

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC141107.11 + phase: 0 /pseudo
(650 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
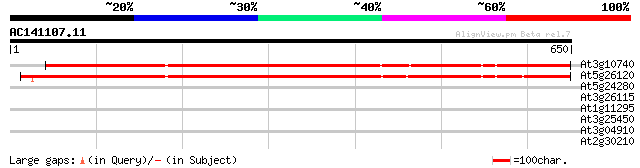
Score E
Sequences producing significant alignments: (bits) Value
At3g10740 putative alpha-L-arabinofuranosidase 746 0.0
At5g26120 arabinosidase - like protein 738 0.0
At5g24280 putative protein 36 0.059
At3g26115 unknown protein 30 4.2
At1g11295 putative s-locus protein kinase (PPC:1.7.2) 29 7.2
At3g25450 hypothetical protein 29 9.4
At3g04910 putative mitogen activated protein kinase kinase 29 9.4
At2g30210 putative laccase 29 9.4
>At3g10740 putative alpha-L-arabinofuranosidase
Length = 678
Score = 746 bits (1927), Expect = 0.0
Identities = 365/612 (59%), Positives = 452/612 (73%), Gaps = 12/612 (1%)
Query: 42 GRPMPNTLFGIFYEEINHAGSGGIWAQLVNNSGFEAAGTRTPSNIFPWTIIGTESSVKLQ 101
GRP+P TLFGIF+EEINHAG+GG+WA+LV+N GFEA G TPSNI+PW+I+G SS+ +
Sbjct: 52 GRPIPETLFGIFFEEINHAGAGGLWAELVSNRGFEAGGQNTPSNIWPWSIVGDHSSIYVA 111
Query: 102 TELSSCFERNKVALRMDVLCDK--CPPDGVGVSNPGFWGMNIVQGKKYKVVFFYRSLGSL 159
T+ SSCFERNK+ALRMDVLCD CP GVGV NPG+WGMNI +GKKYKV + RS G +
Sbjct: 112 TDRSSCFERNKIALRMDVLCDSKGCPSGGVGVYNPGYWGMNIEEGKKYKVALYVRSTGDI 171
Query: 160 DMRVAFRDAISGRILASSHIIRHKASKKKGSKWQRVQTILEARASSSNSNLTLTTTKEGT 219
D+ V+ + R LAS II AS SKW + + +LEA+A+ ++ L LTTTK+G+
Sbjct: 172 DLSVSLTSSNGSRTLASEKII---ASASDVSKWIKKEVLLEAKATDPSARLQLTTTKKGS 228
Query: 220 VWLDQVSAMPTDTFKGHGFRKDLVEMLIQLKPAFLRFPGGCFVEGVQLRNAFRWKDSVGP 279
+W+DQVSAMP DT KGHGFR DL +M+ +KP F+RFPGGCFVEG L NAFRWK++VGP
Sbjct: 229 IWIDQVSAMPVDTHKGHGFRNDLFQMMADIKPRFIRFPGGCFVEGEWLSNAFRWKETVGP 288
Query: 280 WEQRPGHLNDIWNYWTDDGLGFFEGLQLAEDIGALPIWVFNNGISHSDEVDTSVISPFVK 339
WE+RPGH D+W YWTDDGLG FE Q+AEDIGA PIWVFNNGISH+DEV+T+ I PFV+
Sbjct: 289 WEERPGHFGDVWKYWTDDGLGHFEFFQMAEDIGAAPIWVFNNGISHNDEVETASIMPFVQ 348
Query: 340 EALEGIEFARGSSTSKWGSVRASMGHPKPFNLKYVAIGNEDCYKKNYYGNYMAFYKAIKK 399
EAL+GIEFARG + S WGSVRA MG +PF LKYVAIGNEDC K Y GNY+ FY AIKK
Sbjct: 349 EALDGIEFARGDANSTWGSVRAKMGRQEPFELKYVAIGNEDCGKTYYRGNYIVFYDAIKK 408
Query: 400 FYPDIQIISNCPAFKTPLNHPADLYDYHTYPIDARAMFNAYHDFDKSPRNGPKAFVSEYA 459
YPDI+IISNC PL+HPAD YDYH Y A +F+ YH FD++ R GPKAFVSEYA
Sbjct: 409 AYPDIKIISNCDGSSHPLDHPADYYDYHIY-TSASNLFSMYHQFDRTSRKGPKAFVSEYA 467
Query: 460 LIGALQAKYGTLLGAVSEAGFLIGLERNSDHVAMASYAPLLVNANDRNWNPDAIVFNSYQ 519
+ G A G+LL +++EA FLIGLE+NSD V MASYAPL VN NDR WNPDAIVFNS
Sbjct: 468 VTGK-DAGTGSLLASLAEAAFLIGLEKNSDIVEMASYAPLFVNTNDRRWNPDAIVFNSSH 526
Query: 520 AYGTPSYWVTYMFKESNGATFLNSQLQTPDPGSLIASAILCQSPQNNSTYLKIKIANIGS 579
YGTPSYWV F ES+GAT L S L+ + SL+ASAI ++ N Y++IK N G+
Sbjct: 527 LYGTPSYWVQRFFAESSGATLLTSTLK-GNSTSLVASAISWKN--NGKDYIRIKAVNFGA 583
Query: 580 IPVNLKISLQGYVSK--NLAGSTKTVLTSGNILDENTFAAPKKIAPQTSPLQNPGNEMNV 637
N+++ + G ++GS KTVLTS N++DEN+F+ P+K+ P S L+ +M V
Sbjct: 584 NSENMQVLVTGLDPNVMRVSGSKKTVLTSTNVMDENSFSQPEKVVPHESLLELAEEDMTV 643
Query: 638 MIPPVSLTVLDM 649
++PP S + D+
Sbjct: 644 VLPPHSFSSFDL 655
>At5g26120 arabinosidase - like protein
Length = 674
Score = 738 bits (1904), Expect = 0.0
Identities = 372/646 (57%), Positives = 464/646 (71%), Gaps = 18/646 (2%)
Query: 13 LYLIIVSFVAFQC----NANGSQISSLVVNAAQ--GRPMPNTLFGIFYEEINHAGSGGIW 66
L I+ SF +Q +A I +L V+A+ RP+P TLFGIF+EEINHAG+GG+W
Sbjct: 16 LSFILGSFSVYQTLCLVDAQEDAIVTLQVDASNVTRRPIPETLFGIFFEEINHAGAGGLW 75
Query: 67 AQLVNNSGFEAAGTRTPSNIFPWTIIGTESSVKLQTELSSCFERNKVALRMDVLCDK--C 124
A+LV+N GFEA G PSNI+PW+IIG ESS+ + T+ SSCFERNK+ALRM+VLCD C
Sbjct: 76 AELVSNRGFEAGGQIIPSNIWPWSIIGDESSIYVVTDRSSCFERNKIALRMEVLCDSNSC 135
Query: 125 PPDGVGVSNPGFWGMNIVQGKKYKVVFFYRSLGSLDMRVAFRDAISGRILASSHIIRHKA 184
P GVGV NPG+WGMNI +GKKYKVV + RS G +D+ V+F + LAS +II A
Sbjct: 136 PLGGVGVYNPGYWGMNIEEGKKYKVVLYVRSTGDIDVSVSFTSSNGSVTLASENII---A 192
Query: 185 SKKKGSKWQRVQTILEARASSSNSNLTLTTTKEGTVWLDQVSAMPTDTFKGHGFRKDLVE 244
W + + +LEA + + + L TTTK+G++W DQVSAMP DT+KGHGFR DL +
Sbjct: 193 LASDLLNWTKKEMLLEANGTDNGARLQFTTTKKGSIWFDQVSAMPMDTYKGHGFRNDLFQ 252
Query: 245 MLIQLKPAFLRFPGGCFVEGVQLRNAFRWKDSVGPWEQRPGHLNDIWNYWTDDGLGFFEG 304
M++ LKP F+RFPGGCFVEG L NAFRWK++V WE+RPGH D+W YWTDDGLG FE
Sbjct: 253 MMVDLKPRFIRFPGGCFVEGDWLGNAFRWKETVRAWEERPGHYGDVWKYWTDDGLGHFEF 312
Query: 305 LQLAEDIGALPIWVFNNGISHSDEVDTSVISPFVKEALEGIEFARGSSTSKWGSVRASMG 364
QLAED+GA PIWVFNNGISH+D+V+T + PFV+EA++GIEFARG S S WGSVRA+MG
Sbjct: 313 FQLAEDLGASPIWVFNNGISHNDQVETKNVMPFVQEAIDGIEFARGDSNSTWGSVRAAMG 372
Query: 365 HPKPFNLKYVAIGNEDCYKKNYYGNYMAFYKAIKKFYPDIQIISNCPAFKTPLNHPADLY 424
HP+PF LKYVA+GNEDC+K Y GNY+ FY AIKK YPDI+IISNC A PL+HPAD +
Sbjct: 373 HPEPFELKYVAVGNEDCFKSYYRGNYLEFYNAIKKAYPDIKIISNCDASAKPLDHPADYF 432
Query: 425 DYHTYPIDARAMFNAYHDFDKSPRNGPKAFVSEYALIGALQAKYGTLLGAVSEAGFLIGL 484
DYH Y + AR +F+ HDFD +PRNGPKAFVSEYA + AK G LL A+ EA FL+GL
Sbjct: 433 DYHIYTL-ARDLFSKSHDFDNTPRNGPKAFVSEYA-VNKADAKNGNLLAALGEAAFLLGL 490
Query: 485 ERNSDHVAMASYAPLLVNANDRNWNPDAIVFNSYQAYGTPSYWVTYMFKESNGATFLNSQ 544
E+NSD V M SYAPL VN NDR W PDAIVFNS YGTPSYWV + F ES+GAT LNS
Sbjct: 491 EKNSDIVEMVSYAPLFVNTNDRRWIPDAIVFNSSHLYGTPSYWVQHFFTESSGATLLNST 550
Query: 545 LQTPDPGSLIASAILCQSPQNNSTYLKIKIANIGSIPVNLKISLQGYVSKNLAGSTKTVL 604
L+ S+ ASAI Q+ N Y++IK N G VNLK+++ G ++K GS K VL
Sbjct: 551 LK-GKTSSVEASAISFQT--NGKDYIQIKAVNFGEQSVNLKVAVTGLMAK-FYGSKKKVL 606
Query: 605 TSGNILDENTFAAPKKIAPQTSPLQNPGNE-MNVMIPPVSLTVLDM 649
TS +++DEN+F+ P I PQ S L+ E + ++PP S + D+
Sbjct: 607 TSASVMDENSFSNPNMIVPQESLLEMTEQEDLMFVLPPHSFSSFDL 652
>At5g24280 putative protein
Length = 1634
Score = 36.2 bits (82), Expect = 0.059
Identities = 22/75 (29%), Positives = 34/75 (45%), Gaps = 4/75 (5%)
Query: 316 IWVFNNGISHSDEVDTSVISPFVKEALEGIEFARGSSTSKWGSVRASMGHPKPFNLKYVA 375
+W + G VD S V + G++ + G+S KWG + AS+ + K A
Sbjct: 168 VWPYREGARKLISVDISGDHITVFDTGRGMDSSEGNSIDKWGKIGASLHRSQ----KTGA 223
Query: 376 IGNEDCYKKNYYGNY 390
IG Y K Y+G +
Sbjct: 224 IGGNPPYLKPYFGMF 238
>At3g26115 unknown protein
Length = 344
Score = 30.0 bits (66), Expect = 4.2
Identities = 25/109 (22%), Positives = 46/109 (41%), Gaps = 10/109 (9%)
Query: 173 ILASSHIIRHKASKKKGSKWQRVQTILEARASSSNSNLTLTTTKEGTVWLDQVSA-MPTD 231
+L +++H + K + V+ +++A ++ L + G W +++A M D
Sbjct: 168 LLGMFRLVQHLSQDHLLGKKRPVKFVVDAGTGTTAVGLGVAAMSLGLPW--EINAVMLAD 225
Query: 232 TFKGHGFRKDLVEMLIQLKPAFLR-FPGGCFVEGVQLRNAFRWKDSVGP 279
T K + +D L F R FPG F G+ + +W D P
Sbjct: 226 TLKNYKRHED------HLIAEFSRQFPGSVFCSGLDMNQMIKWIDRQHP 268
>At1g11295 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 29.3 bits (64), Expect = 7.2
Identities = 20/61 (32%), Positives = 31/61 (50%), Gaps = 5/61 (8%)
Query: 173 ILASSHIIRHKASKKKGSKWQRVQTILEARASSSNSNLTLTTTKEGTVWLDQVSAMPTDT 232
+L + I+ K +KKKG+ +++ +EA A S L KE ++ QV A TD
Sbjct: 454 VLLARRIVMKKRAKKKGTDAEQIFKRVEALAGGSREKL-----KELPLFEFQVLATATDN 508
Query: 233 F 233
F
Sbjct: 509 F 509
>At3g25450 hypothetical protein
Length = 1343
Score = 28.9 bits (63), Expect = 9.4
Identities = 20/62 (32%), Positives = 30/62 (48%), Gaps = 2/62 (3%)
Query: 210 LTLTTTKEGTVWLDQVSAMPTDTFKGHGFRKDLVEMLIQLKPAFLRFPGGCFVEGVQLRN 269
L LTTT E T+W ++ + +T K +K+LV + P G C G Q R+
Sbjct: 414 LQLTTTNESTIWHARLGHISFETIKAM-IKKELVIGISSSVPQEKETCGSCLF-GKQARH 471
Query: 270 AF 271
+F
Sbjct: 472 SF 473
>At3g04910 putative mitogen activated protein kinase kinase
Length = 700
Score = 28.9 bits (63), Expect = 9.4
Identities = 19/45 (42%), Positives = 24/45 (53%), Gaps = 5/45 (11%)
Query: 249 LKPAFLRFPGGCFVEGVQLRNAFRWKDSVGPWEQRPGHLNDIWNY 293
L FLR G F LR+ +DSVGP ++P HL D +NY
Sbjct: 276 LDDPFLRIDDGEF----DLRSV-DMEDSVGPLYRQPHHLPDYYNY 315
>At2g30210 putative laccase
Length = 570
Score = 28.9 bits (63), Expect = 9.4
Identities = 19/61 (31%), Positives = 30/61 (49%), Gaps = 8/61 (13%)
Query: 395 KAIK-KFYPDIQIISNCPAFKTPLNHPADLYDYHTYPIDARAMFNAYHDFDKSPRNGPKA 453
KA K K+ ++QI+ + TP NHP L+ Y Y + + + +F +PR P
Sbjct: 446 KAYKLKYKSNVQIVLQDTSIVTPENHPMHLHGYQFYVVG-----SGFGNF--NPRTDPAR 498
Query: 454 F 454
F
Sbjct: 499 F 499
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,168,846
Number of Sequences: 26719
Number of extensions: 666059
Number of successful extensions: 1383
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1367
Number of HSP's gapped (non-prelim): 9
length of query: 650
length of database: 11,318,596
effective HSP length: 106
effective length of query: 544
effective length of database: 8,486,382
effective search space: 4616591808
effective search space used: 4616591808
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC141107.11


