

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140916.14 - phase: 0 /pseudo
(541 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
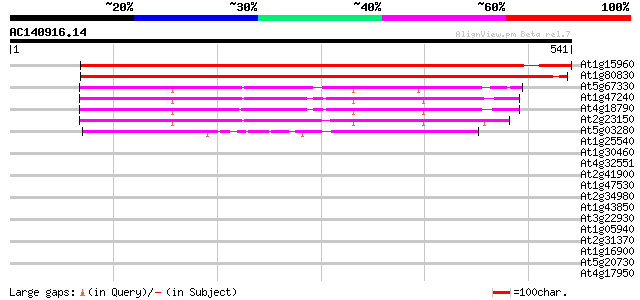
Score E
Sequences producing significant alignments: (bits) Value
At1g15960 Nramp6b 563 e-160
At1g80830 metal ion transporter (PMIT1) 558 e-159
At5g67330 natural resistance-associated macrophage protein 298 5e-81
At1g47240 stress response protein Nramp2 298 7e-81
At4g18790 ion transporter - like protein 293 2e-79
At2g23150 putative metal ion transporter (NRAMP) 293 2e-79
At5g03280 EIN2 145 7e-35
At1g25540 hypothetical protein 36 0.062
At1g30460 hypothetical protein 33 0.40
At4g32551 Leunig protein 33 0.52
At2g41900 CCCH-type zinc finger like protein 33 0.52
At1g47530 unknown protein 32 0.69
At2g34980 putative phosphatidylinositol-glycan synthase 32 0.89
At1g43850 SEUSS transcriptional co-regulator 32 0.89
At3g22930 calmodulin, putative 32 1.2
At1g05940 unknown protein (At1g05940) 31 1.5
At2g31370 bZIP transcription factor PosF21 / AtbZip59 31 2.0
At1g16900 hypothetical protein 31 2.0
At5g20730 auxin response factor 7 (ARF7) 30 2.6
At4g17950 unknown protein 30 2.6
>At1g15960 Nramp6b
Length = 527
Score = 563 bits (1450), Expect = e-160
Identities = 282/473 (59%), Positives = 362/473 (75%), Gaps = 14/473 (2%)
Query: 69 ETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCLW 128
ETDLQ+GA + YELLW++L+ AL+IQSLAANLGV TGKHL+E C EY + LW
Sbjct: 59 ETDLQSGAQYKYELLWIILVASCAALVIQSLAANLGVVTGKHLAEHCRAEYSKVPNFMLW 118
Query: 129 LLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLIT 188
++AE+AV+A DIPEVIGTAFALN+LFNIP+W GVLLTG STL+LL+LQ++G+RKLE LI
Sbjct: 119 VVAEIAVVACDIPEVIGTAFALNMLFNIPVWIGVLLTGLSTLILLALQQYGIRKLEFLIA 178
Query: 189 ILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSA 248
LVF +A CFF E+ Y P VL G+FVP+L G GA AI+LLGA++MPHNLFLHSA
Sbjct: 179 FLVFTIALCFFVELHYSKPDPKEVLYGLFVPQLKGNGATGLAISLLGAMVMPHNLFLHSA 238
Query: 249 LVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERC 308
LVLSRK+P+SV GI EACRY+L ESG AL VAFLINV++ISVSG VC+A+DLS ++ C
Sbjct: 239 LVLSRKIPRSVTGIKEACRYYLIESGLALMVAFLINVSVISVSGAVCNASDLSPEDRASC 298
Query: 309 NDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKR 368
DL LN ASFLL+NV+G+ SS ++AIALLASGQSSTITGTYAGQY+MQGFLD+R++ W R
Sbjct: 299 QDLDLNKASFLLRNVVGKWSSKLFAIALLASGQSSTITGTYAGQYVMQGFLDLRLEPWLR 358
Query: 369 NLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPHK 428
N +TRC+AI PSL VA+IGGS+G+ +LIIIASMILSFELPFAL+PLLKF+SS TKMG H
Sbjct: 359 NFLTRCLAIIPSLIVALIGGSAGAGKLIIIASMILSFELPFALVPLLKFTSSKTKMGSHA 418
Query: 429 NSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIASV 488
NS++I +++WI+G I+ IN+YYL ++F+K ++HS + VA VF+G++ F +A Y+A++
Sbjct: 419 NSLVISSVTWIIGGLIMGINIYYLVSSFIKLLLHSHMNLVAIVFLGVLGFSGIATYLAAI 478
Query: 489 IYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADIPLP 541
YL RK+ + +D + RED+A++ LP
Sbjct: 479 SYLVLRKN--------------RESSSTHFLDFSNSQTEETLPREDIANMQLP 517
>At1g80830 metal ion transporter (PMIT1)
Length = 532
Score = 558 bits (1438), Expect = e-159
Identities = 281/470 (59%), Positives = 360/470 (75%), Gaps = 5/470 (1%)
Query: 69 ETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCLW 128
ETDLQAGA++ YELLW++L+ AL+IQSLAANLGV TGKHL+E C EY + LW
Sbjct: 67 ETDLQAGAHYKYELLWIILVASCAALVIQSLAANLGVVTGKHLAEQCRAEYSKVPNFMLW 126
Query: 129 LLAEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLIT 188
++AE+AV+A DIPEVIGTAFALN+LF+IP+W GVLLTG STL+LL+LQ++GVRKLE LI
Sbjct: 127 VVAEIAVVACDIPEVIGTAFALNMLFSIPVWIGVLLTGLSTLILLALQKYGVRKLEFLIA 186
Query: 189 ILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFLHSA 248
LVF +A CFF E+ Y P VL G+FVP+L G GA AI+LLGA++MPHNLFLHSA
Sbjct: 187 FLVFTIAICFFVELHYSKPDPGEVLHGLFVPQLKGNGATGLAISLLGAMVMPHNLFLHSA 246
Query: 249 LVLSRKVPKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDNVERC 308
LVLSRK+P+S GI EACR++L ESG AL VAFLINV++ISVSG VC+A +LS ++ C
Sbjct: 247 LVLSRKIPRSASGIKEACRFYLIESGLALMVAFLINVSVISVSGAVCNAPNLSPEDRANC 306
Query: 309 NDLTLNSASFLLKNVLGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFLDIRMKRWKR 368
DL LN ASFLL+NV+G+ SS ++AIALLASGQSSTITGTYAGQY+MQGFLD+R++ W R
Sbjct: 307 EDLDLNKASFLLRNVVGKWSSKLFAIALLASGQSSTITGTYAGQYVMQGFLDLRLEPWLR 366
Query: 369 NLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPHK 428
NL+TRC+AI PSL VA+IGGS+G+ +LIIIASMILSFELPFAL+PLLKF+S TKMG H
Sbjct: 367 NLLTRCLAIIPSLIVALIGGSAGAGKLIIIASMILSFELPFALVPLLKFTSCKTKMGSHV 426
Query: 429 NSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFPLMAIYIASV 488
N M I ++W++G I+ IN+YYL ++F+K +IHS + + VF GI+ F +A+Y+A++
Sbjct: 427 NPMAITALTWVIGGLIMGINIYYLVSSFIKLLIHSHMKLILVVFCGILGFAGIALYLAAI 486
Query: 489 IYLTFRKDTVEMFIETKNDTVMQNQVEKGIVDNGQLELSHVPYREDLADI 538
YL FRK+ V + D+ + + + N QL P R +D+
Sbjct: 487 AYLVFRKNRVATSLLISRDSQNVETLPRQDIVNMQL-----PCRVSTSDV 531
>At5g67330 natural resistance-associated macrophage protein
Length = 512
Score = 298 bits (763), Expect = 5e-81
Identities = 180/435 (41%), Positives = 262/435 (59%), Gaps = 24/435 (5%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E+DLQAGA GY L+WL++ L+IQ L+A LGV TG+HL+E+C EYP + + L
Sbjct: 72 LESDLQAGAIAGYSLIWLLMWATAIGLLIQLLSARLGVATGRHLAELCREEYPTWARMVL 131
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W++AE+A+I ADI EVIG+A A+ IL N +PLWAGV++T + L L+ +G+RKLE
Sbjct: 132 WIMAEIALIGADIQEVIGSAIAIKILSNGLVPLWAGVVITALDCFIFLFLENYGIRKLEA 191
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
+ IL+ MA F P + +L G VPKL+ + A+ ++G +IMPHN+FL
Sbjct: 192 VFAILIATMALAFAWMFGQTKPSGTELLVGALVPKLSSR-TIKQAVGIVGCIIMPHNVFL 250
Query: 246 HSALVLSRKV-PKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SR+V PK + EA +Y+ ES AL V+F+INV + +V S
Sbjct: 251 HSALVQSREVDPKKRFRVKEALKYYSIESTGALAVSFIINVFVTTVFAK-------SFYG 303
Query: 305 VERCNDLTLNSASFLLKNVLGRSSST---IYAIALLASGQSSTITGTYAGQYIMQGFLDI 361
E + + L +A L++ G I+AI +LA+GQSSTITGTYAGQ+IM GFL++
Sbjct: 304 TEIADTIGLANAGQYLQDKYGGGFFPILYIWAIGVLAAGQSSTITGTYAGQFIMGGFLNL 363
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGS--SRLIIIASMILSFELPFALIPLLKFSS 419
+MK+W R L+TR AI P++ VA++ SS S L +++ S ++PFA+IPLL S
Sbjct: 364 KMKKWVRALITRSCAIIPTMIVALVFDSSDSMLDELNEWLNVLQSVQIPFAVIPLLCLVS 423
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFP 479
+ MG K ++ TISWI+ +I+IN Y + F S + + +I+F
Sbjct: 424 NEQIMGSFKIQPLVQTISWIVAALVIAINGYLMVDFF------SGAATNLILLVPVIIFA 477
Query: 480 LMAIYIASVIYLTFR 494
+ Y+ V+YL R
Sbjct: 478 I--AYVVFVLYLISR 490
>At1g47240 stress response protein Nramp2
Length = 530
Score = 298 bits (762), Expect = 7e-81
Identities = 176/432 (40%), Positives = 257/432 (58%), Gaps = 24/432 (5%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E DLQAGA GY LLWL++ L+IQ L+A +GV TG+HL+E+C EYP + +Y L
Sbjct: 88 LEGDLQAGAIAGYSLLWLLMWATAMGLLIQMLSARVGVATGRHLAELCRDEYPTWARYVL 147
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W +AE+A+I ADI EVIG+A A+ IL +PLWAGV++T L L L+ +GVRKLE
Sbjct: 148 WSMAELALIGADIQEVIGSAIAIQILSRGFLPLWAGVVITASDCFLFLFLENYGVRKLEA 207
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
+ +L+ M F P ++ G+ +P+L+ + + A+ ++G +IMPHN+FL
Sbjct: 208 VFAVLIATMGLSFAWMFGETKPSGKELMIGILLPRLSSK-TIRQAVGVVGCVIMPHNVFL 266
Query: 246 HSALVLSRKV-PKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SRK+ PK + EA Y+L ES ALF++F+IN+ + T A G
Sbjct: 267 HSALVQSRKIDPKRKSRVQEALNYYLIESSVALFISFMINLFV-----TTVFAKGFYG-- 319
Query: 305 VERCNDLTLNSASFLLKNVLGRSSST---IYAIALLASGQSSTITGTYAGQYIMQGFLDI 361
E+ N++ L +A L+ G I+ I LLA+GQSSTITGTYAGQ+IM GFL++
Sbjct: 320 TEKANNIGLVNAGQYLQEKFGGGLLPILYIWGIGLLAAGQSSTITGTYAGQFIMGGFLNL 379
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLII--IASMILSFELPFALIPLLKFSS 419
R+K+W R ++TR AI P++ VAI+ +S +S ++ +++ S ++PFAL+PLL S
Sbjct: 380 RLKKWMRAVITRSCAIVPTMIVAIVFNTSEASLDVLNEWLNVLQSVQIPFALLPLLTLVS 439
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFP 479
MG K I+ I+W + ++ IN Y L FV +V G+ V
Sbjct: 440 KEEIMGDFKIGPILQRIAWTVAALVMIINGYLLLDFFVS--------EVDGFLFGVTVCV 491
Query: 480 LMAIYIASVIYL 491
YIA ++YL
Sbjct: 492 WTTAYIAFIVYL 503
>At4g18790 ion transporter - like protein
Length = 530
Score = 293 bits (749), Expect = 2e-79
Identities = 172/432 (39%), Positives = 257/432 (58%), Gaps = 24/432 (5%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E DLQAGA GY LLWL+L + L++Q L+A +GV TG+HL+E+C EYP + + L
Sbjct: 90 IEGDLQAGAVAGYSLLWLLLWATLMGLLMQLLSARIGVATGRHLAEICRSEYPSWARILL 149
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W +AEVA+I ADI EVIG+A AL IL +P+W GV++T F L+ L++ G+RKLE
Sbjct: 150 WFMAEVALIGADIQEVIGSAIALQILTRGFLPIWVGVIITSFDCFLISYLEKCGMRKLEG 209
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
L +L+ MA F + P + G+ +PKL G + +A+ ++G +I PHN+FL
Sbjct: 210 LFAVLIATMALSFAWMFNETKPSVEELFIGIIIPKL-GSKTIREAVGVVGCVITPHNVFL 268
Query: 246 HSALVLSRKV-PKSVRGINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SRK PK + + EA Y+ ES ALFV+F+IN+ + T A G
Sbjct: 269 HSALVQSRKTDPKEINRVQEALNYYTIESSAALFVSFMINLFV-----TAVFAKGFYG-- 321
Query: 305 VERCNDLTLNSASFLLKNVLGRSSST---IYAIALLASGQSSTITGTYAGQYIMQGFLDI 361
++ + + L +A + L+ G I+ I LLA+GQSSTITGTYAGQ+IM+GFLD+
Sbjct: 322 TKQADSIGLVNAGYYLQEKYGGGVFPILYIWGIGLLAAGQSSTITGTYAGQFIMEGFLDL 381
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLII--IASMILSFELPFALIPLLKFSS 419
+M++W +TR AI P++ VAI+ +S S ++ +++ S ++PFA+IPLL S
Sbjct: 382 QMEQWLSAFITRSFAIVPTMFVAIMFNTSEGSLDVLNEWLNILQSMQIPFAVIPLLTMVS 441
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANVFIGIIVFP 479
+ MG K + ++W + ++ IN Y L F + +V +G +VF
Sbjct: 442 NEHIMGVFKIGPSLEKLAWTVAVFVMMINGYLLLDFF--------MAEVEGFLVGFLVFG 493
Query: 480 LMAIYIASVIYL 491
+ YI+ +IYL
Sbjct: 494 GVVGYISFIIYL 505
>At2g23150 putative metal ion transporter (NRAMP)
Length = 509
Score = 293 bits (749), Expect = 2e-79
Identities = 174/426 (40%), Positives = 255/426 (59%), Gaps = 19/426 (4%)
Query: 68 VETDLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCL 127
+E DLQAGA GY LLWL++ L++Q L+A LGV TG+HL+E+C EYP + + L
Sbjct: 76 LEGDLQAGAVAGYSLLWLLMWATAMGLLVQLLSARLGVATGRHLAELCRDEYPTWARMVL 135
Query: 128 WLLAEVAVIAADIPEVIGTAFALNILFN--IPLWAGVLLTGFSTLLLLSLQRFGVRKLEL 185
W++AE+A+I +DI EVIG+A A+ IL N +PLWAGV++T + L L+ +G+RKLE
Sbjct: 136 WVMAELALIGSDIQEVIGSAIAIKILSNGILPLWAGVVITALDCFVFLFLENYGIRKLEA 195
Query: 186 LITILVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLFL 245
+ +L+ M F P S +L G+ VPKL+ + A+ ++G +IMPHN+FL
Sbjct: 196 VFAVLIATMGVSFAWMFGQAKPSGSELLIGILVPKLSSR-TIQKAVGVVGCIIMPHNVFL 254
Query: 246 HSALVLSRKVPKSVR-GINEACRYFLYESGFALFVAFLINVAMISVSGTVCSANDLSGDN 304
HSALV SR+V K + + EA Y+ ES ALF++FLIN+ + +V DL+
Sbjct: 255 HSALVQSREVDKRQKYRVQEALNYYTIESTIALFISFLINLFVTTVFAKGFYNTDLA--- 311
Query: 305 VERCNDLTLNSASFLLKNVLGRSSST---IYAIALLASGQSSTITGTYAGQYIMQGFLDI 361
N + L +A L+ G I+AI LLA+GQSSTITGTYAGQ+IM GFL+
Sbjct: 312 ----NSIGLVNAGQYLQEKYGGGVFPILYIWAIGLLAAGQSSTITGTYAGQFIMGGFLNF 367
Query: 362 RMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLII--IASMILSFELPFALIPLLKFSS 419
+MK+W R L+TR AI P++ VA++ SS ++ ++ +++ S ++PFALIPLL S
Sbjct: 368 KMKKWLRALITRSCAIIPTIIVALVFDSSEATLDVLNEWLNVLQSIQIPFALIPLLCLVS 427
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYLSTAF---VKWIIHSSLPKVANVFIGII 476
MG K + TI+W++ +I IN Y L F V I+++ + G
Sbjct: 428 KEQIMGSFKIGPLYKTIAWLVAALVIMINGYLLLEFFSNEVSGIVYTGFVTLFTASYGAF 487
Query: 477 VFPLMA 482
+ L+A
Sbjct: 488 ILYLIA 493
>At5g03280 EIN2
Length = 1294
Score = 145 bits (365), Expect = 7e-35
Identities = 107/393 (27%), Positives = 187/393 (47%), Gaps = 31/393 (7%)
Query: 71 DLQAGANHGYELLWLVLIGLIFALIIQSLAANLGVTTGKHLSEVCEVEYPLFVKYCLWLL 130
+++ GA GY+L+ + L+ A++ Q +AA + V TGKHL+++C EY + L +
Sbjct: 41 NIEGGARFGYDLVAITLLFNFAAILCQYVAARISVVTGKHLAQICNEEYDKWTCMFLGIQ 100
Query: 131 AEVAVIAADIPEVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLITI- 189
AE + I D+ V+G A ALN+LF + L GV L L F + ++I
Sbjct: 101 AEFSAILLDLTMVVGVAHALNLLFGVELSTGVFLAAMDAFLFPVFASFLENGMANTVSIY 160
Query: 190 -----LVFVMAGCFFAEMSYVNPPASGVLKGMFVPKLAGEGAVADAIALLGALIMPHNLF 244
L+ ++G ++ S + +GVL +L GE A A + LLGA I+PHN +
Sbjct: 161 SAGLVLLLYVSGVLLSQ-SEIPLSMNGVL-----TRLNGESAFA-LMGLLGASIVPHNFY 213
Query: 245 LHSALVLSRKVPKSVRGINEACRYFLYESGFALFVAF----LINVAMISVSGTVCSANDL 300
+HS S + C+ L FA+F F L+N +++ + V + L
Sbjct: 214 IHSYFA-GESTSSSDVDKSSLCQDHL----FAIFGVFSGLSLVNYVLMNAAANVFHSTGL 268
Query: 301 SGDNVERCNDLTLNSASFLLKNV-LGRSSSTIYAIALLASGQSSTITGTYAGQYIMQGFL 359
LT + A L++ V + ++ + L S Q + + + G+ ++ FL
Sbjct: 269 V--------VLTFHDALSLMEQVFMSPLIPVVFLMLLFFSSQITALAWAFGGEVVLHDFL 320
Query: 360 DIRMKRWKRNLMTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSS 419
I + W R +A+AP+L G+ G +L+I ++++ LP ++IPL + +S
Sbjct: 321 KIEIPAWLHRATIRILAVAPALYCVWTSGADGIYQLLIFTQVLVAMMLPCSVIPLFRIAS 380
Query: 420 SSTKMGPHKNSMIIITISWILGFGIISINVYYL 452
S MG HK + ++ G + +NV ++
Sbjct: 381 SRQIMGVHKIPQVGEFLALTTFLGFLGLNVVFV 413
>At1g25540 hypothetical protein
Length = 809
Score = 35.8 bits (81), Expect = 0.062
Identities = 14/23 (60%), Positives = 19/23 (81%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQQQQQQQQQHQ+ + +H++
Sbjct: 716 QQQQQQQQQQQHQLTQLQHHHQQ 738
Score = 30.8 bits (68), Expect = 2.0
Identities = 12/23 (52%), Positives = 17/23 (73%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQ QQQQQQQHQ+ + + ++
Sbjct: 694 QQQHQQQQQQQHQLSQLQHHQQQ 716
Score = 28.9 bits (63), Expect = 7.6
Identities = 14/28 (50%), Positives = 16/28 (57%)
Query: 1 MANLIQQQQQQQQQQQHQVMETSSNHEE 28
M L QQQQQ QQQQQ Q + H +
Sbjct: 687 MPQLQQQQQQHQQQQQQQHQLSQLQHHQ 714
>At1g30460 hypothetical protein
Length = 678
Score = 33.1 bits (74), Expect = 0.40
Identities = 15/20 (75%), Positives = 16/20 (80%)
Query: 3 NLIQQQQQQQQQQQHQVMET 22
NL QQQQQQ QQ QHQV +T
Sbjct: 197 NLQQQQQQQPQQSQHQVSQT 216
>At4g32551 Leunig protein
Length = 931
Score = 32.7 bits (73), Expect = 0.52
Identities = 15/29 (51%), Positives = 19/29 (64%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEEP*SWKQ 34
QQQQQQQQQQQ Q + + +P S +Q
Sbjct: 137 QQQQQQQQQQQQQQQQQQQHQNQPPSQQQ 165
Score = 32.0 bits (71), Expect = 0.89
Identities = 14/23 (60%), Positives = 16/23 (68%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQQQQQQQQQHQ S ++
Sbjct: 145 QQQQQQQQQQQHQNQPPSQQQQQ 167
>At2g41900 CCCH-type zinc finger like protein
Length = 716
Score = 32.7 bits (73), Expect = 0.52
Identities = 15/19 (78%), Positives = 15/19 (78%)
Query: 6 QQQQQQQQQQQHQVMETSS 24
QQQQQQQQQQQHQ SS
Sbjct: 582 QQQQQQQQQQQHQFRSLSS 600
Score = 29.3 bits (64), Expect = 5.8
Identities = 12/18 (66%), Positives = 15/18 (82%)
Query: 1 MANLIQQQQQQQQQQQHQ 18
+A ++QQQQQQQQQQ Q
Sbjct: 575 LAQCVKQQQQQQQQQQQQ 592
Score = 28.5 bits (62), Expect = 9.9
Identities = 13/22 (59%), Positives = 15/22 (68%)
Query: 6 QQQQQQQQQQQHQVMETSSNHE 27
QQQQQQQQQQQ + S+ E
Sbjct: 581 QQQQQQQQQQQQHQFRSLSSRE 602
>At1g47530 unknown protein
Length = 484
Score = 32.3 bits (72), Expect = 0.69
Identities = 24/93 (25%), Positives = 44/93 (46%), Gaps = 5/93 (5%)
Query: 84 WLVLIG-LIFALIIQSLAANLGVT--TGKHLSEVCEVEYPLFVKYCL--WLLAEVAVIAA 138
WL++IG L++ LI +S A G + + L ++ + CL W L + V+
Sbjct: 226 WLIVIGQLLYILITKSDGAWTGFSMLAFRDLYGFVKLSLASALMLCLEFWYLMVLVVVTG 285
Query: 139 DIPEVIGTAFALNILFNIPLWAGVLLTGFSTLL 171
+P + A++I NI W ++ GF+ +
Sbjct: 286 LLPNPLIPVDAISICMNIEGWTAMISIGFNAAI 318
>At2g34980 putative phosphatidylinositol-glycan synthase
Length = 303
Score = 32.0 bits (71), Expect = 0.89
Identities = 26/101 (25%), Positives = 47/101 (45%), Gaps = 10/101 (9%)
Query: 371 MTRCIAIAPSLAVAIIGGSSGSSRLIIIASMILSFELPFALIPLLKFSSSSTKMGPHKNS 430
+T CI++ S+ ++ S SRL + A M+ S ++ F PL+ + G H
Sbjct: 182 LTSCISVNASIVASVFVASRLPSRLHVFAVMLFSLQV-FLFAPLVTYCIKKFNFGLH--- 237
Query: 431 MIIITISWILGFGIISINVYYLSTAFVKWIIHSSLPKVANV 471
+ S+ L G+ ++Y L F +++ SL + NV
Sbjct: 238 ---LLFSFAL-MGLTLYSIYALHRLF--FLVFLSLVLLVNV 272
>At1g43850 SEUSS transcriptional co-regulator
Length = 877
Score = 32.0 bits (71), Expect = 0.89
Identities = 13/23 (56%), Positives = 18/23 (77%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQQQQQQQQQ Q + ++N ++
Sbjct: 601 QQQQQQQQQQQQQTVSQNTNSDQ 623
Score = 30.8 bits (68), Expect = 2.0
Identities = 14/21 (66%), Positives = 16/21 (75%)
Query: 6 QQQQQQQQQQQHQVMETSSNH 26
QQQQQQQQQQQ Q +T S +
Sbjct: 598 QQQQQQQQQQQQQQQQTVSQN 618
Score = 30.8 bits (68), Expect = 2.0
Identities = 16/29 (55%), Positives = 17/29 (58%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEEP*SWKQ 34
QQQQQQQQQQQ Q S N S +Q
Sbjct: 599 QQQQQQQQQQQQQQQTVSQNTNSDQSSRQ 627
Score = 29.3 bits (64), Expect = 5.8
Identities = 13/25 (52%), Positives = 16/25 (64%)
Query: 4 LIQQQQQQQQQQQHQVMETSSNHEE 28
L QQQQQQQQQQQ Q + ++
Sbjct: 588 LRQQQQQQQQQQQQQQQQQQQQQQQ 612
Score = 28.9 bits (63), Expect = 7.6
Identities = 12/23 (52%), Positives = 15/23 (65%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEE 28
QQQQQQQQQQQ Q + ++
Sbjct: 591 QQQQQQQQQQQQQQQQQQQQQQQ 613
>At3g22930 calmodulin, putative
Length = 173
Score = 31.6 bits (70), Expect = 1.2
Identities = 14/24 (58%), Positives = 16/24 (66%)
Query: 5 IQQQQQQQQQQQHQVMETSSNHEE 28
IQQQQQQQQQQQ Q + +E
Sbjct: 4 IQQQQQQQQQQQQQQQQQQQQQQE 27
Score = 30.4 bits (67), Expect = 2.6
Identities = 14/27 (51%), Positives = 16/27 (58%)
Query: 1 MANLIQQQQQQQQQQQHQVMETSSNHE 27
M + QQQQQQQQQQQ Q + E
Sbjct: 1 MEEIQQQQQQQQQQQQQQQQQQQQQQE 27
Score = 30.0 bits (66), Expect = 3.4
Identities = 14/31 (45%), Positives = 19/31 (61%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEEP*SWKQDY 36
QQQQQQQQQQQ Q + E+ +K+ +
Sbjct: 10 QQQQQQQQQQQQQQQQQELTQEQIMEFKEAF 40
Score = 28.5 bits (62), Expect = 9.9
Identities = 15/39 (38%), Positives = 21/39 (53%)
Query: 7 QQQQQQQQQQHQVMETSSNHEEP*SWKQDYTYVFFFHLF 45
QQQQQQQQQQ Q + ++ + +Q + F LF
Sbjct: 5 QQQQQQQQQQQQQQQQQQQQQQELTQEQIMEFKEAFCLF 43
>At1g05940 unknown protein (At1g05940)
Length = 569
Score = 31.2 bits (69), Expect = 1.5
Identities = 47/194 (24%), Positives = 84/194 (43%), Gaps = 36/194 (18%)
Query: 125 YCLWLLAEVAVIAADIP-------EVIGTAFALNILFNIPLWAGVLLTGFSTLLLLSLQR 177
Y + LL + IP E++G +LNIL A +LL + +L ++
Sbjct: 155 YAVALLELFPALKGSIPLWMGSGKELLGGLLSLNIL------APILLALLTLVLCQGVRE 208
Query: 178 FG----VRKLELLITILVFVMAGCFFAEMSYVNPPA----SGVLKGMFVP--KLAGEGAV 227
V ++ +LV + AG F +++ +P A VL G V G AV
Sbjct: 209 SSAVNSVMTATKVVIVLVVICAGAFEIDVANWSPFAPNGFKAVLTGATVVFFSYVGFDAV 268
Query: 228 ADA------------IALLGALIMPHNLFLHSALVLSRKVPKSVRGINEACRYFLYESGF 275
A++ I ++G+L++ +L++ LVL+ VP S+ +A + S
Sbjct: 269 ANSAEESKNPQRDLPIGIMGSLLVCISLYIGVCLVLTGMVPFSLLS-EDAPLAEAFSSKG 327
Query: 276 ALFVAFLINVAMIS 289
FV+ LI++ ++
Sbjct: 328 MKFVSILISIGAVA 341
>At2g31370 bZIP transcription factor PosF21 / AtbZip59
Length = 398
Score = 30.8 bits (68), Expect = 2.0
Identities = 13/13 (100%), Positives = 13/13 (100%)
Query: 6 QQQQQQQQQQQHQ 18
QQQQQQQQQQQHQ
Sbjct: 342 QQQQQQQQQQQHQ 354
>At1g16900 hypothetical protein
Length = 919
Score = 30.8 bits (68), Expect = 2.0
Identities = 19/92 (20%), Positives = 51/92 (54%), Gaps = 5/92 (5%)
Query: 150 LNILFNIPLWAGVLLTGFSTLLLLSLQRFGVRKLELLITILVFVMAGCFFAEMSYVNPPA 209
+ + + + L+ G++ T+L+++L R +++ ++ + +GCFFA S++ P+
Sbjct: 473 VRVFYAVRLFLGLVSAVSDTVLVVALSRKYGKRIATYAVAMLCLTSGCFFASTSFL--PS 530
Query: 210 SGVLKGMFVPK---LAGEGAVADAIALLGALI 238
S + + + L + A+A A++++G ++
Sbjct: 531 SFSMYAISLSSGLLLFEKYAMAVAVSVVGVIL 562
>At5g20730 auxin response factor 7 (ARF7)
Length = 1164
Score = 30.4 bits (67), Expect = 2.6
Identities = 13/15 (86%), Positives = 14/15 (92%)
Query: 4 LIQQQQQQQQQQQHQ 18
L+QQQQQQQ QQQHQ
Sbjct: 557 LLQQQQQQQLQQQHQ 571
>At4g17950 unknown protein
Length = 439
Score = 30.4 bits (67), Expect = 2.6
Identities = 13/24 (54%), Positives = 16/24 (66%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEEP 29
QQQQQQQQQQQ Q + ++P
Sbjct: 10 QQQQQQQQQQQQQQQQHLQQQQQP 33
Score = 29.6 bits (65), Expect = 4.4
Identities = 13/24 (54%), Positives = 14/24 (58%)
Query: 6 QQQQQQQQQQQHQVMETSSNHEEP 29
QQQQQQQQQQQ Q + P
Sbjct: 11 QQQQQQQQQQQQQQQHLQQQQQPP 34
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.141 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,733,375
Number of Sequences: 26719
Number of extensions: 417801
Number of successful extensions: 2912
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 2340
Number of HSP's gapped (non-prelim): 245
length of query: 541
length of database: 11,318,596
effective HSP length: 104
effective length of query: 437
effective length of database: 8,539,820
effective search space: 3731901340
effective search space used: 3731901340
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140916.14


