

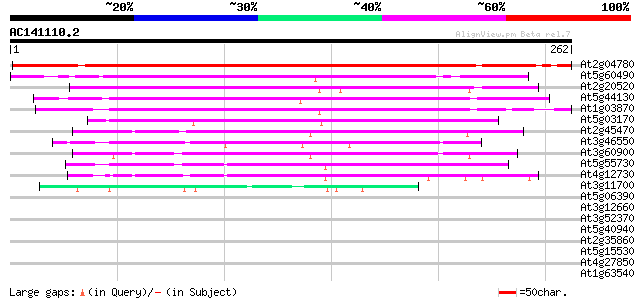
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141110.2 - phase: 0
(262 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g04780 GPI-anchored protein (Fla7) 286 6e-78
At5g60490 fasciclin-like arabinogalactan protein FLA12 145 3e-35
At2g20520 fasciclin-like arabinogalactan-protein 6 (Fla6) 129 2e-30
At5g44130 predicted GPI-anchored protein 126 1e-29
At1g03870 predicted GPI-anchored protein 124 7e-29
At5g03170 fasciclin-like arabinogalactan protein FLA11 120 1e-27
At2g45470 GPI-anchored protein (Fla8) 107 6e-24
At3g46550 predicted GPI-anchored protein 100 1e-21
At3g60900 GPI-anchored protein (Fla10) 97 9e-21
At5g55730 GPI-anchored protein (Fla1) 80 1e-15
At4g12730 fasciclin-like arabinogalactan protein FLA2 71 5e-13
At3g11700 unknown protein 43 1e-04
At5g06390 unknown protein 40 0.001
At3g12660 predicted GPI-anchored protein 40 0.002
At3g52370 unknown protein 37 0.011
At5g40940 unknown protein 37 0.014
At2g35860 unknown protein 35 0.031
At5g15530 biotin carboxyl carrier protein isoform 2 (BCCP2) 35 0.041
At4g27850 putative proline-rich protein 35 0.041
At1g63540 unknown protein 35 0.041
>At2g04780 GPI-anchored protein (Fla7)
Length = 254
Score = 286 bits (733), Expect = 6e-78
Identities = 148/261 (56%), Positives = 196/261 (74%), Gaps = 9/261 (3%)
Query: 2 KVSMIFLVSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHT 61
K+ + ++ + L+ SA AKT SPP+P P PAPAP P+ VNLTELL+VAGPFHT
Sbjct: 3 KMQLSIFIAVVALIVCSASAKTASPPAPVLP---PTPAPAPAPENVNLTELLSVAGPFHT 59
Query: 62 FLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHAL 121
FL YL ST V++TFQNQANNTEEGITIFVPKD +F + K P LS L D++KQ++LFHAL
Sbjct: 60 FLDYLLSTGVIETFQNQANNTEEGITIFVPKDDAFKAQKNPPLSNLTKDQLKQLVLFHAL 119
Query: 122 PHFYSLADFKNLSQTASTPTFAGGDYTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAI 181
PH+YSL++FKNLSQ+ TFAGG Y+L FTD SGTV+I+S W+ TKV+S+V +TDPVA+
Sbjct: 120 PHYYSLSEFKNLSQSGPVSTFAGGQYSLKFTDVSGTVRIDSLWTRTKVSSSVFSTDPVAV 179
Query: 182 YQVDKVLLPEAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSSYKI 241
YQV++VLLPEAIFGTD+PP+ APAP P ++ +DSP+ ADS+ +S S ++S K+
Sbjct: 180 YQVNRVLLPEAIFGTDVPPMPAPAPAPIVSAPSDSPS--VADSEGASSPKSSHKNSGQKL 237
Query: 242 VSYGIWGNLVLATFGLVVVIL 262
+ I ++V++ GLV + L
Sbjct: 238 LLAPI--SMVIS--GLVALFL 254
>At5g60490 fasciclin-like arabinogalactan protein FLA12
Length = 249
Score = 145 bits (365), Expect = 3e-35
Identities = 95/244 (38%), Positives = 137/244 (55%), Gaps = 18/244 (7%)
Query: 1 MKVSMIFLVSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFH 60
M+ S+I L+ T+LLL T +P P+PA APAP P N+T++L AG F
Sbjct: 1 MEHSLIILLFTVLLLL------TTTPGI--LSQPSPAVAPAP-PGPTNVTKILEKAGQFT 51
Query: 61 TFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHA 120
F++ L+ST V + Q NN++ GITIF P DSSF LK +L+ L D++ ++I FH
Sbjct: 52 VFIRLLKSTGVANQLYGQLNNSDNGITIFAPSDSSFTGLKAGTLNSLTDEQQVELIQFHV 111
Query: 121 LPHFYSLADFKNLSQTASTPT--FAGGDYTLNFTDNSGTVKINSGWSITKVTSAVHATDP 178
+P + S ++F+ +S T A G + LN T + TV I SG + T V+ V++
Sbjct: 112 IPSYVSSSNFQTISNPLRTQAGDSADGHFPLNVTTSGNTVNITSGVTNTTVSGNVYSDGQ 171
Query: 179 VAIYQVDKVLLPEAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSS 238
+A+YQVDKVLLP+ +F P APAP AP+ ++ DS SSS S D S +
Sbjct: 172 LAVYQVDKVLLPQQVFDPRPP---APAP----APSVSKSKKKKDDSDSSSDDSPADASFA 224
Query: 239 YKIV 242
+ V
Sbjct: 225 LRNV 228
>At2g20520 fasciclin-like arabinogalactan-protein 6 (Fla6)
Length = 247
Score = 129 bits (324), Expect = 2e-30
Identities = 81/224 (36%), Positives = 123/224 (54%), Gaps = 8/224 (3%)
Query: 29 PPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITI 88
P +S APAP +NLT +L F T +Q L +T+V Q N++++G+TI
Sbjct: 19 PYIQSQPTAPAPTTEKSPINLTAILEAGHQFTTLIQLLNTTQVGFQVSVQLNSSDQGMTI 78
Query: 89 FVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTASTPTFA--GGD 146
F P D++F LK +L+ L + Q++L+H +P +YSL+D S T GG
Sbjct: 79 FAPTDNAFNKLKPGTLNSLTYQQQIQLMLYHIIPKYYSLSDLLLASNPVRTQATGQDGGV 138
Query: 147 YTLNFTD--NSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAP 204
+ LNFT S V +++G T++ +A+ P+A+Y VD VLLPE +FGT P AP
Sbjct: 139 FGLNFTGQAQSNQVNVSTGVVETRINNALRQQFPLAVYVVDSVLLPEELFGTKTTPTGAP 198
Query: 205 APTPEIAPA-ADSPTEQSADSKSSSPSSSPDRSSSYKIVSYGIW 247
AP + + ADSP +AD + S SS R+S +VS+ ++
Sbjct: 199 APKSSTSSSDADSP---AADDEHKSAGSSVKRTSLGIVVSFALF 239
>At5g44130 predicted GPI-anchored protein
Length = 247
Score = 126 bits (316), Expect = 1e-29
Identities = 78/244 (31%), Positives = 131/244 (52%), Gaps = 13/244 (5%)
Query: 12 LLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKV 71
LLLL ++ F T A+ PAP PA +N+T +L G F T ++ L +T++
Sbjct: 7 LLLLLTAVFLST----EITAQRAAPAPGPAGP---INITAILEKGGQFVTLIRLLNTTQI 59
Query: 72 LDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFK 131
+ Q N++ EG+T+ P D++F +LK +L+KL D+ ++IL+H P FY+L D
Sbjct: 60 GNQINIQINSSSEGMTVLAPTDNAFQNLKPGTLNKLSPDDQVKLILYHVSPKFYTLEDLL 119
Query: 132 NLS---QTASTPTFAGGDYTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVL 188
++S +T ++ GG Y LNFT V +++G T++++++ P+A+Y VD VL
Sbjct: 120 SVSNPVRTQASGRDVGGVYGLNFTGQGNQVNVSTGVVETRLSTSLRQERPLAVYVVDMVL 179
Query: 189 LPEAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSSYKIVSYGIWG 248
LPE +FG +AP P + +D + + K+++PS S S G+
Sbjct: 180 LPEEMFGERKISPMAPPPKSKSPDVSD---DSESSKKAAAPSESEKSGSGEMNTGLGLGL 236
Query: 249 NLVL 252
LV+
Sbjct: 237 GLVV 240
>At1g03870 predicted GPI-anchored protein
Length = 247
Score = 124 bits (310), Expect = 7e-29
Identities = 79/252 (31%), Positives = 129/252 (50%), Gaps = 23/252 (9%)
Query: 13 LLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVL 72
LLL ++ T + P A +P PA +NLT +L G F TF+ L T+V
Sbjct: 11 LLLIAAVLLATKATAQPAAPAPEPAGP-------INLTAILEKGGQFTTFIHLLNITQVG 63
Query: 73 DTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKN 132
Q N++ EG+T+F P D++F +LK +L++L D+ ++IL+H P +YS+ D +
Sbjct: 64 SQVNIQVNSSSEGMTVFAPTDNAFQNLKPGTLNQLSPDDQVKLILYHVSPKYYSMDDLLS 123
Query: 133 LSQTASTPTFA--GGDYTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLP 190
+S T G Y LNFT + + +++G+ T++++++ P+A+Y VD VLLP
Sbjct: 124 VSNPVRTQASGRDNGVYGLNFTGQTNQINVSTGYVETRISNSLRQQRPLAVYVVDMVLLP 183
Query: 191 EAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSSYKIVSYGIWGNL 250
+FG +APAP + D + + K++SPS S K V G
Sbjct: 184 GEMFGEHKLSPIAPAPKSKSGGVTD---DSGSTKKAASPSDK--SGSGEKKVGLG----- 233
Query: 251 VLATFGLVVVIL 262
FGL +++L
Sbjct: 234 ----FGLGLIVL 241
>At5g03170 fasciclin-like arabinogalactan protein FLA11
Length = 246
Score = 120 bits (300), Expect = 1e-27
Identities = 68/195 (34%), Positives = 107/195 (54%), Gaps = 4/195 (2%)
Query: 37 APAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEE-GITIFVPKDSS 95
APAP P+ N+T +L AG F F++ L+ST+ D Q N++ G+T+F P D++
Sbjct: 26 APAPGPSGP-TNITAILEKAGQFTLFIRLLKSTQASDQINTQLNSSSSNGLTVFAPTDNA 84
Query: 96 FASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTASTPTFAG--GDYTLNFTD 153
F SLK +L+ L D + Q++ FH LP ++ F+ +S T G G + LN T
Sbjct: 85 FNSLKSGTLNSLSDQQKVQLVQFHVLPTLITMPQFQTVSNPLRTQAGDGQNGKFPLNITS 144
Query: 154 NSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPTPEIAPA 213
+ V I +G V ++V++ +A+YQVD+VLLP A+FG+ + P AP ++
Sbjct: 145 SGNQVNITTGVVSATVANSVYSDKQLAVYQVDQVLLPLAMFGSSVAPAPAPEKGGSVSKG 204
Query: 214 ADSPTEQSADSKSSS 228
+ S + DS SS
Sbjct: 205 SASGGDDGGDSTDSS 219
>At2g45470 GPI-anchored protein (Fla8)
Length = 420
Score = 107 bits (267), Expect = 6e-24
Identities = 73/223 (32%), Positives = 108/223 (47%), Gaps = 16/223 (7%)
Query: 30 PAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIF 89
P +P APAP+ N+T LL AG TF L S+ VL T+++ E+G+T+F
Sbjct: 171 PIIAPGVLTAPAPSASLSNITGLLEKAG-CKTFANLLVSSGVLKTYESAV---EKGLTVF 226
Query: 90 VPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTAST-PTFAGGDYT 148
P D +F + P L+KL E+ ++ +HAL + K ST T G +
Sbjct: 227 APSDEAFKAEGVPDLTKLTQAEVVSLLEYHALAEYKPKGSLKTNKNNISTLATNGAGKFD 286
Query: 149 LNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPTP 208
L + + V +++G + +++ V PV I+ VD VLLP +FG P APAP P
Sbjct: 287 LTTSTSGDEVILHTGVAPSRLADTVLDATPVVIFTVDNVLLPAELFGKSKSPSPAPAPEP 346
Query: 209 EIAP-----------AADSPTEQSADSKSSSPSSSPDRSSSYK 240
AP AA P + +S S+PS SP S++ K
Sbjct: 347 VTAPTPSPADAPSPTAASPPAPPTDESPESAPSDSPTGSANSK 389
Score = 33.9 bits (76), Expect = 0.091
Identities = 43/176 (24%), Positives = 76/176 (42%), Gaps = 12/176 (6%)
Query: 41 APTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLK 100
A T N+T++L + + +F YL TK+ D N+ IT+ V + + ++L
Sbjct: 20 ASTVSSHNITQILADSPDYSSFNSYLSQTKLADEI-----NSRTTITVLVLNNGAMSALA 74
Query: 101 -KPSLSKLKDDEIKQVILFHALPH-FYSLADFKNLSQT--ASTPTFAGGDYTLNFTD-NS 155
K LS +K V+L + P + ++ LS T +T G +N TD
Sbjct: 75 GKHPLSVIKSALSLLVLLDYYDPQKLHKISKGTTLSTTLYQTTGNAPGNLGFVNITDLKG 134
Query: 156 GTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPTPEIA 211
G V S S +K+ S+ + Y + + + I + + APAP+ ++
Sbjct: 135 GKVGFGSAASGSKLDSSYTKSVKQIPYNISILEIDAPIIAPGV--LTAPAPSASLS 188
Score = 33.9 bits (76), Expect = 0.091
Identities = 20/44 (45%), Positives = 28/44 (63%), Gaps = 5/44 (11%)
Query: 5 MIFLVSTLLLLSSSAFAKTVSP---PSP-PAESPTPAPAPAPTP 44
+IF V +LL + F K+ SP P+P P +PTP+PA AP+P
Sbjct: 318 VIFTVDNVLL-PAELFGKSKSPSPAPAPEPVTAPTPSPADAPSP 360
Score = 27.3 bits (59), Expect = 8.5
Identities = 12/20 (60%), Positives = 15/20 (75%), Gaps = 1/20 (5%)
Query: 25 SPPSPPA-ESPTPAPAPAPT 43
SPP+PP ESP AP+ +PT
Sbjct: 364 SPPAPPTDESPESAPSDSPT 383
>At3g46550 predicted GPI-anchored protein
Length = 420
Score = 100 bits (248), Expect = 1e-21
Identities = 70/206 (33%), Positives = 112/206 (53%), Gaps = 16/206 (7%)
Query: 21 AKTVSPPSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQAN 80
++T++PP P + S +P PA +NLT++L F+ L L ++ V+ F+N
Sbjct: 187 SETLTPP-PTSTSLSPPPAG------INLTQILINGHNFNVALSLLVASGVITEFENDER 239
Query: 81 NTEEGITIFVPKDSSFASL-KKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQ---- 135
GIT+FVP DS+F+ L +L L ++ V+ FH L +Y+L ++++
Sbjct: 240 GA--GITVFVPTDSAFSDLPSNVNLQSLPAEQKAFVLKFHVLHSYYTLGSLESITNPVQP 297
Query: 136 TASTPTFAGGDYTLNFTDNSGT-VKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIF 194
T +T G YTLN + +G+ V INSG + VT +PV+++ V KVLLP+ +F
Sbjct: 298 TLATEEMGAGSYTLNISRVNGSIVTINSGVVLAVVTQTAFDQNPVSVFGVSKVLLPKELF 357
Query: 195 GTDIPPVLAPAPTPEIAPAADSPTEQ 220
PV A AP EI+ + +S +EQ
Sbjct: 358 PKSGQPV-ATAPPQEISLSPESSSEQ 382
Score = 36.6 bits (83), Expect = 0.014
Identities = 35/178 (19%), Positives = 68/178 (37%), Gaps = 17/178 (9%)
Query: 47 VNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSK 106
+N+T +L+ +F L S+ + + +T+ +S F+S +
Sbjct: 29 INVTAVLSSFPNLSSFSNLLVSSGIAAELSGR-----NSLTLLAVPNSQFSSASLDLTRR 83
Query: 107 LKDDEIKQVILFHALPHFYSLADFKNLSQTASTPT----------FAGGDYTLNFTDNSG 156
L + ++ FH L F S +D + + + S T F G + SG
Sbjct: 84 LPPSALADLLRFHVLLQFLSDSDLRRIPPSGSAVTTLYEASGRTFFGSGSVNVTRDPASG 143
Query: 157 TVKINSGWSITKVTSAVHATDP--VAIYQVDKVLLPEAIFGTDIPPVLAPAPTPEIAP 212
+V I S + + T P + + VD +++P I T + P + ++P
Sbjct: 144 SVTIGSPATKNVTVLKLLETKPPNITVLTVDSLIVPTGIDITASETLTPPPTSTSLSP 201
>At3g60900 GPI-anchored protein (Fla10)
Length = 422
Score = 97.1 bits (240), Expect = 9e-21
Identities = 71/215 (33%), Positives = 104/215 (48%), Gaps = 13/215 (6%)
Query: 30 PAESPTPAPAPAPTPDFV-NLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITI 88
P +P APAP+ V N+T LL AG TF L S+ V+ TF++ E+G+T+
Sbjct: 171 PIIAPGILTAPAPSSAGVSNITGLLEKAG-CKTFANLLVSSGVIKTFES---TVEKGLTV 226
Query: 89 FVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTAST-PTFAGGDY 147
F P D +F + P L+ L E+ ++ +HAL + K ST T G Y
Sbjct: 227 FAPSDEAFKARGVPDLTNLTQAEVVSLLEYHALAEYKPKGSLKTNKDAISTLATNGAGKY 286
Query: 148 TLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPT 207
L + + V +++G +++ V PV I+ VD VLLP +FG P APAP
Sbjct: 287 DLTTSTSGDEVILHTGVGPSRLADTVVDETPVVIFTVDNVLLPAELFGKSSSP--APAPE 344
Query: 208 PEIAPA-----ADSPTEQSADSKSSSPSSSPDRSS 237
P AP + SP E + + +S P+ D SS
Sbjct: 345 PVSAPTPTPAKSPSPVEAPSPTAASPPAPPVDESS 379
Score = 33.5 bits (75), Expect = 0.12
Identities = 19/42 (45%), Positives = 27/42 (64%), Gaps = 3/42 (7%)
Query: 5 MIFLVSTLLLLSSSAFAKTVSP-PSP-PAESPTPAPAPAPTP 44
+IF V +LL + F K+ SP P+P P +PTP PA +P+P
Sbjct: 319 VIFTVDNVLL-PAELFGKSSSPAPAPEPVSAPTPTPAKSPSP 359
Score = 30.4 bits (67), Expect = 1.0
Identities = 42/165 (25%), Positives = 72/165 (43%), Gaps = 12/165 (7%)
Query: 48 NLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLK-KPSLSK 106
N+T++L+ + +F YL TK+ D N+ IT+ V + + +SL K LS
Sbjct: 27 NITQILSDTPEYSSFNNYLSQTKLADEI-----NSRTTITVLVLNNGAMSSLAGKHPLSV 81
Query: 107 LKDDEIKQVILFHALP-HFYSLADFKNLSQT--ASTPTFAGGDYTLNFTD-NSGTVKINS 162
+K+ V+L + P + L+ L+ T +T G +N TD G V S
Sbjct: 82 VKNALSLLVLLDYYDPLKLHQLSKGTTLTTTLYQTTGHALGNLGFVNVTDLKGGKVGFGS 141
Query: 163 GWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPT 207
+K+ S+ + Y + + + I I + APAP+
Sbjct: 142 AAPGSKLDSSYTKSVKQIPYNISVLEINAPIIAPGI--LTAPAPS 184
>At5g55730 GPI-anchored protein (Fla1)
Length = 424
Score = 79.7 bits (195), Expect = 1e-15
Identities = 59/208 (28%), Positives = 94/208 (44%), Gaps = 11/208 (5%)
Query: 27 PSPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGI 86
PS A +PTPAPA +NLT +++ G L + T+Q + E G+
Sbjct: 171 PSETAAAPTPAPAE------MNLTGIMSAHGCKVFAETLLTNPGASKTYQE---SLEGGM 221
Query: 87 TIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTASTPTFAGGD 146
T+F P D + P L + + + F A+P +YS+A K+ + +T G +
Sbjct: 222 TVFCPGDDAMKGFL-PKYKNLTAPKKEAFLDFLAVPTYYSMAMLKSNNGPMNTLATDGAN 280
Query: 147 -YTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPA 205
+ L ++ V + + + K+ + P+AIY DKVLLP+ +F APA
Sbjct: 281 KFELTVQNDGEKVTLKTRINTVKIVDTLIDEQPLAIYATDKVLLPKELFKASAVEAPAPA 340
Query: 206 PTPEIAPAADSPTEQSADSKSSSPSSSP 233
P PE ADSP +K ++P
Sbjct: 341 PAPEDGDVADSPKAAKGKAKGKKKKAAP 368
>At4g12730 fasciclin-like arabinogalactan protein FLA2
Length = 403
Score = 71.2 bits (173), Expect = 5e-13
Identities = 62/237 (26%), Positives = 110/237 (46%), Gaps = 28/237 (11%)
Query: 28 SPPAESPTPAPAPAPTPDFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGIT 87
SP AE+PT +P+ D + LT +L G F L+ST TFQ+ + G+T
Sbjct: 176 SPEAEAPTASPS-----DLI-LTTILEKQG-CKAFSDILKSTGADKTFQDTVDG---GLT 225
Query: 88 IFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLSQTASTPTFAGGD- 146
+F P DS+ P L ++L+H +P + SL ++ + +T G +
Sbjct: 226 VFCPSDSAVGKFM-PKFKSLSPANKTALVLYHGMPVYQSLQMLRSGNGAVNTLATEGNNK 284
Query: 147 YTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIF---GTDIPPVLA 203
+ ++ V + + KV + +P+ +Y++DKVLLP I+ T P +
Sbjct: 285 FDFTVQNDGEDVTLETDVVTAKVMGTLKDQEPLIVYKIDKVLLPREIYKAVKTSAPAPKS 344
Query: 204 PAPTPEIA------PAADSPTE---QSADSKSSSPSSSPDRSSSYKI----VSYGIW 247
P+ A P+AD+P++ + AD K+ + S+ R+S+ + +G+W
Sbjct: 345 SKKKPKNAEADADGPSADAPSDDDVEVADDKNGAVSAMITRTSNVVTAIVGLCFGVW 401
Score = 33.5 bits (75), Expect = 0.12
Identities = 27/113 (23%), Positives = 48/113 (41%), Gaps = 9/113 (7%)
Query: 48 NLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKL 107
N+T +L F TF YL +T + D N + IT+ +S+ +S+ S
Sbjct: 28 NITRILAKDPDFSTFNHYLSATHLADEI-----NRRQTITVLAVDNSAMSSILSNGYSLY 82
Query: 108 KDDEIKQVILFHALPHFYSLADFKNLSQTASTPTFAGGDYTLNFTDNSGTVKI 160
+I+ ++ H L ++ ++ ST T + T + T SG + I
Sbjct: 83 ---QIRNILSLHVLVDYFGTKKLHQITD-GSTSTASMFQSTGSATGTSGYINI 131
>At3g11700 unknown protein
Length = 462
Score = 43.1 bits (100), Expect = 1e-04
Identities = 55/204 (26%), Positives = 79/204 (37%), Gaps = 34/204 (16%)
Query: 15 LSSSAFAKTVSPPSPP--------AESPTPAPAPAPTPD--FVNLTELLTVAGPFHTFLQ 64
L SA A +V SPP A P+ APAPAP P + V HT L
Sbjct: 221 LKKSATAVSVPAGSPPVLPIESAMAPGPSLAPAPAPGPGGAHKHFNGDAQVKDFIHTLLH 280
Query: 65 YLQSTKVLDTFQNQAN-NTEEG--------ITIFVPKDSSFASLKKPSLSKLKDDEIKQV 115
Y ++ D N + TE G +T+ P D + L LS+ E Q+
Sbjct: 281 YGGYNEMADILVNLTSLATEMGRLVSEGYVLTVLAPNDEAMGKLTTDQLSEPGAPE--QI 338
Query: 116 ILFHALPHFYSLADFKNLSQTASTPTFAGGDY-TLNF------TDNSGTVKINSG-WSIT 167
+ +H +P + + N S F Y TL F + G+VK SG S
Sbjct: 339 MYYHIIPEYQTEESMYN-----SVRRFGKVKYETLRFPHKVGAKEADGSVKFGSGDRSAY 393
Query: 168 KVTSAVHATDPVAIYQVDKVLLPE 191
++ +++ +D VL PE
Sbjct: 394 LFDPDIYTDGRISVQGIDGVLFPE 417
>At5g06390 unknown protein
Length = 458
Score = 40.4 bits (93), Expect = 0.001
Identities = 48/190 (25%), Positives = 75/190 (39%), Gaps = 29/190 (15%)
Query: 25 SPPSPPAES-----PTPAPAPAPTPDFV--NLTELLTVAGPFHTFLQYLQSTKVLDTFQN 77
SPP+ P +S P+ APAPAP P + V HT L Y ++ D N
Sbjct: 231 SPPALPIQSAMAPGPSLAPAPAPGPGGKQHHFDGEAQVKDFIHTLLHYGGYNEMADILVN 290
Query: 78 QAN-NTEEG--------ITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLA 128
+ TE G +T+ P D + A L LS+ E Q++ +H +P + +
Sbjct: 291 LTSLATEMGRLVSEGYVLTVLAPNDEAMAKLTTDQLSEPGAPE--QIVYYHIIPEYQTEE 348
Query: 129 DFKNLSQTASTPTFAGGDYTLNF------TDNSGTVKINSG-WSITKVTSAVHATDPVAI 181
N + F TL F + G+VK G S ++ +++
Sbjct: 349 SMYNSVRRFGKVKFD----TLRFPHKVAAKEADGSVKFGDGEKSAYLFDPDIYTDGRISV 404
Query: 182 YQVDKVLLPE 191
+D VL P+
Sbjct: 405 QGIDGVLFPQ 414
>At3g12660 predicted GPI-anchored protein
Length = 255
Score = 39.7 bits (91), Expect = 0.002
Identities = 47/206 (22%), Positives = 79/206 (37%), Gaps = 26/206 (12%)
Query: 48 NLTELLTVAGPFHTFLQYLQSTKVLDTFQNQANNTEEGITIFVPKDSSFASLKKPSLSKL 107
N+T +L F F Q L T++ T N + IT+ V + + +SL S
Sbjct: 25 NITNILNEHDDFSNFNQLLSETQLASTI-----NKRQTITVLVVSNGALSSLSGQPTSV- 78
Query: 108 KDDEIKQVILFHALPHFYSLADFKNLSQTA--------STPTFAGGDYTLNFT-DNSGTV 158
IK+++ H + +Y KNLS+ S+ G LN T +G V
Sbjct: 79 ----IKKILSLHIVLDYYDQKKLKNLSKKTVLLTTLFQSSGLARGQQGFLNATVMKNGDV 134
Query: 159 KINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAIF----GTDIPPVLAPAPTPEIAPAA 214
S + + + + T + + + + AI G + P +P A
Sbjct: 135 AFGSAVPGSSLDAQLQDTVAALPFNISVLHISSAIMIDVKGDNAPTASPLSPVSSPPRPA 194
Query: 215 DSPTEQSAD---SKSSSPSSSPDRSS 237
+SP + D SS+P ++ D S
Sbjct: 195 ESPNDDGQDFDEPPSSAPGAAADEPS 220
>At3g52370 unknown protein
Length = 436
Score = 37.0 bits (84), Expect = 0.011
Identities = 49/206 (23%), Positives = 79/206 (37%), Gaps = 24/206 (11%)
Query: 25 SPPSPP---AESPTPAPAPAPTP----DFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQN 77
+PP P A SP P+ APAP P + V HT L Y ++ D N
Sbjct: 212 APPVLPVYDAMSPGPSLAPAPAPGPGGPRHHFNGEAQVKDFIHTLLHYGGYNEMADILVN 271
Query: 78 QAN-NTEEG--------ITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLA 128
+ TE G +T+ P D + A L LS+ E Q++ +H +P + +
Sbjct: 272 LTSLATEMGRLVSEGYVLTVLAPNDEAMAKLTTDQLSEPGAPE--QIMYYHIIPEYQTEE 329
Query: 129 DFKNLSQTASTPTFAG--GDYTLNFTDNSGTVKINSG-WSITKVTSAVHATDPVAIYQVD 185
N + + + + + G+VK G S ++ +++ +D
Sbjct: 330 SMYNSVRRFGKIRYDSLRFPHKVEAQEADGSVKFGHGDGSAYLFDPDIYTDGRISVQGID 389
Query: 186 KVLLPEAIFGTDIP---PVLAPAPTP 208
VL PE + PV+ AP P
Sbjct: 390 GVLFPEEKTPVEKKTGVPVVKKAPKP 415
>At5g40940 unknown protein
Length = 424
Score = 36.6 bits (83), Expect = 0.014
Identities = 47/195 (24%), Positives = 80/195 (40%), Gaps = 25/195 (12%)
Query: 77 NQANNTEEG--ITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLADFKNLS 134
NQ N E+ +T+F P D SF+ +PSL +K + P +NL
Sbjct: 92 NQDLNLEDWQELTLFAPSDQSFSKFGQPSLLDMK---------YQLSPTRLPGETLRNLP 142
Query: 135 QTASTPTFAGGDYTLNFTDNS---GTVKINSGWSITKVTSAVHATDPVAIYQVDKV---- 187
A PT +Y+L T++S G IN ++ S V V IY D+
Sbjct: 143 NGAKIPTLR-SNYSLTVTNSSRFGGKTSIN---NVVVQDSPVFDDGYVVIYGSDEFFTSP 198
Query: 188 --LLPEAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSSYKIVSYG 245
+ ++ + IP + + I P++ + T S + S S + P+RS +
Sbjct: 199 TKISDDSSSSSSIPSTTSSTGSIPI-PSSATQTPPSPNIASDSTRNLPNRSKPVNRFNIF 257
Query: 246 IWGNLVLATFGLVVV 260
+ +L + G V++
Sbjct: 258 ESASRLLMSRGFVII 272
>At2g35860 unknown protein
Length = 445
Score = 35.4 bits (80), Expect = 0.031
Identities = 49/209 (23%), Positives = 78/209 (36%), Gaps = 32/209 (15%)
Query: 25 SPPSPP---AESPTPAPAPAPTP----DFVNLTELLTVAGPFHTFLQYLQSTKVLDTFQN 77
+PP P A SP P+ APAP P + V HT L Y ++ D N
Sbjct: 220 APPVLPIYDAMSPGPSLAPAPAPGPGGPRGHFNGDAQVKDFIHTLLHYGGYNEMADILVN 279
Query: 78 QAN-NTEEG--------ITIFVPKDSSFASLKKPSLSKLKDDEIKQVILFHALPHFYSLA 128
+ TE G +T+ P D + A L LS+ E Q++ +H +P + +
Sbjct: 280 LTSLATEMGRLVSEGYVLTVLAPNDEAMAKLTTDQLSEPGAPE--QIMYYHIIPEYQTEE 337
Query: 129 DFKNLSQTASTPTFAG--GDYTLNFTDNSGTVKINSG-WSITKVTSAVHATDPVAIYQVD 185
N + + + + + G+VK G S ++ +++ +D
Sbjct: 338 SMYNAVRRFGKVKYDSLRFPHKVLAQEADGSVKFGHGDGSAYLFDPDIYTDGRISVQGID 397
Query: 186 KVLLPEAIFGTDIPPVLAPAPTPEIAPAA 214
VL P+ P EI PAA
Sbjct: 398 GVLFPK-----------EETPATEIKPAA 415
>At5g15530 biotin carboxyl carrier protein isoform 2 (BCCP2)
Length = 255
Score = 35.0 bits (79), Expect = 0.041
Identities = 23/75 (30%), Positives = 31/75 (40%), Gaps = 2/75 (2%)
Query: 166 ITKVTSAVHATDPVAIYQVDKVLLPEAIFGTDIPPVLAPAPTPEIAPAADSPTEQSADSK 225
I K + A P +Y ++ + P L P+PTP PA PT SA S
Sbjct: 119 IRKKEALQQAVPPAPVYHSMPPVMADFSMPPAQPVALPPSPTPTSTPATAKPT--SAPSS 176
Query: 226 SSSPSSSPDRSSSYK 240
S P SP + Y+
Sbjct: 177 SHPPLKSPMAGTFYR 191
>At4g27850 putative proline-rich protein
Length = 577
Score = 35.0 bits (79), Expect = 0.041
Identities = 16/43 (37%), Positives = 24/43 (55%)
Query: 3 VSMIFLVSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAPTPD 45
V+++ L+S ++L SS PP P P P+P+P P PD
Sbjct: 145 VAVLLLLSVIVLWSSDPPLPPPPPPYPSPLPPPPSPSPTPGPD 187
Score = 32.3 bits (72), Expect = 0.26
Identities = 19/41 (46%), Positives = 24/41 (58%), Gaps = 4/41 (9%)
Query: 8 LVSTLLLLSSSAFAKT---VSPPSPPAESP-TPAPAPAPTP 44
LV+ LLLLS + + PP PP SP P P+P+PTP
Sbjct: 144 LVAVLLLLSVIVLWSSDPPLPPPPPPYPSPLPPPPSPSPTP 184
Score = 30.4 bits (67), Expect = 1.0
Identities = 14/36 (38%), Positives = 19/36 (51%)
Query: 7 FLVSTLLLLSSSAFAKTVSPPSPPAESPTPAPAPAP 42
+L+ +LLL S + PP PP P P+P P P
Sbjct: 142 YLLVAVLLLLSVIVLWSSDPPLPPPPPPYPSPLPPP 177
Score = 29.3 bits (64), Expect = 2.2
Identities = 13/24 (54%), Positives = 16/24 (66%), Gaps = 3/24 (12%)
Query: 25 SPPSPPAESPTP---APAPAPTPD 45
SP PP+ SPTP +P P+P PD
Sbjct: 248 SPGPPPSPSPTPGPDSPLPSPGPD 271
Score = 29.3 bits (64), Expect = 2.2
Identities = 16/50 (32%), Positives = 23/50 (46%), Gaps = 5/50 (10%)
Query: 190 PEAIFGTDIPPVLAPAPTPE-----IAPAADSPTEQSADSKSSSPSSSPD 234
P + + +PP +P+PTP +P DSP S SP+ PD
Sbjct: 166 PPPPYPSPLPPPPSPSPTPGPDSPLPSPGPDSPLPLPGPPPSPSPTPGPD 215
Score = 28.9 bits (63), Expect = 2.9
Identities = 10/20 (50%), Positives = 14/20 (70%)
Query: 25 SPPSPPAESPTPAPAPAPTP 44
S P+P +SP P+P P P+P
Sbjct: 236 SSPTPGPDSPLPSPGPPPSP 255
Score = 28.9 bits (63), Expect = 2.9
Identities = 15/39 (38%), Positives = 19/39 (48%), Gaps = 3/39 (7%)
Query: 199 PPVLAPAPTPEI---APAADSPTEQSADSKSSSPSSSPD 234
PP +P P P+ +P DSP SSSP+ PD
Sbjct: 205 PPSPSPTPGPDSPLPSPGPDSPLPLPGPPPSSSPTPGPD 243
Score = 28.9 bits (63), Expect = 2.9
Identities = 15/41 (36%), Positives = 18/41 (43%), Gaps = 1/41 (2%)
Query: 195 GTDIP-PVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPD 234
G D P P+ P P+ P DSP S SP+ PD
Sbjct: 222 GPDSPLPLPGPPPSSSPTPGPDSPLPSPGPPPSPSPTPGPD 262
Score = 28.5 bits (62), Expect = 3.8
Identities = 12/24 (50%), Positives = 15/24 (62%), Gaps = 1/24 (4%)
Query: 23 TVSPPSP-PAESPTPAPAPAPTPD 45
T P SP P+ P P+P+P P PD
Sbjct: 239 TPGPDSPLPSPGPPPSPSPTPGPD 262
Score = 28.1 bits (61), Expect = 5.0
Identities = 10/18 (55%), Positives = 12/18 (66%)
Query: 27 PSPPAESPTPAPAPAPTP 44
PSP +SP P P P P+P
Sbjct: 191 PSPGPDSPLPLPGPPPSP 208
Score = 27.7 bits (60), Expect = 6.5
Identities = 12/23 (52%), Positives = 15/23 (65%), Gaps = 3/23 (13%)
Query: 26 PPSPPAESPTP---APAPAPTPD 45
P PP+ SPTP +P P+P PD
Sbjct: 202 PGPPPSPSPTPGPDSPLPSPGPD 224
Score = 27.3 bits (59), Expect = 8.5
Identities = 11/20 (55%), Positives = 14/20 (70%), Gaps = 1/20 (5%)
Query: 26 PPSPPAESPTPAP-APAPTP 44
P PP+ SPTP P +P P+P
Sbjct: 230 PGPPPSSSPTPGPDSPLPSP 249
>At1g63540 unknown protein
Length = 635
Score = 35.0 bits (79), Expect = 0.041
Identities = 34/105 (32%), Positives = 47/105 (44%), Gaps = 11/105 (10%)
Query: 134 SQTASTPTFAGGDYTLNFTDNSGTVKINSGWSITKVTSAVHATDPVAIYQVDKVLLPEAI 193
S A +PTF G L T SG S S + +S +H++ P + P AI
Sbjct: 106 SGPAPSPTF-GEPRILLATSGSGA----SATSTSSTSSPLHSSSPFSFGSA-----PAAI 155
Query: 194 FGTDIPPVLAPAPTPEIAPAADSPTEQSADSKSSSPSSSPDRSSS 238
P +PA +P + + T SA + SSS +SSP SSS
Sbjct: 156 TSVSSGPAQSPASSPRLWIDRFA-TSSSASATSSSSTSSPFHSSS 199
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.369
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,115,713
Number of Sequences: 26719
Number of extensions: 292967
Number of successful extensions: 4814
Number of sequences better than 10.0: 189
Number of HSP's better than 10.0 without gapping: 122
Number of HSP's successfully gapped in prelim test: 68
Number of HSP's that attempted gapping in prelim test: 2708
Number of HSP's gapped (non-prelim): 1583
length of query: 262
length of database: 11,318,596
effective HSP length: 97
effective length of query: 165
effective length of database: 8,726,853
effective search space: 1439930745
effective search space used: 1439930745
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC141110.2


