

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.8 + phase: 0
(672 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
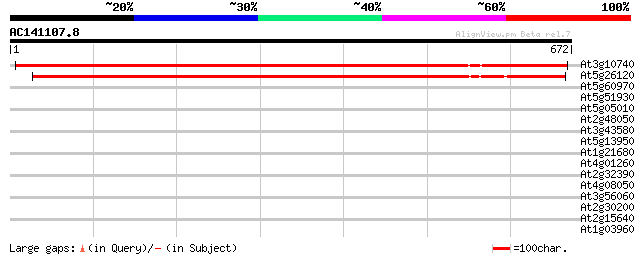
Sequences producing significant alignments: (bits) Value
At3g10740 putative alpha-L-arabinofuranosidase 969 0.0
At5g26120 arabinosidase - like protein 893 0.0
At5g60970 DNA binding protein - like 32 1.5
At5g51930 mandelonitrile lyase-like protein 32 1.5
At5g05010 coatomer delta subunit (delta-coat protein) (delta-COP) 32 1.5
At2g48050 hypothetical protein 32 1.5
At3g43580 hypothetical protein 31 2.0
At5g13950 unknown protein 31 2.6
At1g21680 unknown protein 31 2.6
At4g01260 hypothetical protein 30 4.4
At2g32390 ionotropic glutamate receptor ortholog GLR6 (GLR6) 29 7.5
At4g08050 29 9.8
At3g56060 mandelonitrile lyase-like protein 29 9.8
At2g30200 putative malonyl-CoA:Acyl carrier protein transacylase 29 9.8
At2g15640 hypothetical protein 29 9.8
At1g03960 protein phosphatase 2A like protein 29 9.8
>At3g10740 putative alpha-L-arabinofuranosidase
Length = 678
Score = 969 bits (2506), Expect = 0.0
Identities = 468/662 (70%), Positives = 549/662 (82%), Gaps = 4/662 (0%)
Query: 8 CCVFMLQLLIVVYLVVQCFDVQVQADLNATLVVDASQASGRRIPETLFGIFFEEINHAGA 67
C + L VY ++ D Q TL VDAS GR IPETLFGIFFEEINHAGA
Sbjct: 13 CVLSFLLGSCFVYQSLRVVDAQEDPKPAVTLQVDASNGGGRPIPETLFGIFFEEINHAGA 72
Query: 68 GGLWAELVSNRGFEAGGPNIPSNIDPWSIIGNATYINVETDRTSCFERNKVALRLEVLCD 127
GGLWAELVSNRGFEAGG N PSNI PWSI+G+ + I V TDR+SCFERNK+ALR++VLCD
Sbjct: 73 GGLWAELVSNRGFEAGGQNTPSNIWPWSIVGDHSSIYVATDRSSCFERNKIALRMDVLCD 132
Query: 128 GT-CPTDGVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSLASTVIT 186
CP+ GVGVYNPG+WGMNIE+GKKYKV Y RSTG ++L VSLT SNG +LAS I
Sbjct: 133 SKGCPSGGVGVYNPGYWGMNIEEGKKYKVALYVRSTGDIDLSVSLTSSNGSRTLASEKII 192
Query: 187 GSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGFRSDLL 246
SASD S W K E +LEAKAT+P++RLQLTTT KG IW+DQVSAMP+DT+KGHGFR+DL
Sbjct: 193 ASASDVSKWIKKEVLLEAKATDPSARLQLTTTKKGSIWIDQVSAMPVDTHKGHGFRNDLF 252
Query: 247 QMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDDGLGYYE 306
QM+ D+KP FIRFPGGCFVEG++L NAFRWK VGPWEERPGHFGDVWKYWTDDGLG++E
Sbjct: 253 QMMADIKPRFIRFPGGCFVEGEWLSNAFRWKETVGPWEERPGHFGDVWKYWTDDGLGHFE 312
Query: 307 FLQLSEDLGALPIWVFNNGVSHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWGSMRAAM 366
F Q++ED+GA PIWVFNNG+SHNDEV+T++++PFVQEALDG+EFARGD S WGS+RA M
Sbjct: 313 FFQMAEDIGAAPIWVFNNGISHNDEVETASIMPFVQEALDGIEFARGDANSTWGSVRAKM 372
Query: 367 GHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPLDHPADM 426
G EPF LKYVA+GNEDCGK YRGNY+ FYDAI++AYPDI+IISNCDGSS PLDHPAD
Sbjct: 373 GRQEPFELKYVAIGNEDCGKTYYRGNYIVFYDAIKKAYPDIKIISNCDGSSHPLDHPADY 432
Query: 427 YDYHIYTNANDMFSRSTTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAGFLIGLE 486
YDYHIYT+A+++FS F+R +R GPKAFVSEYAVTG DAG GSLLA+LAEA FLIGLE
Sbjct: 433 YDYHIYTSASNLFSMYHQFDRTSRKGPKAFVSEYAVTGKDAGTGSLLASLAEAAFLIGLE 492
Query: 487 KNSDIVHMASYAPLFVNANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSL 546
KNSDIV MASYAPLFVN NDRRWNPDAIVFNS LYGTPSYW+Q FF+ES+GATLL S+L
Sbjct: 493 KNSDIVEMASYAPLFVNTNDRRWNPDAIVFNSSHLYGTPSYWVQRFFAESSGATLLTSTL 552
Query: 547 QTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGSTKTV 606
+ S SLVASAI+W+N + K+YIRIKAVNFG ++ N+++ GLDPN ++ SGS KTV
Sbjct: 553 KGN-STSLVASAISWKN--NGKDYIRIKAVNFGANSENMQVLVTGLDPNVMRVSGSKKTV 609
Query: 607 LTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLLKESSNLKMLESDS 666
LTSTN+MDENSFSQP+KV+P +SLL+ +DM V++PPHSF+SFDLLKES+ ++M SDS
Sbjct: 610 LTSTNVMDENSFSQPEKVVPHESLLELAEEDMTVVLPPHSFSSFDLLKESAKIRMPISDS 669
Query: 667 SS 668
SS
Sbjct: 670 SS 671
>At5g26120 arabinosidase - like protein
Length = 674
Score = 893 bits (2308), Expect = 0.0
Identities = 444/642 (69%), Positives = 516/642 (80%), Gaps = 9/642 (1%)
Query: 28 VQVQADLNATLVVDASQASGRRIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGPNI 87
V Q D TL VDAS + R IPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGG I
Sbjct: 32 VDAQEDAIVTLQVDASNVTRRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQII 91
Query: 88 PSNIDPWSIIGNATYINVETDRTSCFERNKVALRLEVLCDG-TCPTDGVGVYNPGFWGMN 146
PSNI PWSIIG+ + I V TDR+SCFERNK+ALR+EVLCD +CP GVGVYNPG+WGMN
Sbjct: 92 PSNIWPWSIIGDESSIYVVTDRSSCFERNKIALRMEVLCDSNSCPLGGVGVYNPGYWGMN 151
Query: 147 IEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSLASTVITGSASDFSNWTKVETVLEAKA 206
IE+GKKYKVV Y RSTG +++ VS T SNG +LAS I ASD NWTK E +LEA
Sbjct: 152 IEEGKKYKVVLYVRSTGDIDVSVSFTSSNGSVTLASENIIALASDLLNWTKKEMLLEANG 211
Query: 207 TNPNSRLQLTTTTKGVIWLDQVSAMPLDTYKGHGFRSDLLQMLVDLKPSFIRFPGGCFVE 266
T+ +RLQ TTT KG IW DQVSAMP+DTYKGHGFR+DL QM+VDLKP FIRFPGGCFVE
Sbjct: 212 TDNGARLQFTTTKKGSIWFDQVSAMPMDTYKGHGFRNDLFQMMVDLKPRFIRFPGGCFVE 271
Query: 267 GDYLRNAFRWKAAVGPWEERPGHFGDVWKYWTDDGLGYYEFLQLSEDLGALPIWVFNNGV 326
GD+L NAFRWK V WEERPGH+GDVWKYWTDDGLG++EF QL+EDLGA PIWVFNNG+
Sbjct: 272 GDWLGNAFRWKETVRAWEERPGHYGDVWKYWTDDGLGHFEFFQLAEDLGASPIWVFNNGI 331
Query: 327 SHNDEVDTSAVLPFVQEALDGLEFARGDPTSKWGSMRAAMGHPEPFNLKYVAVGNEDCGK 386
SHND+V+T V+PFVQEA+DG+EFARGD S WGS+RAAMGHPEPF LKYVAVGNEDC K
Sbjct: 332 SHNDQVETKNVMPFVQEAIDGIEFARGDSNSTWGSVRAAMGHPEPFELKYVAVGNEDCFK 391
Query: 387 KNYRGNYLRFYDAIRRAYPDIQIISNCDGSSRPLDHPADMYDYHIYTNANDMFSRSTTFN 446
YRGNYL FY+AI++AYPDI+IISNCD S++PLDHPAD +DYHIYT A D+FS+S F+
Sbjct: 392 SYYRGNYLEFYNAIKKAYPDIKIISNCDASAKPLDHPADYFDYHIYTLARDLFSKSHDFD 451
Query: 447 RVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAGFLIGLEKNSDIVHMASYAPLFVNAND 506
R+GPKAFVSEYAV DA G+LLAAL EA FL+GLEKNSDIV M SYAPLFVN ND
Sbjct: 452 NTPRNGPKAFVSEYAVNKADAKNGNLLAALGEAAFLLGLEKNSDIVEMVSYAPLFVNTND 511
Query: 507 RRWNPDAIVFNSFQLYGTPSYWMQLFFSESNGATLLNSSLQTTASNSLVASAITWQNSVD 566
RRW PDAIVFNS LYGTPSYW+Q FF+ES+GATLLNS+L+ S S+ ASAI++Q +
Sbjct: 512 RRWIPDAIVFNSSHLYGTPSYWVQHFFTESSGATLLNSTLKGKTS-SVEASAISFQ--TN 568
Query: 567 KKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIP 626
K+YI+IKAVNFG +VNLK++ GL + GS K VLTS ++MDENSFS P ++P
Sbjct: 569 GKDYIQIKAVNFGEQSVNLKVAVTGL---MAKFYGSKKKVLTSASVMDENSFSNPNMIVP 625
Query: 627 IQSLLQ-SVGKDMNVIVPPHSFTSFDLLKESSN-LKMLESDS 666
+SLL+ + +D+ ++PPHSF+SFDLL ES N +KM SDS
Sbjct: 626 QESLLEMTEQEDLMFVLPPHSFSSFDLLTESENVIKMPISDS 667
>At5g60970 DNA binding protein - like
Length = 360
Score = 31.6 bits (70), Expect = 1.5
Identities = 35/133 (26%), Positives = 55/133 (41%), Gaps = 8/133 (6%)
Query: 542 LNSSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSG 601
LN LQ +S V+S+ W ++ +R+ G + + GL ++ S
Sbjct: 27 LNHMLQQQQPSS-VSSSRQWTSAFRNPRIVRVSRTFGGKDRHSKVCTVRGLRDRRIRLS- 84
Query: 602 STKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVP---PHSFTSFDLLKESSN 658
T + +L D SQP KV I LL++ D++ + P PH F N
Sbjct: 85 -VPTAIQLYDLQDRLGLSQPSKV--IDWLLEAAKDDVDKLPPLQFPHGFNQMYPNLIFGN 141
Query: 659 LKMLESDSSSWSS 671
ES SS+ S+
Sbjct: 142 SGFGESPSSTTST 154
>At5g51930 mandelonitrile lyase-like protein
Length = 586
Score = 31.6 bits (70), Expect = 1.5
Identities = 28/126 (22%), Positives = 45/126 (35%), Gaps = 23/126 (18%)
Query: 443 TTFNRVTRSGPKAFVSEYAVTGNDAGQGSLLAALAEAGFLIGLEKNSDIVHMASYAPLFV 502
TTF + PK +Y + G +L A L++ ++ LE+ + +
Sbjct: 58 TTFMKDATLAPKNASFDYIIIGGGTAGCALAATLSQNASVLVLERGGSPYENPTATDMGN 117
Query: 503 NANDRRWNPDAIVFNSFQLYGTPSYWMQLFFSES----------NGATLLNSSLQTTASN 552
+ N L TP+ W QLF SE G +++N + A N
Sbjct: 118 SVNTL-------------LNNTPNSWSQLFISEDGVYNTRPRVLGGGSVINGGFYSRAGN 164
Query: 553 SLVASA 558
V A
Sbjct: 165 DYVEEA 170
>At5g05010 coatomer delta subunit (delta-coat protein) (delta-COP)
Length = 527
Score = 31.6 bits (70), Expect = 1.5
Identities = 23/80 (28%), Positives = 38/80 (46%), Gaps = 5/80 (6%)
Query: 66 GAGGLWAELVSNRGFEAGGPNIPSNIDPW-SIIGNATYINVETDRTSCFERNKVALRLEV 124
G GG L+ R A +P I+ W S+ GN TY+++E + +S F+ V + + +
Sbjct: 367 GQGGDGVGLLRWRMQRADESMVPLTINCWPSVSGNETYVSLEYEASSMFDLTNVIISVPL 426
Query: 125 LCDGTCPT----DGVGVYNP 140
P+ DG Y+P
Sbjct: 427 PALREAPSVRQCDGEWRYDP 446
>At2g48050 hypothetical protein
Length = 1500
Score = 31.6 bits (70), Expect = 1.5
Identities = 22/54 (40%), Positives = 29/54 (52%), Gaps = 7/54 (12%)
Query: 134 GVGVYNPGFWGMNIEQGKKYKVVFYARSTGPLNLKVSLTGSNGVGSLASTVITG 187
G+ VY PGFWG IE G + KV+ A T N+ L ++G+ TVI G
Sbjct: 256 GLRVYEPGFWG--IESGLRGKVLVVAACTLQYNVFRWLERTSGL-----TVIKG 302
>At3g43580 hypothetical protein
Length = 204
Score = 31.2 bits (69), Expect = 2.0
Identities = 30/88 (34%), Positives = 43/88 (48%), Gaps = 13/88 (14%)
Query: 176 GVGSLASTV--ITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKGVIWLDQVSAMPL 233
G+ +L S++ I S S FS W ET+ E K SR+ L T ++ L + A
Sbjct: 87 GISALPSSLSRIASSLSVFSKWFTPETI-ETKEHFSKSRILLIVT---IVKLPEFFAKVS 142
Query: 234 DTYKGHGF-----RSDLLQMLVDLKPSF 256
T+ H F RS L++L+DL P F
Sbjct: 143 LTH--HDFACDMLRSSCLRVLMDLSPMF 168
>At5g13950 unknown protein
Length = 939
Score = 30.8 bits (68), Expect = 2.6
Identities = 20/68 (29%), Positives = 32/68 (46%), Gaps = 11/68 (16%)
Query: 341 VQEALDGLEFARGDPTSKWG-SMRAAMGHPEPFNLKYVAVGNEDCGKKNYRGNYLRFYDA 399
VQ LDG F G+P+ WG ++ + HP+ V E+C R + R+Y
Sbjct: 134 VQALLDGENFHFGNPSLDWGTAVCSGKAHPDQI------VSREEC----LRADKRRYYSN 183
Query: 400 IRRAYPDI 407
+ + + DI
Sbjct: 184 LEKYHQDI 191
>At1g21680 unknown protein
Length = 706
Score = 30.8 bits (68), Expect = 2.6
Identities = 25/76 (32%), Positives = 41/76 (53%), Gaps = 12/76 (15%)
Query: 162 TGPLNLKVSLTGSNGVGSLASTVITGSASDFSNWTKVETVLEAKATNPNSRLQLTTTTKG 221
+ PL+L + +T NG SL ++ G SDF TK +VLEA SR+Q+ ++
Sbjct: 99 SSPLHL-IYVTERNGTSSLYYDLVYGGNSDFK--TKRRSVLEAP-----SRVQVPLLSR- 149
Query: 222 VIWLDQVSAMPLDTYK 237
D +S M ++++K
Sbjct: 150 ---FDHLSGMTVNSFK 162
>At4g01260 hypothetical protein
Length = 325
Score = 30.0 bits (66), Expect = 4.4
Identities = 26/110 (23%), Positives = 42/110 (37%), Gaps = 2/110 (1%)
Query: 533 FSESNGATLLNSSLQTTASNSLVASAIT-WQNSVDKKNYIRIKAVNFGTSAVNLKISFNG 591
F+E + A LL L + I + S+ + T NLK FNG
Sbjct: 111 FTEEDEAILLQGFLDFATKKENPSDHIDDFYESIKNSISFDVTKPQLVTKIGNLKKKFNG 170
Query: 592 LDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVI 641
L+ G + V+ + D+N F +K+ +L S K+M +
Sbjct: 171 RVSKGLKK-GKNEEVMVFSKASDQNCFDLSRKIWGSNGVLYSKSKNMRQV 219
>At2g32390 ionotropic glutamate receptor ortholog GLR6 (GLR6)
Length = 895
Score = 29.3 bits (64), Expect = 7.5
Identities = 33/148 (22%), Positives = 59/148 (39%), Gaps = 16/148 (10%)
Query: 516 FNSFQLYGTPSYW-----MQLFFSESNGATLLNS-SLQTTASNSLVASAITWQNSVDKKN 569
FNS+ LY S W + +FFS+ N T N SL+ T + + S + N ++
Sbjct: 273 FNSYALYAYDSVWLVARALDVFFSQGNTVTFSNDPSLRNTNDSGIKLSKLHIFNEGERFL 332
Query: 570 YIRIKAVNFG-TSAVNLKISFNGLDPN----SLQSSGSTKTVLTSTN-----LMDENSFS 619
+ ++ G T + N ++P +++S+G + S + E +S
Sbjct: 333 QVILEMNYTGLTGQIEFNSEKNRINPAYDILNIKSTGPLRVGYWSNHTGFSVAPPETLYS 392
Query: 620 QPKKVIPIQSLLQSVGKDMNVIVPPHSF 647
+P L + VI PP +
Sbjct: 393 KPSNTSAKDQRLNEIIWPGEVIKPPRGW 420
>At4g08050
Length = 1428
Score = 28.9 bits (63), Expect = 9.8
Identities = 27/95 (28%), Positives = 39/95 (40%), Gaps = 16/95 (16%)
Query: 388 NYRGNYLRFYDAIRRAYPDIQ---IISNCDG-SSRPLDHPADMYD--------YHIYTNA 435
N+ + D IR Y D+ I+SN G SS D+ D + H+Y +
Sbjct: 777 NFSRENAEYDDDIREDYADLLYSLIMSNYSGESSMDPDYNVDEVESWSARPREQHVYQSF 836
Query: 436 NDMFSRSTTFNRVTRS----GPKAFVSEYAVTGND 466
D F RST + R+ G +A S Y + D
Sbjct: 837 RDEFERSTARHNQRRAEIARGKRAMSSRYELIDED 871
>At3g56060 mandelonitrile lyase-like protein
Length = 577
Score = 28.9 bits (63), Expect = 9.8
Identities = 28/116 (24%), Positives = 43/116 (36%), Gaps = 23/116 (19%)
Query: 453 PKAFVSEYAVTGNDAGQGSLLAALAEAGFLIGLEKNSDIVHMASYAPLFVNANDRRWNPD 512
PK +Y + G +L A L++ ++ LE+ + D D
Sbjct: 41 PKLSHFDYIIIGGGTAGCALAATLSQNATVLVLERGG-------------SPYDDPAATD 87
Query: 513 AIVFNSFQLYGTPSYWMQLFFSES----------NGATLLNSSLQTTASNSLVASA 558
F + L TP+ W QLF SE G T++N+ + A VA A
Sbjct: 88 IGNFANTLLNITPNSWSQLFISEDGVFNSRARVLGGGTVINAGFYSRAEEDFVAEA 143
>At2g30200 putative malonyl-CoA:Acyl carrier protein transacylase
Length = 393
Score = 28.9 bits (63), Expect = 9.8
Identities = 31/116 (26%), Positives = 49/116 (41%), Gaps = 16/116 (13%)
Query: 544 SSLQTTASNSLVASAITWQNSVDKKNYIRIKAVNFGTSAVNLKISFNGLDPNSLQSSGST 603
S++ TTAS+SL+ +I+ N KN +FG +A NL S + S+GS
Sbjct: 25 SAMATTASSSLLLPSISLNNLSSSKN------ASFGFAAKNLSRSRISMS----VSAGSQ 74
Query: 604 KTVLTSTNLMDENSFS------QPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFDLL 653
T + + D S Q + + + QSVG + + +DLL
Sbjct: 75 STTVHDSLFADYKPTSAFLFPGQGAQAVGMGKESQSVGAAGELYKKANDILGYDLL 130
>At2g15640 hypothetical protein
Length = 426
Score = 28.9 bits (63), Expect = 9.8
Identities = 23/87 (26%), Positives = 38/87 (43%), Gaps = 7/87 (8%)
Query: 238 GHGFRSDLLQMLVDLKPSFIRFPGGCFVEGDYLRNAFRWKAAVGPWEERPGHFGDVWKYW 297
GHG SD + + D++ F VE ++ K AV WE+ D++K +
Sbjct: 236 GHGIVSDYVIVCFDIRSEKFTFID---VERFCRLINYKGKLAVIYWEDDV----DIYKLY 288
Query: 298 TDDGLGYYEFLQLSEDLGALPIWVFNN 324
D Y E+ +D+ L +WV +
Sbjct: 289 YSDVDEYVEYNINDDDINELRVWVLED 315
>At1g03960 protein phosphatase 2A like protein
Length = 529
Score = 28.9 bits (63), Expect = 9.8
Identities = 23/79 (29%), Positives = 38/79 (47%)
Query: 592 LDPNSLQSSGSTKTVLTSTNLMDENSFSQPKKVIPIQSLLQSVGKDMNVIVPPHSFTSFD 651
LDP LQ G + + L T+L+ ++ FSQ + +L++S+ D P + +
Sbjct: 13 LDPELLQLPGLSPSPLKPTSLIADDLFSQWLSLPETATLVKSLIDDAKSGTPTNKSKNLP 72
Query: 652 LLKESSNLKMLESDSSSWS 670
+ SS+ L SSS S
Sbjct: 73 SVFLSSSTPPLSPRSSSGS 91
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,699,798
Number of Sequences: 26719
Number of extensions: 700103
Number of successful extensions: 1534
Number of sequences better than 10.0: 16
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1523
Number of HSP's gapped (non-prelim): 17
length of query: 672
length of database: 11,318,596
effective HSP length: 106
effective length of query: 566
effective length of database: 8,486,382
effective search space: 4803292212
effective search space used: 4803292212
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC141107.8


