKAREVSIAVQGKMVEQQRTKGKGENSFSNAELWLLFSVA 624
Query: 584 SLRKLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQ 643
SLRKLDEQPEWL GG LRDYQLEGLNFLVNSW NDTNV+LADEMGLGKTVQSVSMLGFLQ
Sbjct: 625 SLRKLDEQPEWLIGGTLRDYQLEGLNFLVNSWLNDTNVILADEMGLGKTVQSVSMLGFLQ 684
Query: 644 NAQQIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGK 703
N QQI GPFLVVVPLSTL+NWAKEFRKWLP +N+IVYVGTR+SREV + + K G+
Sbjct: 685 NTQQIPGPFLVVVPLSTLANWAKEFRKWLPGMNIIVYVGTRASREVRNKTN--DVHKVGR 742
Query: 704 QIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLIT 763
IKFNALLTTYEVVLKDKAVLSKIKW YLMVDEAHRLKNSEAQLYTAL EF+TKNKLLIT
Sbjct: 743 PIKFNALLTTYEVVLKDKAVLSKIKWIYLMVDEAHRLKNSEAQLYTALLEFSTKNKLLIT 802
Query: 764 GTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVI 823
GTPLQNSVEELWALLHFLD KFK+KDEF +NYKNLSSFNE+EL+NLH+ELRPH+LRRVI
Sbjct: 803 GTPLQNSVEELWALLHFLDPGKFKNKDEFVENYKNLSSFNESELANLHLELRPHILRRVI 862
Query: 824 KDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEEV 879
KDVEKSLPPKIERILRV+MSPLQKQYYKWILERNF DLNKGVRGNQ + + E+
Sbjct: 863 KDVEKSLPPKIERILRVEMSPLQKQYYKWILERNFHDLNKGVRGNQVSLLNIVVEL 918
Score = 1005 bits (2598), Expect = 0.0
Identities = 529/773 (68%), Positives = 610/773 (78%), Gaps = 39/773 (5%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DILA+Y+SLRGFQFQRLDGSTK+ELRQQAMDHFNAP SDDFCFLLSTRAGGLGINLATAD
Sbjct: 983 DILAEYLSLRGFQFQRLDGSTKAELRQQAMDHFNAPASDDFCFLLSTRAGGLGINLATAD 1042
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TV+IFDSDWNPQNDLQAMSRAHRIGQ+EVVNIYRFVTSKSVEE+ILERAK+KMVLDHLVI
Sbjct: 1043 TVVIFDSDWNPQNDLQAMSRAHRIGQQEVVNIYRFVTSKSVEEEILERAKRKMVLDHLVI 1102
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDEIL 1117
QKLNAEG+LEK+E KKG +F DKNELSAILRFGAEELFKE++NDEESKKRLLSMDIDEIL
Sbjct: 1103 QKLNAEGRLEKRETKKGSNF-DKNELSAILRFGAEELFKEDKNDEESKKRLLSMDIDEIL 1161
Query: 1118 ERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAAR 1177
ERAE+VEEK E HELL AFKVANFCN EDDGSFWSRWIK DSV AE ALAPRAAR
Sbjct: 1162 ERAEQVEEKHTD-ETEHELLGAFKVANFCNAEDDGSFWSRWIKPDSVVTAEEALAPRAAR 1220
Query: 1178 NIKSYAEADQSERSKKRKKKENEP---TERIPKRRKADYSAHVISMIDGASAQVRSWSYG 1234
N KSY + +R+ KRKKK +EP TER KRRK +Y +++G SAQVR WSYG
Sbjct: 1221 NTKSYVDPSHPDRTSKRKKKGSEPPEHTERSQKRRKTEYFVPSTPLLEGTSAQVRGWSYG 1280
Query: 1235 NLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVG 1294
NL KRDA RF R+VMKFGN +Q+ I EVGG +EAAP +AQVELF+ALIDGC+E+VE G
Sbjct: 1281 NLPKRDAQRFYRTVMKFGNHNQMACIAEEVGGVVEAAPEEAQVELFDALIDGCKESVETG 1340
Query: 1295 SLDLKGPLLDFYGVPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGC 1354
+ + KGP+LDF+GVP+KANELL RVQ LQLL+KRISRY DPI+QFRVL+YLKPSNWSKGC
Sbjct: 1341 NFEPKGPVLDFFGVPVKANELLKRVQGLQLLSKRISRYNDPISQFRVLSYLKPSNWSKGC 1400
Query: 1355 GWNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRAN 1414
GWNQIDDARLLLG+ YHG+GNWE IRLDE LGLTKKIAPVELQHHETFLPRAPNL++RA
Sbjct: 1401 GWNQIDDARLLLGILYHGFGNWEKIRLDESLGLTKKIAPVELQHHETFLPRAPNLKERAT 1460
Query: 1415 ALLEQELAVLGVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKKKNIGSSKVNVQMRK 1474
ALLE ELA G KN ++K RK SKK +++L++ + ++++ G + V++ K
Sbjct: 1461 ALLEMELAAAGGKNTNAKASRKNSKK---VKDNLINQFKAPARDRRGKSGPANVSLLSTK 1517
Query: 1475 DRLQKPLNVEPIVKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHRLQTTSA 1534
D +K EP+VKEEGEMSDD +VYEQFKE KW EWC+D++ +E+KTL RL RLQTTSA
Sbjct: 1518 DGPRKTQKAEPLVKEEGEMSDDGEVYEQFKEQKWMEWCEDVLADEIKTLGRLQRLQTTSA 1577
Query: 1535 SLPKEKVLSKIRNYLQLLGRRIDQIVSEQEDEPHKQDRMTTRLWKYVSTFSHLSGERLHQ 1594
LPKEKVL KIR YL++LGRRID IV E E++ +KQDRMT RLW YVSTFS+LSG+RL+Q
Sbjct: 1578 DLPKEKVLFKIRRYLEILGRRIDAIVLEHEEDLYKQDRMTMRLWNYVSTFSNLSGDRLNQ 1637
Query: 1595 IYSKLKLE-QNAVGVGSSLPNGSVSGPFSRNGNPNSSFPRPMERQTRFQNVTAHPMREQT 1653
IYSKLK E + GVG S NGS R +RQ +F+ +Q
Sbjct: 1638 IYSKLKQEKEEEEGVGPSHLNGS----------------RNFQRQQKFKTAGNSQGSQQV 1681
Query: 1654 Y---DTGMSEAWKRRRRAENDGCFQGQPPPQRITSNGIRPLDPNSLGILGAGP 1703
+ DT EAWKRRRR END Q + P IT++ NSLGILG GP
Sbjct: 1682 HKGIDTAKFEAWKRRRRTEND--VQTERP--TITNS-------NSLGILGPGP 1723
>At2g25170 putative chromodomain-helicase-DNA-binding protein
Length = 1359
Score = 308 bits (788), Expect = 2e-83
Identities = 172/411 (41%), Positives = 259/411 (62%), Gaps = 38/411 (9%)
Query: 479 IRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLVKWQGLS 538
++ R+ ++E + S++ + I + + V+RI+A R + G + EYLVK++ LS
Sbjct: 150 LKTRVNNFHRQMESFNNSEDDFVAIRPEWTTVDRILACR---EEDGEL--EYLVKYKELS 204
Query: 539 YAEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKG- 597
Y E WE + DI+ Q+ I +K S + D +++ ++ D PE+LK
Sbjct: 205 YDECYWESESDISTFQNEIQRFKD---VNSRTRRSKDVDHKRNPRDFQQFDHTPEFLKDV 261
Query: 598 ---------GKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQI 648
G L YQLEGLNFL SW T+V+LADEMGLGKT+QS+++L L I
Sbjct: 262 MIYLFPAIEGLLHPYQLEGLNFLRFSWSKQTHVILADEMGLGKTIQSIALLASLFEENLI 321
Query: 649 HGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEF--------CNEKK 700
P LV+ PLSTL NW +EF W P +NV++Y GT +R V +++EF +KK
Sbjct: 322 --PHLVIAPLSTLRNWEREFATWAPQMNVVMYFGTAQARAVIREHEFYLSKDQKKIKKKK 379
Query: 701 AG--------KQIKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALS 752
+G K+IKF+ LLT+YE++ D AVL IKW ++VDE HRLKN +++L+++L+
Sbjct: 380 SGQISSESKQKRIKFDVLLTSYEMINLDSAVLKPIKWECMIVDEGHRLKNKDSKLFSSLT 439
Query: 753 EFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHM 812
++++ +++L+TGTPLQN+++EL+ L+HFLD+ KF S +EF + +K+++ E ++S LH
Sbjct: 440 QYSSNHRILLTGTPLQNNLDELFMLMHFLDAGKFGSLEEFQEEFKDIN--QEEQISRLHK 497
Query: 813 ELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNK 863
L PH+LRRV KDV K +PPK E ILRVD+S LQK+YYK I RN++ L K
Sbjct: 498 MLAPHLLRRVKKDVMKDMPPKKELILRVDLSSLQKEYYKAIFTRNYQVLTK 548
Score = 196 bits (498), Expect = 1e-49
Identities = 158/497 (31%), Positives = 246/497 (48%), Gaps = 71/497 (14%)
Query: 932 YSDF--LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGL 989
Y+ F + D+L Y + + +Q++R+DG RQ +D FNA S+ FCFLLSTRAGGL
Sbjct: 617 YTQFQHMLDLLEDYCTHKKWQYERIDGKVGGAERQIRIDRFNAKNSNKFCFLLSTRAGGL 676
Query: 990 GINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKK 1049
GINLATADTVII+DSDWNP DLQAM+RAHR+GQ V IYR + ++EE +++ KKK
Sbjct: 677 GINLATADTVIIYDSDWNPHADLQAMARAHRLGQTNKVMIYRLINRGTIEERMMQLTKKK 736
Query: 1050 MVLDHLVIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLL 1109
MVL+HLV+ KL + ++ EL I+R+G++ELF E +DE K +
Sbjct: 737 MVLEHLVVGKLKTQN-------------INQEELDDIIRYGSKELFASE-DDEAGKSGKI 782
Query: 1110 SMD---IDEILER-AEKVEEKENGGEQAHELLSAFKVANF--CNDEDDGSFWSRWIKADS 1163
D ID++L+R + EE E+ + L AFKVANF ++ + + ++ + A+S
Sbjct: 783 HYDDAAIDKLLDRDLVEAEEVSVDDEEENGFLKAFKVANFEYIDENEAAALEAQRVAAES 842
Query: 1164 VAQAENA-LAPRAARNIKSYAEADQSER-----SKKRKKKENEPTERIPKRRKADYSAHV 1217
+ A N+ A +K E Q+E +KR +K+ E D S+
Sbjct: 843 KSSAGNSDRASYWEELLKDKFELHQAEELNALGKRKRSRKQLVSIEEDDLAGLEDVSS-- 900
Query: 1218 ISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPL-KAQ 1276
DG + + G + + R + G ++ +E PL + +
Sbjct: 901 ----DGDESYEAESTDGEAAGQGVQTGRRPYRRKGRDN------------LEPTPLMEGE 944
Query: 1277 VELFNAL-IDGCREAVEVGSLDLKGPLLDFY--------------GVP---MKANELLIR 1318
F L + + A+ V +L G L + GVP ++ ++L+R
Sbjct: 945 GRSFRVLGFNQSQRAIFVQTLMRYGILFLKHIAEEIDENSPTFSDGVPKEGLRIEDVLVR 1004
Query: 1319 VQELQLLAKRISRYED----PIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLGVHYHGYG 1374
+ L L+ +++ ED P+ R+L + G W + D ++ V HGYG
Sbjct: 1005 IALLILVQEKVKFVEDHPGKPVFPSRILE--RFPGLRSGKIWKEEHDKIMIRAVLKHGYG 1062
Query: 1375 NWEVIRLDERLGLTKKI 1391
W+ I D+ LG+ + I
Sbjct: 1063 RWQAIVDDKELGIQELI 1079
>At5g44800 helicase-like protein
Length = 2228
Score = 280 bits (716), Expect = 5e-75
Identities = 193/584 (33%), Positives = 306/584 (52%), Gaps = 72/584 (12%)
Query: 318 SDSESDEDFKSLKKRGVRVRKNNGRSSAATSFSRPSNEVRSSSRTIRKVSYVESDESEGA 377
SD ++ ++ +R V+ N A S+E SSR R + + D+S
Sbjct: 399 SDDLCSDNLQATDQRDSLVQDTNAELVVAEDRIDSSSETGKSSRDSR-LRDKDMDDSALG 457
Query: 378 DEGTKKSQKEEIE-----------VDDGD--------SVEKVLW---HQPKGMAAEAQRN 415
EG + ++E + VDD D SVE+ L HQ G +
Sbjct: 458 TEGMVEVKEEMLSEDISNATLSRHVDDEDMKVSETHVSVERELLEEAHQETGEKSTVA-- 515
Query: 416 NQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYTKRV 475
++ +E + + D + + EFL+KW +S++H W S +L+ L+ KR
Sbjct: 516 DEEIEEPVAAKTSDLIGETVSYEFLVKWVDKSNIHNTWISEAELKGLA---------KRK 566
Query: 476 TEEIRNRMGISREEIEVNDVSKEMDIDIIKQN-SQVERIIADRISNDNSGNVFPEYLVKW 534
E + + G + I+I + Q +RI+A R+S + + + VKW
Sbjct: 567 LENYKAKYGTAV-------------INICEDKWKQPQRIVALRVSKEGNQEAY----VKW 609
Query: 535 QGLSYAEVTWE--KDIDIAFAQHTID---EYKTREAAMSVQGKMVDFQRRQSKGSLRKLD 589
GL+Y E TWE ++ + + H ID +Y+ + + +G + +G + L
Sbjct: 610 TGLAYDECTWESLEEPILKHSSHLIDLFHQYEQKTLERNSKGNPT-----RERGEVVTLT 664
Query: 590 EQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIH 649
EQP+ L+GG L +QLE LN+L W NV+LADEMGLGKTV + + L L +
Sbjct: 665 EQPQELRGGALFAHQLEALNWLRRCWHKSKNVILADEMGLGKTVSASAFLSSLYFEFGVA 724
Query: 650 GPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGK-----Q 704
P LV+VPLST+ NW EF W P LNV+ Y G+ R + + YE+ + G
Sbjct: 725 RPCLVLVPLSTMPNWLSEFSLWAPLLNVVEYHGSAKGRAIIRDYEWHAKNSTGTTKKPTS 784
Query: 705 IKFNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITG 764
KFN LLTTYE+VL D + L + W L+VDE HRLKNSE++L++ L+ F+ ++++L+TG
Sbjct: 785 YKFNVLLTTYEMVLADSSHLRGVPWEVLVVDEGHRLKNSESKLFSLLNTFSFQHRVLLTG 844
Query: 765 TPLQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENELSNLHMELRPHMLRRVIK 824
TPLQN++ E++ LL+FL F S F + + +L+S + E L + PHMLRR+ K
Sbjct: 845 TPLQNNIGEMYNLLNFLQPSSFPSLSSFEERFHDLTSAEKVE--ELKKLVAPHMLRRLKK 902
Query: 825 DVEKSLPPKIERILRVDMSPLQKQYYKWILERNF---RDLNKGV 865
D +++PPK ER++ V+++ +Q +YY+ +L +N+ R++ KGV
Sbjct: 903 DAMQNIPPKTERMVPVELTSIQAEYYRAMLTKNYQILRNIGKGV 946
Score = 137 bits (344), Expect = 7e-32
Identities = 84/196 (42%), Positives = 114/196 (57%), Gaps = 20/196 (10%)
Query: 936 LSDILAQYMSLR--GFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINL 993
L DIL Y+++ F+R+DGS RQ A+ FN + F FLLSTRA GLGINL
Sbjct: 1018 LLDILEDYLNIEFGPKTFERVDGSVAVADRQAAIARFNQD-KNRFVFLLSTRACGLGINL 1076
Query: 994 ATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLD 1053
ATADTVII+DSD+NP D+QAM+RAHRIGQ + + +YR V SVEE IL+ AKKK++LD
Sbjct: 1077 ATADTVIIYDSDFNPHADIQAMNRAHRIGQSKRLLVYRLVVRASVEERILQLAKKKLMLD 1136
Query: 1054 HLVIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDI 1113
L + K ++ E ILR+G EELF + + + + ++
Sbjct: 1137 QLFVNKSGSQ-----------------KEFEDILRWGTEELFNDSAGENKKDTAESNGNL 1179
Query: 1114 DEILERAEKVEEKENG 1129
D I++ K +K G
Sbjct: 1180 DVIMDLESKSRKKGGG 1195
Score = 37.4 bits (85), Expect = 0.077
Identities = 16/38 (42%), Positives = 22/38 (57%)
Query: 1356 WNQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAP 1393
W++ + L +G+ HGYGNWE I D RL +K P
Sbjct: 1710 WSEDELDSLWIGIRRHGYGNWETILRDPRLKFSKFKTP 1747
>At3g06010 putative transcriptional regulator
Length = 1132
Score = 233 bits (593), Expect = 1e-60
Identities = 133/331 (40%), Positives = 194/331 (58%), Gaps = 26/331 (7%)
Query: 545 EKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGKLRDYQ 604
++DIDI + + D E + Q K+ EQP L+GG+LR YQ
Sbjct: 386 DQDIDITESDNNDDSNDLLEGQRQYNSAIHSIQE--------KVTEQPSLLEGGELRSYQ 437
Query: 605 LEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNW 664
LEGL ++V+ + N+ N +LADEMGLGKT+Q++S++ +L + + GP+L+V P + L NW
Sbjct: 438 LEGLQWMVSLFNNNLNGILADEMGLGKTIQTISLIAYLLENKGVPGPYLIVAPKAVLPNW 497
Query: 665 AKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVL 724
EF W+P + +Y G R+ + EK AG+ KFN L+T Y+++++DKA L
Sbjct: 498 VNEFATWVPSIAAFLYDGRLEERKAIR------EKIAGEG-KFNVLITHYDLIMRDKAFL 550
Query: 725 SKIKWNYLMVDEAHRLKNSEAQL-YTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDS 783
KI+W Y++VDE HRLKN E+ L T L+ + K +LL+TGTP+QNS++ELW+LL+FL
Sbjct: 551 KKIEWYYMIVDEGHRLKNHESALAKTLLTGYRIKRRLLLTGTPIQNSLQELWSLLNFLLP 610
Query: 784 DKFKSKDEFAQNYK-------NLSSFNENEL---SNLHMELRPHMLRRVIKDVEKSLPPK 833
F S F + + N+S +E EL LH +RP +LRR +VEK LP K
Sbjct: 611 HIFNSVQNFEEWFNAPFADRGNVSLTDEEELLIIHRLHHVIRPFILRRKKDEVEKFLPGK 670
Query: 834 IERILRVDMSPLQKQYYKWILERNFRDLNKG 864
+ IL+ DMS QK YYK + + L G
Sbjct: 671 TQVILKCDMSAWQKVYYKQVTDMGRVGLQTG 701
Score = 149 bits (375), Expect = 2e-35
Identities = 92/216 (42%), Positives = 127/216 (58%), Gaps = 20/216 (9%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLAT 995
L D+L Y++L +++ RLDG+TK++ R + FN P S F FLLSTRAGGLG+NL T
Sbjct: 771 LIDVLEIYLTLNDYKYLRLDGTTKTDQRGLLLKQFNEPDSPYFMFLLSTRAGGLGLNLQT 830
Query: 996 ADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHL 1055
ADTVIIFDSDWNPQ D QA RAHRIGQ++ V ++ V+ SVEE ILERAK+KM +D
Sbjct: 831 ADTVIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSVGSVEEVILERAKQKMGIDAK 890
Query: 1056 VIQ------KLNAEGKLEKKE--AKKGGSFF-----DKNELSAILRFGAEELFKEERNDE 1102
VIQ A+ + E E +KG S + E++ + +E + ER DE
Sbjct: 891 VIQAGLFNTTSTAQDRREMLEEIMRKGTSSLGTDVPSEREINRLAARSEDEFWMFERMDE 950
Query: 1103 ESKKR-------LLSMDIDEILERAEKVEEKENGGE 1131
E +++ + ++ E + EEK N G+
Sbjct: 951 ERRRKENYRARLMQEQEVPEWAYTTQTQEEKLNNGK 986
>At5g18620 chromatin remodelling complex ATPase chain ISWI -like
protein
Length = 1069
Score = 231 bits (590), Expect = 2e-60
Identities = 123/280 (43%), Positives = 183/280 (64%), Gaps = 11/280 (3%)
Query: 587 KLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
+L QP ++G KLRDYQL GLN+L+ ++N N +LADEMGLGKT+Q++S+L +L +
Sbjct: 182 RLLTQPACIQG-KLRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLAYLHEYR 240
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIK 706
I+GP +VV P STL NW E R++ P L + ++G R ++ AG K
Sbjct: 241 GINGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLGNPEERRHIREELLV----AG---K 293
Query: 707 FNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTP 766
F+ +T++E+ +K+K L + W Y+++DEAHR+KN + L + F+T +LLITGTP
Sbjct: 294 FDICVTSFEMAIKEKTTLRRFSWRYIIIDEAHRIKNENSLLSKTMRLFSTNYRLLITGTP 353
Query: 767 LQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENE--LSNLHMELRPHMLRRVIK 824
LQN++ ELWALL+FL + F S + F + ++ +S N+ + + LH LRP +LRR+
Sbjct: 354 LQNNLHELWALLNFLLPEVFSSAETFDEWFQ-ISGENDQQEVVQQLHKVLRPFLLRRLKS 412
Query: 825 DVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKG 864
DVEK LPPK E IL+V MS +QKQYYK +L+++ +N G
Sbjct: 413 DVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEVVNGG 452
Score = 179 bits (455), Expect = 1e-44
Identities = 146/498 (29%), Positives = 239/498 (47%), Gaps = 97/498 (19%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLAT 995
L DIL Y+ RG+Q+ R+DG+T + R +++ +N PGS+ F FLLSTRAGGLGINLAT
Sbjct: 523 LLDILEDYLMYRGYQYCRIDGNTGGDERDASIEAYNKPGSEKFVFLLSTRAGGLGINLAT 582
Query: 996 ADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHL 1055
AD VI++DSDWNPQ DLQA RAHRIGQ++ V ++RF T ++E ++ERA KK+ LD L
Sbjct: 583 ADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTENAIEAKVIERAYKKLALDAL 642
Query: 1056 VIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
VIQ +G+L +++ +K+EL ++R+GAE +F + + DID
Sbjct: 643 VIQ----QGRLAEQKT------VNKDELLQMVRYGAEMVFSSK------DSTITDEDIDR 686
Query: 1116 ILERAEK-VEEKENGGEQAHELLSAFKV---ANFCNDEDDGSFWSRWIKAD-SVAQAENA 1170
I+ + E+ E + ++ E FK+ A+F + +DD S K D +EN
Sbjct: 687 IIAKGEEATAELDAKMKKFTEDAIQFKMDDSADFYDFDDDNKDES---KVDFKKIVSENW 743
Query: 1171 LAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRS 1230
P ++Y+E + +++ ++ RIP+ Q+
Sbjct: 744 NDPPKRERKRNYSEVEYFKQTLRQGAPAKPKEPRIPR-----------------MPQLHD 786
Query: 1231 WSYGNLSKRDAL--RFSRSVMKFGNESQINLIV-----AEVGGAIEAAPLKAQVEL---- 1279
+ + N+ + L + R +M+ ++Q+ + EVG + A ++ + L
Sbjct: 787 FQFFNIQRLTELYEKEVRYLMQAHQKTQMKDTIEVDEPEEVGDPLTAEEVEEKELLLEEG 846
Query: 1280 --------FNALIDGCREAVEVGSLDLKGPLLDFYG-----VPMKANELLIRVQEL---- 1322
FNA I C + G D+K + G V A +R +EL
Sbjct: 847 FSTWSRRDFNAFIRACE---KYGRNDIKSIASEMEGKTEEEVERYAQVFQVRYKELNDYD 903
Query: 1323 --------------------QLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDA 1362
+ + K++ RY +P + ++ +KG +N+ D
Sbjct: 904 RIIKNIERGEARISRKDEIMKAIGKKLDRYRNPWLELKI-----QYGQNKGKLYNEECDR 958
Query: 1363 RLLLGVHYHGYGNWEVIR 1380
++ VH GYGNW+ ++
Sbjct: 959 FMICMVHKLGYGNWDELK 976
>At3g06400 putative ATPase (ISW2-like)
Length = 1057
Score = 231 bits (590), Expect = 2e-60
Identities = 122/280 (43%), Positives = 184/280 (65%), Gaps = 11/280 (3%)
Query: 587 KLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
+L QP ++G K+RDYQL GLN+L+ ++N N +LADEMGLGKT+Q++S+L +L +
Sbjct: 177 RLLTQPSCIQG-KMRDYQLAGLNWLIRLYENGINGILADEMGLGKTLQTISLLAYLHEYR 235
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIK 706
I+GP +VV P STL NW E R++ P L + ++G R ++ + AG K
Sbjct: 236 GINGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLGNPEERRHIRE----DLLVAG---K 288
Query: 707 FNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTP 766
F+ +T++E+ +K+K L + W Y+++DEAHR+KN + L + F+T +LLITGTP
Sbjct: 289 FDICVTSFEMAIKEKTALRRFSWRYIIIDEAHRIKNENSLLSKTMRLFSTNYRLLITGTP 348
Query: 767 LQNSVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENE--LSNLHMELRPHMLRRVIK 824
LQN++ ELWALL+FL + F S + F + ++ +S N+ + + LH LRP +LRR+
Sbjct: 349 LQNNLHELWALLNFLLPEIFSSAETFDEWFQ-ISGENDQQEVVQQLHKVLRPFLLRRLKS 407
Query: 825 DVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKG 864
DVEK LPPK E IL+V MS +QKQYYK +L+++ +N G
Sbjct: 408 DVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEAVNAG 447
Score = 175 bits (443), Expect = 2e-43
Identities = 140/493 (28%), Positives = 237/493 (47%), Gaps = 86/493 (17%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLAT 995
L DIL Y+ RG+ + R+DG+T + R +++ +N PGS+ F FLLSTRAGGLGINLAT
Sbjct: 518 LLDILEDYLMYRGYLYCRIDGNTGGDERDASIEAYNKPGSEKFVFLLSTRAGGLGINLAT 577
Query: 996 ADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHL 1055
AD VI++DSDWNPQ DLQA RAHRIGQ++ V ++RF T ++EE ++ERA KK+ LD L
Sbjct: 578 ADVVILYDSDWNPQVDLQAQDRAHRIGQKKEVQVFRFCTESAIEEKVIERAYKKLALDAL 637
Query: 1056 VIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMDIDE 1115
VIQ +G+L ++++K +K+EL ++R+GAE +F + + DID
Sbjct: 638 VIQ----QGRLAEQKSKS----VNKDELLQMVRYGAEMVFSSK------DSTITDEDIDR 683
Query: 1116 ILERAEK-VEEKENGGEQAHELLSAFKV---ANFCNDEDD-------------------- 1151
I+ + E+ E + ++ E FK+ A+F + +DD
Sbjct: 684 IIAKGEEATAELDAKMKKFTEDAIQFKMDDSADFYDFDDDNKDENKLDFKKIVSDNWNDP 743
Query: 1152 -----------GSFWSRWIKADSVAQAENALAPRAAR-------NIKSYAEADQSE-RSK 1192
++ + ++ + A+ + PR + NI+ E + E R
Sbjct: 744 PKRERKRNYSESEYFKQTLRQGAPAKPKEPRIPRMPQLHDFQFFNIQRLTELYEKEVRYL 803
Query: 1193 KRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSYGNLSKRDALRFSRSVMKFG 1252
+ ++N+ + I ++ + + S+RD F R+ K+G
Sbjct: 804 MQTHQKNQLKDTIDVEEPEGGDPLTTEEVEEKEGLLEE-GFSTWSRRDFNTFLRACEKYG 862
Query: 1253 NESQINLIVAEVGGAIEAAP---LKAQVELFNALIDGCR--EAVEVGSLDLKGPLLDFYG 1307
+ I I +E+ G E K E + L D R + +E G +
Sbjct: 863 -RNDIKSIASEMEGKTEEEVERYAKVFKERYKELNDYDRIIKNIERGEARIS-------- 913
Query: 1308 VPMKANELLIRVQELQLLAKRISRYEDPIAQFRVLTYLKPSNWSKGCGWNQIDDARLLLG 1367
+ +E+ ++ + K++ RY +P + ++ +KG +N+ D ++
Sbjct: 914 ---RKDEI------MKAIGKKLDRYRNPWLELKI-----QYGQNKGKLYNEECDRFMICM 959
Query: 1368 VHYHGYGNWEVIR 1380
+H GYGNW+ ++
Sbjct: 960 IHKLGYGNWDELK 972
Score = 32.7 bits (73), Expect = 1.9
Identities = 34/152 (22%), Positives = 58/152 (37%), Gaps = 29/152 (19%)
Query: 197 KSRILDDAEDDDDDADY--EEDEPDEDDPDDADFEPATSGRGANKYKDWEGEDSDEVDDS 254
+ R+ D+ E+D+++ + D+D+ AD P + G A D+E E+ +E +
Sbjct: 17 EERVKDNEEEDEEELEAVARSSGSDDDEVAAADESPVSDGEAAPVEDDYEDEEDEEKAEI 76
Query: 255 D------------------EDIDVSDNDDLYFDKKAKGRQRGK--------FGPSVRSTR 288
+++ S N + D KG+ R K F +S
Sbjct: 77 SKREKARLKEMQKLKKQKIQEMLESQNASIDADMNNKGKGRLKYLLQQTELFAHFAKSDG 136
Query: 289 DCKAFTASSRQRRV-KSSFEDEDENSTAEDSD 319
A R R K + E+EDE E+ D
Sbjct: 137 SSSQKKAKGRGRHASKITEEEEDEEYLKEEED 168
>At5g19310 homeotic gene regulator - like protein
Length = 1041
Score = 227 bits (579), Expect = 4e-59
Identities = 122/289 (42%), Positives = 181/289 (62%), Gaps = 19/289 (6%)
Query: 587 KLDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQ 646
K+ +QP L+GG+LR YQLEGL ++V+ + ND N +LADEMGLGKT+Q+++++ +L ++
Sbjct: 350 KVTKQPSLLQGGELRSYQLEGLQWMVSLYNNDYNGILADEMGLGKTIQTIALIAYLLESK 409
Query: 647 QIHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIK 706
+HGP L++ P + L NW EF W P ++ +Y G++ R + + AG K
Sbjct: 410 DLHGPHLILAPKAVLPNWENEFALWAPSISAFLYDGSKEKRTEIR------ARIAGG--K 461
Query: 707 FNALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTAL-SEFNTKNKLLITGT 765
FN L+T Y+++++DKA L KI WNY++VDE HRLKN E L L + + K +LL+TGT
Sbjct: 462 FNVLITHYDLIMRDKAFLKKIDWNYMIVDEGHRLKNHECALAKTLGTGYRIKRRLLLTGT 521
Query: 766 PLQNSVEELWALLHFLDSDKFKSKDEFAQNYK-------NLSSFNENEL---SNLHMELR 815
P+QNS++ELW+LL+FL F S F + + + S +E EL + LH +R
Sbjct: 522 PIQNSLQELWSLLNFLLPHIFNSIHNFEEWFNTPFAECGSASLTDEEELLIINRLHHVIR 581
Query: 816 PHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWILERNFRDLNKG 864
P +LRR +VEK LP K + IL+ DMS QK YYK + + L+ G
Sbjct: 582 PFLLRRKKSEVEKFLPGKTQVILKCDMSAWQKLYYKQVTDVGRVGLHSG 630
Score = 152 bits (385), Expect = 1e-36
Identities = 106/305 (34%), Positives = 153/305 (49%), Gaps = 30/305 (9%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLAT 995
L D+L Y+SL + + RLDGSTK++ R + FN P S F FLLSTRAGGLG+NL T
Sbjct: 700 LIDLLEIYLSLNDYMYLRLDGSTKTDQRGILLKQFNEPDSPYFMFLLSTRAGGLGLNLQT 759
Query: 996 ADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHL 1055
ADT+IIFDSDWNPQ D QA RAHRIGQ++ V ++ V+ S+EE ILERAK+KM +D
Sbjct: 760 ADTIIIFDSDWNPQMDQQAEDRAHRIGQKKEVRVFVLVSIGSIEEVILERAKQKMGIDAK 819
Query: 1056 VIQ-------------KLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDE 1102
VIQ + E + K + G + E++ + EE + E+ DE
Sbjct: 820 VIQAGLFNTTSTAQDRREMLEEIMSKGTSSLGEDVPSEREINRLAARTEEEFWMFEQMDE 879
Query: 1103 ESKKR-------LLSMDIDEILERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFW 1155
E +K+ + ++ E +E E+K N L K +
Sbjct: 880 ERRKKENYKTRLMEEKEVPEWAYTSETQEDKTNAKNHFGSLTGKRKRKEAVYSDSLSDL- 938
Query: 1156 SRWIK--------ADSVAQAENALAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPK 1207
+W+K A V+Q + + S AEA SE ++++++E E E K
Sbjct: 939 -QWMKAMESEDEDASKVSQKRKRTDTKTRMSNGSKAEAVLSESDEEKEEEEEERKEESGK 997
Query: 1208 RRKAD 1212
+ +
Sbjct: 998 ESEEE 1002
Score = 31.2 bits (69), Expect = 5.5
Identities = 35/146 (23%), Positives = 60/146 (40%), Gaps = 12/146 (8%)
Query: 55 VDRLSTRESNVETTSRNPSASERWGSSYLKDCQPMSPQNGSESGDDSKSGSDYRNEDEFE 114
+D ++ N +T W +Y + Q + +++ S +G R E +
Sbjct: 877 MDEERRKKENYKTRLMEEKEVPEW--AYTSETQ--EDKTNAKNHFGSLTGKRKRKEAVYS 932
Query: 115 DNSSEGRGEK-LGSEDEDGQKDSGKGQRGDSDV------PAEEMLSDDSYGQDGEEQGES 167
D+ S+ + K + SEDED K S K +R D+ AE +LS+ ++ EE+ E
Sbjct: 933 DSLSDLQWMKAMESEDEDASKVSQKRKRTDTKTRMSNGSKAEAVLSESDEEKE-EEEEER 991
Query: 168 VHSRGFRPSTGSNSCLQPTSTNVNRR 193
G + L TN +R
Sbjct: 992 KEESGKESEEENEKPLHSWKTNKKKR 1017
>At2g28290 putative SNF2 subfamily transcription regulator
Length = 1331
Score = 223 bits (569), Expect = 6e-58
Identities = 121/278 (43%), Positives = 173/278 (61%), Gaps = 21/278 (7%)
Query: 588 LDEQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQ 647
++EQP L GGKLR+YQ+ GL +LV+ + N N +LADEMGLGKTVQ +S++ +L +
Sbjct: 781 INEQPSSLVGGKLREYQMNGLRWLVSLYNNHLNGILADEMGLGKTVQVISLICYLMETKN 840
Query: 648 IHGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKF 707
GPFLVVVP S L W E W P ++ IVY GT R + + ++ KF
Sbjct: 841 DRGPFLVVVPSSVLPGWQSEINFWAPSIHKIVYCGTPDERRKLFKEQIVHQ-------KF 893
Query: 708 NALLTTYEVVLK--DKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGT 765
N LLTTYE ++ D+ LSKI W+Y+++DE HR+KN+ +L L + + ++LL+TGT
Sbjct: 894 NVLLTTYEYLMNKHDRPKLSKIHWHYIIIDEGHRIKNASCKLNADLKHYVSSHRLLLTGT 953
Query: 766 PLQNSVEELWALLHFLDSDKFKSKDEFAQ--------NYKNLSSFNENE----LSNLHME 813
PLQN++EELWALL+FL + F S ++F+Q N ++ + +E E ++ LH
Sbjct: 954 PLQNNLEELWALLNFLLPNIFNSSEDFSQWFNKPFQSNGESSALLSEEENLLIINRLHQV 1013
Query: 814 LRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYK 851
LRP +LRR+ VE LP KIER++R + S QK K
Sbjct: 1014 LRPFVLRRLKHKVENELPEKIERLIRCEASAYQKLLMK 1051
Score = 130 bits (327), Expect = 7e-30
Identities = 77/202 (38%), Positives = 109/202 (53%), Gaps = 28/202 (13%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLAT 995
L D++ Y++L+G+++ RLDG T R +D FN GS F FLLS RAGG+G+NL
Sbjct: 1130 LLDVMEDYLTLKGYKYLRLDGQTSGGDRGALIDGFNKSGSPFFIFLLSIRAGGVGVNLQA 1189
Query: 996 ADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHL 1055
ADTVI+FD+DWNPQ DLQA +RAHRIGQ++ V + RF T SVEE +
Sbjct: 1190 ADTVILFDTDWNPQVDLQAQARAHRIGQKKDVLVLRFETVNSVEEQV------------- 1236
Query: 1056 VIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEES----------- 1104
+ +AE KL FFD N + + E L +E + +E++
Sbjct: 1237 ---RASAEHKLGVANQSITAGFFDNNTSAEDRKEYLESLLRESKKEEDAPVLDDDALNDL 1293
Query: 1105 -KKRLLSMDIDEILERAEKVEE 1125
+R +DI E +++ K E
Sbjct: 1294 IARRESEIDIFESIDKQRKENE 1315
>At2g46020 putative SNF2 subfamily transcriptional activator
Length = 1245
Score = 213 bits (541), Expect = 1e-54
Identities = 114/280 (40%), Positives = 168/280 (59%), Gaps = 26/280 (9%)
Query: 591 QPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHG 650
QP L+ G LRDYQL GL ++++ + N N +LADEMGLGKTVQ ++++ +L + +G
Sbjct: 25 QPSMLQAGTLRDYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYG 84
Query: 651 PFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSR-EVCQQYEFCNEKKAGKQIKFNA 709
P L++VP + L NW E WLP ++ I YVGT+ R ++ Q +F KFN
Sbjct: 85 PHLIIVPNAVLVNWKSELHTWLPSVSCIYYVGTKDQRSKLFSQVKF---------EKFNV 135
Query: 710 LLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQN 769
L+TTYE ++ D++ LSK+ W Y+++DEA R+K+ E+ L L + + +LL+TGTPLQN
Sbjct: 136 LVTTYEFIMYDRSKLSKVDWKYIIIDEAQRMKDRESVLARDLDRYRCQRRLLLTGTPLQN 195
Query: 770 SVEELWALLHFLDSDKFKSK----DEFAQNYKNLSSFNENE------------LSNLHME 813
++ELW+LL+ L D F ++ D FAQ ++ + E + LH
Sbjct: 196 DLKELWSLLNLLLPDVFDNRKAFHDWFAQPFQKEGPAHNIEDDWLETEKKVIVIHRLHQI 255
Query: 814 LRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWI 853
L P MLRR ++DVE SLP K+ +LR MS +Q Y WI
Sbjct: 256 LEPFMLRRRVEDVEGSLPAKVSVVLRCRMSAIQSAVYDWI 295
Score = 104 bits (259), Expect = 5e-22
Identities = 72/209 (34%), Positives = 111/209 (52%), Gaps = 21/209 (10%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLAT 995
L DIL +Y+ R ++R+DG+T E R+ A+ FN P +D F FLLS RA G G+NL T
Sbjct: 388 LLDILEEYLQWRRLVYRRIDGTTSLEDRESAIVDFNDPDTDCFIFLLSIRAAGRGLNLQT 447
Query: 996 ADTVIIFDSDWNPQNDLQAMSRAHRIGQ-REVVNIYRFVTSKSVEEDILERAKKKMVLDH 1054
ADTV+I+D D NP+N+ QA++RAHRIGQ REV IY E ++E+ + H
Sbjct: 448 ADTVVIYDPDPNPKNEEQAVARAHRIGQTREVKVIYM--------EAVVEK-----LSSH 494
Query: 1055 LVIQKLNAEGKLEKKEAKKG-----GSF--FDKNELSAILRFGAEELFKEERNDEESKKR 1107
+L + G ++ ++ G GS +N + A+E+ R D+ +
Sbjct: 495 QKEDELRSGGSVDLEDDMAGKDRYIGSIEGLIRNNIQQYKIDMADEVINAGRFDQRTTHE 554
Query: 1108 LLSMDIDEILERAEKVEEKENGGEQAHEL 1136
M ++ +L E+ +E + HE+
Sbjct: 555 ERRMTLETLLHDEERYQETVHDVPSLHEV 583
>At5g66750 SWI2/SNF2-like protein (gb|AAD28303.1)
Length = 764
Score = 206 bits (525), Expect = 7e-53
Identities = 121/322 (37%), Positives = 189/322 (58%), Gaps = 24/322 (7%)
Query: 551 AFAQHTIDEYKTREAAM----SVQGKMVDFQRRQSKGSLRKLDEQPEWLKGGKLRDYQLE 606
A +Q+ + K AAM G+ ++ + + ++ +E L GG+L+ YQL+
Sbjct: 149 AASQYNNTKAKRAVAAMISRSKEDGETINSDLTEEETVIKLQNELCPLLTGGQLKSYQLK 208
Query: 607 GLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAK 666
G+ +L++ W+N N +LAD+MGLGKT+Q++ L L+ + GP+LV+ PLSTLSNW
Sbjct: 209 GVKWLISLWQNGLNGILADQMGLGKTIQTIGFLSHLKG-NGLDGPYLVIAPLSTLSNWFN 267
Query: 667 EFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKD-KAVLS 725
E ++ P +N I+Y G ++ R+ ++ K G KF ++T+YEV + D K +L
Sbjct: 268 EIARFTPSINAIIYHGDKNQRDELRRKHM--PKTVGP--KFPIVITSYEVAMNDAKRILR 323
Query: 726 KIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDK 785
W Y+++DE HRLKN + +L L NKLL+TGTPLQN++ ELW+LL+F+ D
Sbjct: 324 HYPWKYVVIDEGHRLKNHKCKLLRELKHLKMDNKLLLTGTPLQNNLSELWSLLNFILPDI 383
Query: 786 FKSKDEFAQNYKNLSSFNENE-------------LSNLHMELRPHMLRRVIKDVEKSLPP 832
F S DEF +++ + S N+NE +S LH LRP +LRR+ DVE SLP
Sbjct: 384 FTSHDEF-ESWFDFSEKNKNEATKEEEEKRRAQVVSKLHGILRPFILRRMKCDVELSLPR 442
Query: 833 KIERILRVDMSPLQKQYYKWIL 854
K E I+ M+ QK++ + ++
Sbjct: 443 KKEIIMYATMTDHQKKFQEHLV 464
Score = 138 bits (347), Expect = 3e-32
Identities = 77/188 (40%), Positives = 116/188 (60%), Gaps = 15/188 (7%)
Query: 936 LSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLAT 995
L DI+ Y S +GF+ R+DGS K + R++ + F+ S FLLSTRAGGLGINL
Sbjct: 552 LLDIMDYYFSEKGFEVCRIDGSVKLDERRRQIKDFSDEKSSCSIFLLSTRAGGLGINLTA 611
Query: 996 ADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHL 1055
ADT I++DSDWNPQ DLQAM R HRIGQ + V++YR T++S+E +L+RA K+ L+H+
Sbjct: 612 ADTCILYDSDWNPQMDLQAMDRCHRIGQTKPVHVYRLSTAQSIETRVLKRAYSKLKLEHV 671
Query: 1056 VIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEES--KKRLLSMDI 1113
VI +G+ ++ AK +++ L+ L KE+ E+ + + D+
Sbjct: 672 VI----GQGQFHQERAKSSTPLEEEDILA---------LLKEDETAEDKLIQTDISDADL 718
Query: 1114 DEILERAE 1121
D +L+R++
Sbjct: 719 DRLLDRSD 726
>At3g57300 helicase-like protein
Length = 1507
Score = 200 bits (508), Expect = 7e-51
Identities = 104/272 (38%), Positives = 168/272 (61%), Gaps = 10/272 (3%)
Query: 590 EQPEWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIH 649
+ PE KG L++YQ++GL +LVN ++ N +LADEMGLGKT+Q+++ L L + I
Sbjct: 577 QTPELFKG-TLKEYQMKGLQWLVNCYEQGLNGILADEMGLGKTIQAMAFLAHLAEEKNIW 635
Query: 650 GPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNA 709
GPFLVV P S L+NWA E ++ PDL + Y G R + ++ ++ + F+
Sbjct: 636 GPFLVVAPASVLNNWADEISRFCPDLKTLPYWGGLQERTILRK-NINPKRMYRRDAGFHI 694
Query: 710 LLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQN 769
L+T+Y++++ D+ ++KW Y+++DEA +K+S + + L FN +N+LL+TGTP+QN
Sbjct: 695 LITSYQLLVTDEKYFRRVKWQYMVLDEAQAIKSSSSIRWKTLLSFNCRNRLLLTGTPIQN 754
Query: 770 SVEELWALLHFLDSDKFKSKDEFAQ--------NYKNLSSFNENELSNLHMELRPHMLRR 821
++ ELWALLHF+ F + D+F + + ++ + NE++L+ LH L+P MLRR
Sbjct: 755 NMAELWALLHFIMPMLFDNHDQFNEWFSKGIENHAEHGGTLNEHQLNRLHAILKPFMLRR 814
Query: 822 VIKDVEKSLPPKIERILRVDMSPLQKQYYKWI 853
V KDV L K E + +S Q+ +Y+ I
Sbjct: 815 VKKDVVSELTTKTEVTVHCKLSSRQQAFYQAI 846
Score = 133 bits (335), Expect = 8e-31
Identities = 95/270 (35%), Positives = 140/270 (51%), Gaps = 19/270 (7%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
+IL YM+ R +++ RLDGS+ R+ + F SD F FLLSTRAGGLGINL AD
Sbjct: 1236 NILEDYMNYRKYKYLRLDGSSTIMDRRDMVRDFQHR-SDIFVFLLSTRAGGLGINLTAAD 1294
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVI ++SDWNP DLQAM RAHR+GQ + V +YR + ++VEE IL RA +K + LV+
Sbjct: 1295 TVIFYESDWNPTLDLQAMDRAHRLGQTKDVTVYRLICKETVEEKILHRASQKNTVQQLVM 1354
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAI-LRFGAEELFKEERNDEESKKRLLSMDIDEI 1116
+ +G A D E + + +F L ++R +++K+ + + D
Sbjct: 1355 TGGHVQGDDFLGAADVVSLLMDDAEAAQLEQKFRELPLQVKDRQKKKTKRIRIDAEGDAT 1414
Query: 1117 LERAEKVEEKENGGEQAHELLSAFKVANFCNDEDDGSFWSRWIKADSVAQAENALAPRAA 1176
LE E V+ ++NG E E K +N K A A AP+ A
Sbjct: 1415 LEELEDVDRQDNGQEPLEEPEKP-KSSN---------------KKRRAASNPKARAPQKA 1458
Query: 1177 RNIKSYAEADQ-SERSKKRKKKENEPTERI 1205
+ + + Q ++R K++ K NE E +
Sbjct: 1459 KEEANGEDTPQRTKRVKRQTKSINESLEPV 1488
>At3g12810 Snf2-related CBP activator protein, putative
Length = 1048
Score = 197 bits (500), Expect = 6e-50
Identities = 109/308 (35%), Positives = 171/308 (55%), Gaps = 18/308 (5%)
Query: 555 HTIDEYKTREAAMSVQGKMVDFQRRQSKG---SLRKLDEQPEWLKGGKLRDYQLEGLNFL 611
H + E ++++ + + Q G S K+ + +L LR+YQ GL++L
Sbjct: 73 HEVAEDNDKDSSDKIADAAAAARSAQPTGFTYSTTKVRTKLPFLLKHSLREYQHIGLDWL 132
Query: 612 VNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKW 671
V ++ N +LADEMGLGKT+ ++++L L + I GP L+VVP S + NW EF KW
Sbjct: 133 VTMYEKKLNGILADEMGLGKTIMTIALLAHLACDKGIWGPHLIVVPTSVMLNWETEFLKW 192
Query: 672 LPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKIKWNY 731
P ++ Y G+ R++ +Q K F+ +TTY +V++D + + KW Y
Sbjct: 193 CPAFKILTYFGSAKERKLKRQGWM-------KLNSFHVCITTYRLVIQDSKMFKRKKWKY 245
Query: 732 LMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDE 791
L++DEAH +KN ++Q + L FN+K ++L+TGTPLQN + ELW+L+HFL F+S E
Sbjct: 246 LILDEAHLIKNWKSQRWQTLLNFNSKRRILLTGTPLQNDLMELWSLMHFLMPHVFQSHQE 305
Query: 792 FAQNYKN--------LSSFNENELSNLHMELRPHMLRRVIKDVEKSLPPKIERILRVDMS 843
F + N N+ + LH LRP +LRR+ +DVEK LP K E ++ +S
Sbjct: 306 FKDWFCNPIAGMVEGQEKINKEVIDRLHNVLRPFLLRRLKRDVEKQLPSKHEHVIFCRLS 365
Query: 844 PLQKQYYK 851
Q+ Y+
Sbjct: 366 KRQRNLYE 373
Score = 134 bits (336), Expect = 6e-31
Identities = 83/201 (41%), Positives = 115/201 (56%), Gaps = 14/201 (6%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+L +++L G+ + RLDGST E RQ M FN F F+LSTR+GG+GINL AD
Sbjct: 690 DVLEAFINLYGYTYMRLDGSTPPEERQTLMQRFNT-NPKIFLFILSTRSGGVGINLVGAD 748
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVI +DSDWNP D QA R HRIGQ V+IYR ++ ++EE+IL++A +K VLD+LVI
Sbjct: 749 TVIFYDSDWNPAMDQQAQDRCHRIGQTREVHIYRLISESTIEENILKKANQKRVLDNLVI 808
Query: 1058 Q--KLNAE--GKLEKKEAKKG-GSFFDKNELSAILRFGAE--------ELFKEERNDEES 1104
Q + N E KL+ E G + K+E GA+ E ++ DE
Sbjct: 809 QNGEYNTEFFKKLDPMELFSGHKALTTKDEKETSKHCGADIPLSNADVEAALKQAEDEAD 868
Query: 1105 KKRLLSMDIDEILERAEKVEE 1125
L ++ +E ++ E EE
Sbjct: 869 YMALKRVEQEEAVDNQEFTEE 889
>At2g44980 putative transcription activator
Length = 861
Score = 170 bits (431), Expect = 6e-42
Identities = 106/273 (38%), Positives = 156/273 (56%), Gaps = 15/273 (5%)
Query: 600 LRDYQLEGLNFLVNSWKNDTNVVLA-DEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPL 658
L+ +Q+EG+++L+ + NVVL D+MGLGKT+Q++S L +L+ Q + GPFLV+ PL
Sbjct: 51 LKPHQVEGVSWLIQKYLLGVNVVLELDQMGLGKTLQAISFLSYLKFRQGLPGPFLVLCPL 110
Query: 659 STLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVL 718
S W E ++ P+L V+ YVG + R ++ + + G + F+ LLTTY++ L
Sbjct: 111 SVTDGWVSEINRFTPNLEVLRYVGDKYCRLDMRKSMYDH----GHFLPFDVLLTTYDIAL 166
Query: 719 KDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSE-FNTKNKLLITGTPLQNSVEELWAL 777
D+ LS+I W Y ++DEA RLKN + LY L E F +LLITGTP+QN++ ELWAL
Sbjct: 167 VDQDFLSQIPWQYAIIDEAQRLKNPNSVLYNVLLEQFLIPRRLLITGTPIQNNLTELWAL 226
Query: 778 LHFLDSDKFKSKDEFAQNYK----NLSSFNENE-LSNLHMELRPHMLRR----VIKDVEK 828
+HF F + D+F +K L N+ E +L L MLRR +I+
Sbjct: 227 MHFCMPLVFGTLDQFLSAFKETGDGLDVSNDKETYKSLKFILGAFMLRRTKSLLIESGNL 286
Query: 829 SLPPKIERILRVDMSPLQKQYYKWILERNFRDL 861
LPP E + V + LQK+ Y IL + L
Sbjct: 287 VLPPLTELTVMVPLVSLQKKIYTSILRKELPGL 319
Score = 134 bits (336), Expect = 6e-31
Identities = 99/300 (33%), Positives = 157/300 (52%), Gaps = 46/300 (15%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAP----------GSDDFCFLLSTRAG 987
DIL +M LR + ++RLDGS ++E R A+ +F+A GS+ F F++STRAG
Sbjct: 398 DILQDFMELRRYSYERLDGSVRAEERFAAIKNFSAKTERGLDSEVDGSNAFVFMISTRAG 457
Query: 988 GLGINLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAK 1047
G+G+NL ADTVI ++ DWNPQ D QA+ RAHRIGQ V VT SVEE IL RA+
Sbjct: 458 GVGLNLVAADTVIFYEQDWNPQVDKQALQRAHRIGQISHVLSINLVTEHSVEEVILRRAE 517
Query: 1048 KKMVLDHLVIQKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKR 1107
+K+ L H V+ + EK+E D +L +++ FG + EE ++EES
Sbjct: 518 RKLQLSHNVV----GDNMEEKEE--------DGGDLRSLV-FGLQRFDPEEIHNEESD-- 562
Query: 1108 LLSMDIDEILERAEKV-----------EEKENGGEQAHELLSAFKVANFCNDEDDGSFWS 1156
++ + EI AEKV EE+ + LL A+ ++ D+ S+ S
Sbjct: 563 --NLKMVEISSLAEKVVAIRQNVEPDKEERRFEINSSDTLLGNTSSASLDSELDEASYLS 620
Query: 1157 RWIKADSVAQAENALAPRAARNIKSYAEADQSERSKKRKKKENEPTERIPKRRKADYSAH 1216
W++ + A R++++ K ++ S++R + ++ +++ A + AH
Sbjct: 621 -WVE-------KLKEAARSSKDEKIIELGNRKNLSEERNLRIEAARKKAEEKKLATWGAH 672
>At4g31900 putative protein
Length = 1067
Score = 166 bits (421), Expect = 8e-41
Identities = 173/655 (26%), Positives = 280/655 (42%), Gaps = 134/655 (20%)
Query: 939 ILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATADT 998
+L Y + + + ++R+DG RQ +D FNA S+ FCFLLSTRAGG+GINLATADT
Sbjct: 445 LLEDYFTFKNWNYERIDGKISGPERQVRIDRFNAENSNRFCFLLSTRAGGIGINLATADT 504
Query: 999 VIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVIQ 1058
VII+DSDWNP DLQAM+R HR+GQ V IYR + +VEE ++E K KM+L+HLV+
Sbjct: 505 VIIYDSDWNPHADLQAMARVHRLGQTNKVMIYRLIHKGTVEERMMEITKNKMLLEHLVV- 563
Query: 1059 KLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKEERNDEESKKRLLSMD---IDE 1115
G ++EL I+++G++ELF EE NDE + + D I++
Sbjct: 564 ---------------GKQHLCQDELDDIIKYGSKELFSEE-NDEAGRSGKIHYDDAAIEQ 607
Query: 1116 ILERAE-KVEEKENGGEQAHELLSAFKVANF--CNDEDDG-------------------- 1152
+L+R E E+ + L FKVA+F +DE++
Sbjct: 608 LLDRNHVDAVEVSLDDEEETDFLKNFKVASFEYVDDENEAAALEEAQAIENNSSVRNADR 667
Query: 1153 -SFWSRWIKAD-SVAQAENALA-PRAARNIKS--YA------------------------ 1183
S W +K V QAE A + RN K YA
Sbjct: 668 TSHWKDLLKDKYEVQQAEELSALGKRKRNGKQVMYAEDDLDGLEEISDEEDEYCLDDLKV 727
Query: 1184 ------EADQSERSKKRKKKENEPTERIPKRRKADYSAHVISMIDGASAQVRSWSYGNLS 1237
EAD+ E +++RK + T P R++A ++ I +++G + + N +
Sbjct: 728 TSDEEEEADEPEAARQRKPR----TVTRPYRKRARDNSEEIPLMEGEGRYLMVLGF-NET 782
Query: 1238 KRDALRFSRSVMKFGNESQINLIVAEVGGAIEAAPLKAQVELFNALIDGCREAVEVGSLD 1297
+RD F R+ ++G ++ + + A V +G
Sbjct: 783 ERDI--FLRTFKRYGILFLKHIAENPTDNSTNFKVITAMVYADGVPKEG----------- 829
Query: 1298 LKGPLLDFYGVPMKANELLIRVQELQLLAKRISRYED-PIAQFRVLTYLKPSNWSKGCGW 1356
+ ++ELL+ + + L+ ++ ++ P A + N G
Sbjct: 830 ------------ISSDELLVSMTFMMLVKEKCQFLDNHPTAPVFSNYVISKYNLRNGAFS 877
Query: 1357 NQIDDARLLLGVHYHGYGNWEVIRLDERLGLTKKIAPVELQHHETFLPRAPNLRDRANAL 1416
+ D L+ V HGYG W I DE +G +++A +L +P P+ +
Sbjct: 878 KEEHDRILIPAVSKHGYGRWVAIVEDEEIGF-QEVACKDLN-----IPFPPDTKSAR--- 928
Query: 1417 LEQELAVLGVKNASSKVGRKTSKKEREEREHLVDISLSRGQEKKKNIGSSKVNVQ---MR 1473
K VG++ K E + + L+ Q K + G+S V+ + ++
Sbjct: 929 ----------KRICDHVGKRVKKMEDAIKYEYAEKILAE-QAKAETKGTSFVDAEKEMLK 977
Query: 1474 KDRLQKPLNVEPIVKEEGEMSDDDDVYEQFKEGKWKEWCQDLMVEEMKTLKRLHR 1528
D + N V + + Y+Q K E Q + +++ L R+ R
Sbjct: 978 NDPITSKKNSATAVDNKQGRVEMAQSYDQSVNEKSGESFQTYL--DIQPLNRMPR 1030
Score = 165 bits (418), Expect = 2e-40
Identities = 118/343 (34%), Positives = 176/343 (50%), Gaps = 35/343 (10%)
Query: 357 RSSSRTIRKVSYVESDESEGADEGTKKSQKEEIEVDDGD----SVEKVLWHQPKGMAAEA 412
R RT K Y+E + DE ++ Q EE + D +EK+L + + A+
Sbjct: 7 RLRRRTGPKPDYIE----DKLDEYIREEQVEETGGSNQDCPLGEIEKILDREWRPTASNN 62
Query: 413 QRNNQSMEPVLMSHLFDSEPDWNNMEFLIKWKGQSHLHCQWKSFVDLQNLSGFKKVLNYT 472
++ + P L+ ++L+KWKG S+LHC W + + L
Sbjct: 63 PNSSDNGTPTLVVV----------KQYLVKWKGLSYLHCSWVPEQEFEKAYKSHPHLKLK 112
Query: 473 KRVTEEIRNRMGISREEIEVNDVSKEMDIDIIKQNSQVERIIADRISNDNSGNVFPEYLV 532
RVT R + + + + E I I + V+RIIA R +D EYLV
Sbjct: 113 LRVT-----RFNAAMDVFIAENGAHEF-IAIRPEWKTVDRIIACREGDDGE-----EYLV 161
Query: 533 KWQGLSYAEVTWEKDIDIAFAQHTIDEYKTREAAMSVQGKMVDFQRRQSKGSLRKLDEQP 592
K++ LSY WE + DI+ Q+ I +K ++ S + K V+ +R + + ++ D P
Sbjct: 162 KYKELSYRNSYWESESDISDFQNEIQRFKDINSS-SRRDKYVENERNREE--FKQFDLTP 218
Query: 593 EWLKGGKLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPF 652
E+L G L YQLEGLNFL SW TNV+LADEMGLGKT+QS++ L L ++ P
Sbjct: 219 EFLTG-TLHTYQLEGLNFLRYSWSKKTNVILADEMGLGKTIQSIAFLASL--FEENLSPH 275
Query: 653 LVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEF 695
LVV PLST+ NW +EF W P +NV++Y G +R+V ++EF
Sbjct: 276 LVVAPLSTIRNWEREFATWAPHMNVVMYTGDSEARDVIWEHEF 318
Score = 43.1 bits (100), Expect = 0.001
Identities = 25/61 (40%), Positives = 36/61 (58%), Gaps = 1/61 (1%)
Query: 820 RRVIKDVEKS-LPPKIERILRVDMSPLQKQYYKWILERNFRDLNKGVRGNQSKYCSRIEE 878
+R+ KDV K +PPK E ILRVDMS QK+ YK ++ N++ L K S ++ +
Sbjct: 325 KRLKKDVLKDKVPPKKELILRVDMSSQQKEVYKAVITNNYQVLTKKRDAKISNVLMKLRQ 384
Query: 879 V 879
V
Sbjct: 385 V 385
>At2g02090 putative helicase
Length = 763
Score = 161 bits (408), Expect = 3e-39
Identities = 100/267 (37%), Positives = 146/267 (54%), Gaps = 17/267 (6%)
Query: 600 LRDYQLEGLNFLVNSWKNDTN-VVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPL 658
L+ YQL G+NFL+ +K +LADEMGLGKT+Q+++ L L GP LVV P
Sbjct: 213 LKPYQLVGVNFLLLLYKKGIEGAILADEMGLGKTIQAITYLTLLSRLNNDPGPHLVVCPA 272
Query: 659 STLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVL 718
S L NW +E RKW P V+ Y G + E + KAGK FN LL Y +
Sbjct: 273 SVLENWERELRKWCPSFTVLQYHG---AARAAYSRELNSLSKAGKPPPFNVLLVCYSLFE 329
Query: 719 K-------DKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEF--NTKNKLLITGTPLQN 769
+ D+ VL + +W+ +++DEAH LK+ + + L N +L++TGTPLQN
Sbjct: 330 RHSEQQKDDRKVLKRWRWSCVLMDEAHALKDKNSYRWKNLMSVARNANQRLMLTGTPLQN 389
Query: 770 SVEELWALLHFLDSDKFKSKDEFAQNYKNLSSFNENEL-SNLHMELRPHMLRRVIKDVEK 828
+ ELW+LL F+ D F +++ + K L + + EL + + L P +LRR+ DV +
Sbjct: 390 DLHELWSLLEFMLPDIFTTEN---VDLKKLLNAEDTELITRMKSILGPFILRRLKSDVMQ 446
Query: 829 SLPPKIERILRVDMSPLQKQYYKWILE 855
L PKI+R+ V M Q+ YK +E
Sbjct: 447 QLVPKIQRVEYVLMERKQEDAYKEAIE 473
Score = 112 bits (281), Expect = 1e-24
Identities = 64/121 (52%), Positives = 81/121 (66%), Gaps = 1/121 (0%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
DIL + + G ++RLDGST+ RQ +D FN S C LLSTRAGG G+NL AD
Sbjct: 618 DILEWTLDVIGVTYRRLDGSTQVTDRQTIVDTFNNDKSIFAC-LLSTRAGGQGLNLTGAD 676
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
TVII D D+NPQ D QA R HRIGQ + V I+R VT +V+E+I E AK+K+VLD V+
Sbjct: 677 TVIIHDMDFNPQIDRQAEDRCHRIGQTKPVTIFRLVTKSTVDENIYEIAKRKLVLDAAVL 736
Query: 1058 Q 1058
+
Sbjct: 737 E 737
>At3g54280 TATA box binding protein (TBP) associated factor (TAF)
-like protein
Length = 2049
Score = 134 bits (336), Expect = 6e-31
Identities = 86/276 (31%), Positives = 154/276 (55%), Gaps = 35/276 (12%)
Query: 599 KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHG-------- 650
+LR YQ EG+N+L + + +L D+MGLGKT+Q+ +++ +A + G
Sbjct: 1443 QLRRYQQEGINWLGFLKRFKLHGILCDDMGLGKTLQASAIVA--SDAAERRGSTDELDVF 1500
Query: 651 PFLVVVPLSTLSNWAKEFRKW--LPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFN 708
P ++V P + + +WA E K+ L L+V+ YVG+ R V + +F N N
Sbjct: 1501 PSIIVCPSTLVGHWAFEIEKYIDLSLLSVLQYVGSAQDR-VSLREQFNNH---------N 1550
Query: 709 ALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQ 768
++T+Y+VV KD L++ WNY ++DE H +KN+++++ A+ + +++L+++GTP+Q
Sbjct: 1551 VIITSYDVVRKDVDYLTQFSWNYCILDEGHIIKNAKSKITAAVKQLKAQHRLILSGTPIQ 1610
Query: 769 NSVEELWALLHFLDSDKFKSKDEFAQNY-KNLSSFNENELS------------NLHMELR 815
N++ ELW+L FL ++ +F +Y K L + + + S LH ++
Sbjct: 1611 NNIMELWSLFDFLMPGFLGTERQFQASYGKPLLAARDPKCSAKDAEAGVLAMEALHKQVM 1670
Query: 816 PHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYK 851
P +LRR ++V LP KI + D+SP+Q + Y+
Sbjct: 1671 PFLLRRTKEEVLSDLPEKIIQDRYCDLSPVQLKLYE 1706
Score = 89.7 bits (221), Expect = 1e-17
Identities = 49/131 (37%), Positives = 74/131 (56%), Gaps = 12/131 (9%)
Query: 946 LRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATADTVIIFDSD 1005
++ + RLDGS E R + + FN+ + D LL+T GGLG+NL +ADT++ + D
Sbjct: 1848 MKSVTYMRLDGSVVPEKRFEIVKAFNSDPTIDV-LLLTTHVGGLGLNLTSADTLVFMEHD 1906
Query: 1006 WNPQND-----------LQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDH 1054
WNP D QAM RAHR+GQ+ VVN++R + ++EE ++ K K+ + +
Sbjct: 1907 WNPMRDHQFANIELNKLWQAMDRAHRLGQKRVVNVHRLIMRGTLEEKVMSLQKFKVSVAN 1966
Query: 1055 LVIQKLNAEGK 1065
VI NA K
Sbjct: 1967 TVINAENASMK 1977
>At1g03750 hypothetical protein
Length = 874
Score = 125 bits (315), Expect = 2e-28
Identities = 80/281 (28%), Positives = 142/281 (50%), Gaps = 34/281 (12%)
Query: 599 KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNA----------QQI 648
+L ++Q EG+ F+ N +KN+ +L D+MGLGKT+Q+++ L + +
Sbjct: 150 RLLEHQREGVKFMYNLYKNNHGGILGDDMGLGKTIQTIAFLAAVYGKDGDAGESCLLESD 209
Query: 649 HGPFLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFN 708
GP L++ P S + NW EF +W V VY G S+R++ + K G ++
Sbjct: 210 KGPVLIICPSSIIHNWESEFSRWASFFKVSVYHG--SNRDMILE----KLKARGVEV--- 260
Query: 709 ALLTTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQ 768
L+T+++ VLS I W ++ DEAHRLKN +++LY A E TK ++ +TGT +Q
Sbjct: 261 -LVTSFDTFRIQGPVLSGINWEIVIADEAHRLKNEKSKLYEACLEIKTKKRIGLTGTVMQ 319
Query: 769 NSVEELWALLHFLDSDKFKSKDEFAQNY-------------KNLSSFNENELSNLHMELR 815
N + EL+ L ++ +++ F Y + + +L LR
Sbjct: 320 NKISELFNLFEWVAPGSLGTREHFRDFYDEPLKLGQRATAPERFVQIADKRKQHLGSLLR 379
Query: 816 PHMLRRVIKD-VEKSLPPKIERILRVDMSPLQKQYYKWILE 855
+MLRR ++ + + K + ++ MS LQ++ Y+ +++
Sbjct: 380 KYMLRRTKEETIGHLMMGKEDNVVFCQMSQLQRRVYQRMIQ 420
Score = 120 bits (301), Expect = 7e-27
Identities = 87/279 (31%), Positives = 135/279 (48%), Gaps = 18/279 (6%)
Query: 931 SYSDFLSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLG 990
SYS + DIL +++ +G+ F RLDGST + LRQ +D FNA S FL+ST+AGGLG
Sbjct: 563 SYSVRMLDILEKFLIRKGYSFARLDGSTPTNLRQSLVDDFNASPSKQV-FLISTKAGGLG 621
Query: 991 INLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKM 1050
+NL +A+ V+IFD +WNP +DLQA R+ R GQ+ V ++R +++ S+EE + R K
Sbjct: 622 LNLVSANRVVIFDPNWNPSHDLQAQDRSFRYGQKRHVVVFRLLSAGSLEELVYTRQVYKQ 681
Query: 1051 VLDHLVI----QKLNAEGKLEKKEAKKGGSFFDKNEL-----SAILRFGAEELFKEERND 1101
L ++ + + EG + KE + G F + L + EL ++ D
Sbjct: 682 QLSNIAVAGKMETRYFEGVQDCKEFQ--GELFGISNLFRDLSDKLFTSDIVELHRDSNID 739
Query: 1102 EESKKRLLSMDIDEILERAEKVEEK--ENGGEQAHELLSAFKVANFCNDEDDGSFWSRWI 1159
E K+ LL + E EK EE E +L + +ED +
Sbjct: 740 ENKKRSLLETGVSE----DEKEEEVMCSYKPEMEKPILKDLGIVYAHRNEDIINIGETTT 795
Query: 1160 KADSVAQAENALAPRAARNIKSYAEADQSERSKKRKKKE 1198
+ A R + K +E + S + +K+E
Sbjct: 796 STSQRLNGDGNSADRKKKKRKGCSEEEDMSSSNREQKRE 834
>At2g18760 putative SNF2/RAD54 family DNA repair and recombination
protein
Length = 1187
Score = 115 bits (289), Expect = 2e-25
Identities = 80/295 (27%), Positives = 135/295 (45%), Gaps = 40/295 (13%)
Query: 599 KLRDYQLEGLNFLVNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPL 658
KL DYQ G+ +L ++ DEMGLGKT+Q +S LG L + +++ P +++ P+
Sbjct: 384 KLFDYQRVGVQWLWELHCQRAGGIIGDEMGLGKTIQVLSFLGSL-HFSKMYKPSIIICPV 442
Query: 659 STLSNWAKEFRKWLPDLNVIVY--------------VGTRSSREVCQQYEFCNEKKAGKQ 704
+ L W +E +KW PD +V + + S + + +E K+
Sbjct: 443 TLLRQWRREAQKWYPDFHVEILHDSAQDSGHGKGQGKASESDYDSESSVDSDHEPKSKNT 502
Query: 705 IKFNALL------------TTYEVVLKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALS 752
K+++LL TTYE + L I+W Y ++DE HR++N + +
Sbjct: 503 KKWDSLLNRVLNSESGLLITTYEQLRLQGEKLLNIEWGYAVLDEGHRIRNPNSDITLVCK 562
Query: 753 EFNTKNKLLITGTPLQNSVEELWALLHFLDSDKFKSKDEF---------AQNYKNLSSFN 803
+ T +++++TG P+QN + ELW+L F+ K F Y N S
Sbjct: 563 QLQTVHRIIMTGAPIQNKLTELWSLFDFVFPGKLGVLPVFEAEFSVPITVGGYANASPLQ 622
Query: 804 ENELSNLHMELR----PHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYKWIL 854
+ + LR P++LRR+ DV L K E +L ++ Q+ Y+ L
Sbjct: 623 VSTAYRCAVVLRDLIMPYLLRRMKADVNAHLTKKTEHVLFCSLTVEQRSTYRAFL 677
Score = 103 bits (257), Expect = 9e-22
Identities = 52/128 (40%), Positives = 78/128 (60%), Gaps = 1/128 (0%)
Query: 931 SYSDFLSDILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLG 990
S + + DIL ++ + ++R+DG T + R +D FN D F F+L+T+ GGLG
Sbjct: 750 SQTQQMLDILESFLVANEYSYRRMDGLTPVKQRMALIDEFNN-SEDMFVFVLTTKVGGLG 808
Query: 991 INLATADTVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKM 1050
NL A+ VIIFD DWNP ND+QA RA RIGQ++ V +YR +T ++EE + R K
Sbjct: 809 TNLTGANRVIIFDPDWNPSNDMQARERAWRIGQKKDVTVYRLITRGTIEEKVYHRQIYKH 868
Query: 1051 VLDHLVIQ 1058
L + +++
Sbjct: 869 FLTNKILK 876
>At5g63950 unknown protein
Length = 1090
Score = 108 bits (271), Expect = 2e-23
Identities = 96/333 (28%), Positives = 154/333 (45%), Gaps = 46/333 (13%)
Query: 559 EYKTREAAMSVQGKMVDFQRR--QSKGSLRKLDEQPEWLKGGK----LRDYQLEGLNFLV 612
E K + A S K + R + +GS+ + GK L +Q EGLN+L
Sbjct: 330 EMKINKPARSYNAKRHGYDERSLEDEGSITLTGLNLSYTLPGKIATMLYPHQREGLNWLW 389
Query: 613 NSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGPFLVVVPLSTLSNWAKEFRKWL 672
+ +L D+MGLGKT+Q S L L +++ I LVV P + L +W KE
Sbjct: 390 SLHTQGKGGILGDDMGLGKTMQICSFLAGLFHSKLIKRA-LVVAPKTLLPHWMKELATVG 448
Query: 673 PDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFNALLTTYEVVLKDKAVLSKI----- 727
Y GT + ++Y+ + GK I LLTTY++V + L
Sbjct: 449 LSQMTREYYGTSTK---AREYDL-HHILQGKGI----LLTTYDIVRNNTKALQGDDHYTD 500
Query: 728 -------KWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPLQNSVEELWALLHF 780
KW+Y+++DE H +KN Q +L E + ++++I+GTP+QN+++ELWAL +F
Sbjct: 501 EDDEDGNKWDYMILDEGHLIKNPNTQRAKSLLEIPSSHRIIISGTPIQNNLKELWALFNF 560
Query: 781 LDSDKFKSKDEFAQNYKN--LSSFNENE-----------LSNLHMELRPHMLRRVIKDV- 826
K+ F QNY++ L ++N NL ++P LRR+ +V
Sbjct: 561 SCPGLLGDKNWFKQNYEHYILRGTDKNATDREQRIGSTVAKNLREHIQPFFLRRLKSEVF 620
Query: 827 -----EKSLPPKIERILRVDMSPLQKQYYKWIL 854
L K E ++ + ++ Q+Q Y+ L
Sbjct: 621 GDDGATSKLSKKDEIVVWLRLTACQRQLYEAFL 653
Score = 85.9 bits (211), Expect = 2e-16
Identities = 51/160 (31%), Positives = 84/160 (51%), Gaps = 25/160 (15%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
+++ ++ G+ F R+DG+TK+ R + ++ F G FLL+++ GGLG+ L AD
Sbjct: 762 NLIQDSLTSNGYSFLRIDGTTKAPDRLKTVEEFQE-GHVAPIFLLTSQVGGLGLTLTKAD 820
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLVI 1057
VI+ D WNP D Q++ RA+RIGQ + V +YR +TS +VEE
Sbjct: 821 RVIVVDPAWNPSTDNQSVDRAYRIGQTKDVIVYRLMTSATVEE----------------- 863
Query: 1058 QKLNAEGKLEKKEAKKGGSFFDKNELSAILRFGAEELFKE 1097
K+ +K+ KGG F E +R+ +++ +E
Sbjct: 864 -------KIYRKQVYKGGLFKTATEHKEQIRYFSQQDLRE 896
>At3g19210 DNA repair protein, putative
Length = 688
Score = 107 bits (268), Expect = 5e-23
Identities = 81/289 (28%), Positives = 137/289 (47%), Gaps = 37/289 (12%)
Query: 600 LRDYQLEGLNFL------VNSWKNDTNVVLADEMGLGKTVQSVSMLGFLQNAQQIHGP-- 651
LR +Q EG+ F+ ++ N +LAD+MGLGKT+QS+++L L Q G
Sbjct: 76 LRPHQREGVQFMFDCVSGLHGSANINGCILADDMGLGKTLQSITLLYTLL-CQGFDGTPM 134
Query: 652 ---FLVVVPLSTLSNWAKEFRKWLPDLNVIVYVGTRSSREVCQQYEFCNEKKAGKQIKFN 708
++V P S +SNW E +KW+ D ++ + + +V + ++ Q+
Sbjct: 135 VKKAIIVTPTSLVSNWEAEIKKWVGDRIQLIALCESTRDDVLSGIDSFTRPRSALQV--- 191
Query: 709 ALLTTYEVV-LKDKAVLSKIKWNYLMVDEAHRLKNSEAQLYTALSEFNTKNKLLITGTPL 767
L+ +YE + + L+ DEAHRLKN + AL+ K ++L++GTP+
Sbjct: 192 -LIISYETFRMHSSKFCQSESCDLLICDEAHRLKNDQTLTNRALASLTCKRRVLLSGTPM 250
Query: 768 QNSVEELWALLHFLDSDKFKSKDEFAQNY----------------KNLSSFNENELSNLH 811
QN +EE +A+++F + F Y KNL++ ELS+
Sbjct: 251 QNDLEEFFAMVNFTNPGSLGDAAHFRHYYEAPIICGREPTATEEEKNLAADRSAELSS-- 308
Query: 812 MELRPHMLRRVIKDVEKSLPPKIERILRVDMSPLQKQYYK-WILERNFR 859
++ +LRR + LPPKI ++ M+ LQ Y +I +N +
Sbjct: 309 -KVNQFILRRTNALLSNHLPPKIIEVVCCKMTTLQSTLYNHFISSKNLK 356
Score = 107 bits (267), Expect = 6e-23
Identities = 51/119 (42%), Positives = 77/119 (63%)
Query: 938 DILAQYMSLRGFQFQRLDGSTKSELRQQAMDHFNAPGSDDFCFLLSTRAGGLGINLATAD 997
D+ AQ R + F RLDGST RQ+ ++ N P D+F FLLS++AGG G+NL A+
Sbjct: 461 DLFAQLCRERRYPFLRLDGSTTISKRQKLVNRLNDPTKDEFAFLLSSKAGGCGLNLIGAN 520
Query: 998 TVIIFDSDWNPQNDLQAMSRAHRIGQREVVNIYRFVTSKSVEEDILERAKKKMVLDHLV 1056
+++FD DWNP ND QA +R R GQ++ V +YRF+++ ++EE + +R K L ++
Sbjct: 521 RLVLFDPDWNPANDKQAAARVWRDGQKKRVYVYRFLSTGTIEEKVYQRQMSKEGLQKVI 579
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.133 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,676,519
Number of Sequences: 26719
Number of extensions: 1955566
Number of successful extensions: 19213
Number of sequences better than 10.0: 623
Number of HSP's better than 10.0 without gapping: 180
Number of HSP's successfully gapped in prelim test: 462
Number of HSP's that attempted gapping in prelim test: 11492
Number of HSP's gapped (non-prelim): 3694
length of query: 1730
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1617
effective length of database: 8,299,349
effective search space: 13420047333
effective search space used: 13420047333
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC141111.7


