

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.2 + phase: 0
(413 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
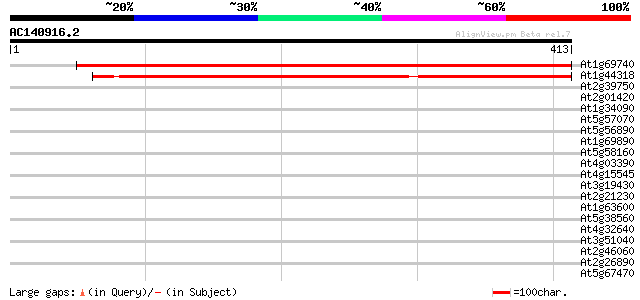
Score E
Sequences producing significant alignments: (bits) Value
At1g69740 aminolevulinate dehydratase like protein 662 0.0
At1g44318 delta-aminolevulinic acid dehydratase (Alad), putative 494 e-140
At2g39750 unknown protein 34 0.17
At2g01420 auxin transporter splice variant b (PIN4) 33 0.29
At1g34090 putative protein 32 0.64
At5g57070 putative protein 31 1.1
At5g56890 unknown protein 31 1.1
At1g69890 unknown protein 31 1.4
At5g58160 strong similarity to unknown protein (gb|AAD23008.1) 30 1.9
At4g03390 SRF3 (LRR receptor-like protein kinase like protein) 30 2.4
At4g15545 unknown protein 30 3.2
At3g19430 putative late embryogenesis abundant protein 30 3.2
At2g21230 bZIP transcription factor AtbZip30 30 3.2
At1g63600 unknown protein 30 3.2
At5g38560 putative protein 29 4.2
At4g32640 putative protein 29 4.2
At3g51040 unknown protein 29 4.2
At2g46060 unknown protein 29 4.2
At2g26890 unknown protein 29 4.2
At5g67470 formin-like protein 29 5.4
>At1g69740 aminolevulinate dehydratase like protein
Length = 430
Score = 662 bits (1707), Expect = 0.0
Identities = 323/364 (88%), Positives = 342/364 (93%)
Query: 50 SDADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTL 109
SDA+ EAAV AGNVP+APPVPP PAAP GTP++ L + RRPRRNR S R+AFQET +
Sbjct: 67 SDAECEAAVAAGNVPEAPPVPPKPAAPVGTPIIKPLNLSRRPRRNRASPVTRAAFQETDI 126
Query: 110 SPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDAL 169
SPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGL++EVAKAR VGVNS+VLFPK+P+AL
Sbjct: 127 SPANFVYPLFIHEGEEDTPIGAMPGCYRLGWRHGLVQEVAKARAVGVNSIVLFPKVPEAL 186
Query: 170 KTPTGDEAYNENGLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDET 229
K TGDEAYN+NGLVPRTIRLLKDKYPDL+IYTDVALDPYSSDGHDGIVREDGVIMNDET
Sbjct: 187 KNSTGDEAYNDNGLVPRTIRLLKDKYPDLIIYTDVALDPYSSDGHDGIVREDGVIMNDET 246
Query: 230 VHQLCKQAVAQARAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGP 289
VHQLCKQAV+QARAGADVVSPSDMMDGRVGA+R+ALDAEGFQ+VSIMSYTAKYASSFYGP
Sbjct: 247 VHQLCKQAVSQARAGADVVSPSDMMDGRVGAIRSALDAEGFQNVSIMSYTAKYASSFYGP 306
Query: 290 FREALDSNPRFGDKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDN 349
FREALDSNPRFGDKKTYQMNPANYREAL E REDE EGADILLVKPGLPYLDIIRLLRD
Sbjct: 307 FREALDSNPRFGDKKTYQMNPANYREALIEAREDEAEGADILLVKPGLPYLDIIRLLRDK 366
Query: 350 SPLPIAAYQVSGEYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLC 409
SPLPIAAYQVSGEYSMIKAGG LKMIDEEKVMMESL+CLRRAGADIILTYFALQAA CLC
Sbjct: 367 SPLPIAAYQVSGEYSMIKAGGVLKMIDEEKVMMESLMCLRRAGADIILTYFALQAATCLC 426
Query: 410 GEKR 413
GEKR
Sbjct: 427 GEKR 430
>At1g44318 delta-aminolevulinic acid dehydratase (Alad), putative
Length = 406
Score = 494 bits (1272), Expect = e-140
Identities = 251/352 (71%), Positives = 288/352 (81%), Gaps = 9/352 (2%)
Query: 62 NVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIH 121
N +APPVP PA P + L + RR RRNR+ T R+AFQET +SPANF+YPLFIH
Sbjct: 59 NALEAPPVPSKPATPL---IDQPLQLSRRARRNRKCPTQRAAFQETNISPANFIYPLFIH 115
Query: 122 EGEEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPDALKTPTGDEAYNEN 181
EGE D PI +MPG Y LGWRHGLIEEVA+A DVGVNSV L+PK+P+ALK+PTG+EA+N+N
Sbjct: 116 EGEVDIPITSMPGRYMLGWRHGLIEEVARALDVGVNSVKLYPKVPEALKSPTGEEAFNDN 175
Query: 182 GLVPRTIRLLKDKYPDLVIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQA 241
GL+PRT+RLLKD++PDLVIYTDV D YS+ GH GIV EDGVI+NDET+HQL KQAV+QA
Sbjct: 176 GLIPRTVRLLKDRFPDLVIYTDVNFDEYSTTGHGGIVGEDGVILNDETIHQLRKQAVSQA 235
Query: 242 RAGADVVSPSDMMDGRVGAMRAALDAEGFQHVSIMSYTAKYASSFYGPFREALDSNPRFG 301
RAGADVV S+M+DGRVGA+RAALDAEGFQ VSIMSY+ KY SS YG FR+
Sbjct: 236 RAGADVVCTSEMLDGRVGAVRAALDAEGFQDVSIMSYSVKYTSSLYGRFRKVQ------L 289
Query: 302 DKKTYQMNPANYREALTEMREDETEGADILLVKPGLPYLDIIRLLRDNSPLPIAAYQVSG 361
DKKTYQ+NPAN REAL E REDE EGADIL+VKP LP LDIIRLL++ + LPI A QVSG
Sbjct: 290 DKKTYQINPANSREALLEAREDEAEGADILMVKPALPSLDIIRLLKNQTLLPIGACQVSG 349
Query: 362 EYSMIKAGGALKMIDEEKVMMESLLCLRRAGADIILTYFALQAARCLCGEKR 413
EYSMIKA G LKMIDEEKVMMESLLC+RRAGAD+ILTYFALQAA LCGE +
Sbjct: 350 EYSMIKAAGLLKMIDEEKVMMESLLCIRRAGADLILTYFALQAATKLCGENK 401
>At2g39750 unknown protein
Length = 694
Score = 33.9 bits (76), Expect = 0.17
Identities = 23/79 (29%), Positives = 37/79 (46%), Gaps = 10/79 (12%)
Query: 3 SSSTIPNTPLTLNSHTYVDLKPALPLKNYLSF---SSSKRRPPCLFTVRASDADFEAAVV 59
S S+IP ++ NS+ +L +P N+ ++ +++PP + AD E V
Sbjct: 51 SGSSIPEVSVSPNSNRVFNLSAIIPT-NHTQIEIPATIRQQPPSVV------ADTEKVKV 103
Query: 60 AGNVPDAPPVPPTPAAPAG 78
N P PP P+P P G
Sbjct: 104 EANPPPPPPPSPSPPPPPG 122
>At2g01420 auxin transporter splice variant b (PIN4)
Length = 612
Score = 33.1 bits (74), Expect = 0.29
Identities = 26/106 (24%), Positives = 48/106 (44%), Gaps = 8/106 (7%)
Query: 190 LLKDKYPDL---VIYTDVALDPYSSDGHDGIVREDGVIMNDETVHQLCKQAVAQARAGAD 246
L+ +++P+ ++ V D S DGHD + D I ND +H +++ A R+
Sbjct: 160 LIMEQFPETGASIVSFKVESDVVSLDGHD-FLETDAEIGNDGKLHVTVRKSNASRRSLMM 218
Query: 247 VVSPSDMMDGRVGAMRAALDAEGFQH---VSIMSYTAKYASSFYGP 289
PS++ + ++ + F H S+M + S+F GP
Sbjct: 219 TPRPSNLTGAEIYSLSSTPRGSNFNHSDFYSVMGFPGGRLSNF-GP 263
>At1g34090 putative protein
Length = 761
Score = 32.0 bits (71), Expect = 0.64
Identities = 34/164 (20%), Positives = 60/164 (35%), Gaps = 10/164 (6%)
Query: 5 STIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSKRRPPCLFTVRASDADFEAAVVAGNVP 64
++IP+TP T + + + S S P + +S + + + VA N
Sbjct: 101 TSIPSTPTTPQRSSTESTETPMSSPVITQPSPSASSIPTMHAATSSASTSQQSSVASNKS 160
Query: 65 DAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQ-ETTLSPANFVYPLFIHEG 123
V A+P T P++++P TH + + + N Y L +H+
Sbjct: 161 TTDVVQIQEASPKSTAPCIEAPVEKQP-------THTMVTRSKAGIHKPNPRYALMLHKV 213
Query: 124 EEDTPIGAMPGCYRLGWRHGLIEEVAKARDVGVNSVVLFPKIPD 167
P GW + + EE+ ++ S L P PD
Sbjct: 214 SYPEPKTVTAAMKDEGWNNAMHEEMDNYKEAQTWS--LIPYTPD 255
>At5g57070 putative protein
Length = 607
Score = 31.2 bits (69), Expect = 1.1
Identities = 16/51 (31%), Positives = 23/51 (44%), Gaps = 1/51 (1%)
Query: 64 PDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANF 114
P +PP P PA P P + + ++PRR RS +R + S F
Sbjct: 241 PSSPPQQP-PATPPPPPPPPPVEVPQKPRRTHRSVRNRDLQENAKRSETKF 290
>At5g56890 unknown protein
Length = 1113
Score = 31.2 bits (69), Expect = 1.1
Identities = 25/74 (33%), Positives = 34/74 (45%), Gaps = 6/74 (8%)
Query: 66 APPVPPT--PAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSP---ANFV-YPLF 119
APPV P P+ +PV TS P + PRRN + + +SP AN V +P
Sbjct: 209 APPVHPVIPKLTPSSSPVPTSTPTKGSPRRNPPTTHPVFPIESPAVSPDHAANPVKHPPP 268
Query: 120 IHEGEEDTPIGAMP 133
G++ GA P
Sbjct: 269 SDNGDDSKSPGAAP 282
>At1g69890 unknown protein
Length = 279
Score = 30.8 bits (68), Expect = 1.4
Identities = 29/124 (23%), Positives = 53/124 (42%), Gaps = 20/124 (16%)
Query: 51 DADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLS 110
D D +V P P P T P P T P+ R+++ +S + T
Sbjct: 140 DVDVVEILVGTTTPAPAPAPVTTPPPHRRPSYTYSPVS-------RTSSEKSEEELTVPP 192
Query: 111 PANFVYPLFIH----EG--EEDTPIGAMPGCYRLGWRHGLIEEVAKA--RDVGVNSVVLF 162
P + P++ H EG EE++ +G Y L ++ +E++ +A + ++ VV+
Sbjct: 193 PKSEGRPIYYHIADEEGHVEEESAVG-----YALTFKGNSVEQLTQALQEETSMDDVVVC 247
Query: 163 PKIP 166
+ P
Sbjct: 248 TRHP 251
>At5g58160 strong similarity to unknown protein (gb|AAD23008.1)
Length = 1307
Score = 30.4 bits (67), Expect = 1.9
Identities = 26/80 (32%), Positives = 28/80 (34%), Gaps = 14/80 (17%)
Query: 1 MASSSTIPNTPLTLNSHTYVDLKPALPLKNYLSFSSSKRRPPCLFTVRASDADFEAAVVA 60
M SS P P L +H+ P P PP L RA A
Sbjct: 750 MKSSPPAPPAPPRLPTHSASPPPPTAP------------PPPPLGQTRAPSAPPPPPPKL 797
Query: 61 GNV--PDAPPVPPTPAAPAG 78
G P P VPPTPA P G
Sbjct: 798 GTKLSPSGPNVPPTPALPTG 817
>At4g03390 SRF3 (LRR receptor-like protein kinase like protein)
Length = 776
Score = 30.0 bits (66), Expect = 2.4
Identities = 19/63 (30%), Positives = 27/63 (42%), Gaps = 5/63 (7%)
Query: 39 RRPPCLFTVRASDADFEAAVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSA 98
RRP + + D DF + P PP PP P P VT +PI R ++++
Sbjct: 407 RRPAPISRQESQDIDFSMLM-----PPPPPPPPPPPPPPLDEKVTVMPIISPERPVKKTS 461
Query: 99 THR 101
R
Sbjct: 462 PKR 464
>At4g15545 unknown protein
Length = 337
Score = 29.6 bits (65), Expect = 3.2
Identities = 20/45 (44%), Positives = 24/45 (52%), Gaps = 3/45 (6%)
Query: 67 PPVPPTPAAPAGTPVVTSLPIQRRPRRNRRS-ATHRSAFQETTLS 110
PP P + +GTP TS PI PRR+ S AT R F +T S
Sbjct: 214 PPGSPPILSASGTPKTTSRPIS--PRRHSVSFATTRGMFDDTRSS 256
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 29.6 bits (65), Expect = 3.2
Identities = 12/24 (50%), Positives = 15/24 (62%)
Query: 63 VPDAPPVPPTPAAPAGTPVVTSLP 86
VP P PPTP+ P+ TP V+ P
Sbjct: 78 VPPVSPPPPTPSVPSPTPPVSPPP 101
>At2g21230 bZIP transcription factor AtbZip30
Length = 519
Score = 29.6 bits (65), Expect = 3.2
Identities = 21/68 (30%), Positives = 28/68 (40%), Gaps = 5/68 (7%)
Query: 66 APPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANFVYPLFIHEGEE 125
APP P P +P + +P +PR +R + S F +L P N P EE
Sbjct: 52 APPPPIPPISP-----YSQIPATLQPRHSRSMSQPSSFFSFDSLPPLNPSAPSVSVSVEE 106
Query: 126 DTPIGAMP 133
T G P
Sbjct: 107 KTGAGFSP 114
>At1g63600 unknown protein
Length = 302
Score = 29.6 bits (65), Expect = 3.2
Identities = 12/36 (33%), Positives = 21/36 (58%)
Query: 57 AVVAGNVPDAPPVPPTPAAPAGTPVVTSLPIQRRPR 92
A + P P PP P++P +P++ S P+ +RP+
Sbjct: 250 ATALSSPPPPYPSPPPPSSPLFSPLLPSPPLFKRPQ 285
>At5g38560 putative protein
Length = 681
Score = 29.3 bits (64), Expect = 4.2
Identities = 22/81 (27%), Positives = 33/81 (40%), Gaps = 14/81 (17%)
Query: 20 VDLKPALPLKNYLSFSSSKRRPPCLFTVRASDADFEAAVVAGNVPDAPPV---------- 69
+ L P LP+ + S +SS PP L T + + + P +PP
Sbjct: 1 MSLVPPLPILSPPSSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPV 60
Query: 70 ----PPTPAAPAGTPVVTSLP 86
PP+ + P PV+TS P
Sbjct: 61 VSSPPPSSSPPPSPPVITSPP 81
>At4g32640 putative protein
Length = 1069
Score = 29.3 bits (64), Expect = 4.2
Identities = 19/54 (35%), Positives = 24/54 (44%), Gaps = 7/54 (12%)
Query: 40 RPPCLFTVRASDADFEAAVVAGNVP----DAPPVPPT---PAAPAGTPVVTSLP 86
RPP R + ++GN P AP +PP P AP G P V+ LP
Sbjct: 225 RPPSAPYARTPPQPLGSHSLSGNPPLTPFTAPSMPPPATFPGAPHGRPAVSGLP 278
>At3g51040 unknown protein
Length = 231
Score = 29.3 bits (64), Expect = 4.2
Identities = 17/46 (36%), Positives = 27/46 (57%), Gaps = 6/46 (13%)
Query: 185 PRTIRLLKDKYPDLVIYTDVA----LDPYSSDGHDGIVREDGVIMN 226
P I +D++P +++T + L P+ GH GI REDGVI++
Sbjct: 19 PMKIDPKRDRFPCCIVWTPLPFISWLVPFI--GHVGICREDGVILD 62
>At2g46060 unknown protein
Length = 766
Score = 29.3 bits (64), Expect = 4.2
Identities = 16/43 (37%), Positives = 22/43 (50%)
Query: 72 TPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAFQETTLSPANF 114
TPAA G+P + SL ++R PR R AF L+ +F
Sbjct: 497 TPAASRGSPTLVSLSLERCPRGCSSYGQCRYAFDANGLTSYSF 539
>At2g26890 unknown protein
Length = 2535
Score = 29.3 bits (64), Expect = 4.2
Identities = 17/44 (38%), Positives = 26/44 (58%), Gaps = 2/44 (4%)
Query: 62 NVP-DAPPVPPTPAAPAGTPVVTSLPIQRRPRRNRRSATHRSAF 104
N+P DAPP+PP P+ A V+ + RR +R +A+H +F
Sbjct: 465 NLPVDAPPLPP-PSPKAAATVIGFVTCLRRLLSSRSAASHIMSF 507
>At5g67470 formin-like protein
Length = 899
Score = 28.9 bits (63), Expect = 5.4
Identities = 16/48 (33%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query: 67 PPVPPTPAAPAGTPVVTSLPIQRRPR--RNRRSATHRSAFQETTLSPA 112
PP+ P P P+ P Q+RPR + R T+ A +T SP+
Sbjct: 374 PPLQTPPPPPPPPPLAPPPPPQKRPRDFQMLRKVTNSEATTNSTTSPS 421
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,663,486
Number of Sequences: 26719
Number of extensions: 435750
Number of successful extensions: 1672
Number of sequences better than 10.0: 35
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1594
Number of HSP's gapped (non-prelim): 80
length of query: 413
length of database: 11,318,596
effective HSP length: 102
effective length of query: 311
effective length of database: 8,593,258
effective search space: 2672503238
effective search space used: 2672503238
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC140916.2


