

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140915.5 - phase: 0
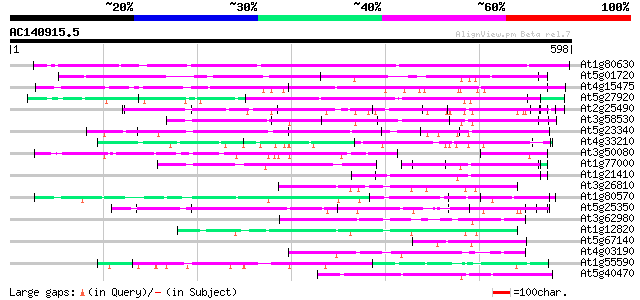
(598 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g80630 unknown protein 270 2e-72
At5g01720 putative F-box protein family, AtFBL3 110 2e-24
At4g15475 unknown protein 99 7e-21
At5g27920 unknown protein 88 2e-17
At2g25490 putative glucose regulated repressor protein 73 5e-13
At3g58530 unknown protein 67 3e-11
At5g23340 unknown protein 65 1e-10
At4g33210 unknown protein 65 1e-10
At3g50080 unknown protein 65 1e-10
At1g77000 unknown protein 63 4e-10
At1g21410 unknown protein 63 4e-10
At3g26810 transport inhibitor response 1-like protein 60 3e-09
At1g80570 unknown protein 60 3e-09
At5g25350 leucine-rich repeats containing protein 59 1e-08
At3g62980 transport inhibitor response 1 (TIR1) 57 3e-08
At1g12820 transport inhibitor response 1, putative 56 5e-08
At5g67140 unknown protein 52 7e-07
At4g03190 putative homolog of transport inhibitor response 1 52 7e-07
At1g55590 unknown protein 52 9e-07
At5g40470 unknown protein 50 4e-06
>At1g80630 unknown protein
Length = 578
Score = 270 bits (690), Expect = 2e-72
Identities = 192/573 (33%), Positives = 299/573 (51%), Gaps = 24/573 (4%)
Query: 26 LDLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTILYPALPVLP 85
+DLP++ WE + + + D+DD+R +L+S+S+ S FLS+TNR + + A+P+L
Sbjct: 1 MDLPEECWELICKAI---DEDDYR--FLESVSLVSTLFLSITNRVRSTFVVTDRAVPLLN 55
Query: 86 GLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLKLTSLNLSNQLILPANGLRAFSQNIT 145
L RF L + S + DL+++L Q+S L S+++S P ++
Sbjct: 56 RHLLRFRNLKRIRFSDFTQDLNSILLQVSRSG-LDFESVDVSQTRYFPDFEMK------- 107
Query: 146 TLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTGLEALSLALIK 205
+ L C + + +D+ I FPLLE+LD++YP+ + +G+ LS L
Sbjct: 108 NVKELKCYGVGGFSDSDLVSIGVNFPLLEKLDISYPNSSPSRVS---DSGVIELSSNLKG 164
Query: 206 LRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSITC 265
L K+N+S + ++ L L +NC L+E+I C+ ++ + L L SL+I
Sbjct: 165 LLKINISGNSFITDKSLIALSQNCLLLREIIFRDCDFISSDCIKFVLRNSRNLESLAIN- 223
Query: 266 TVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYT 325
GL S D+ L + LT L L+ +SD L IA LPL++L+LS C G+T
Sbjct: 224 --GIGLRPRESLLTDAFLFARCLTELDLSDSFLSDDLLCLIASAKLPLKKLLLSDCHGFT 281
Query: 326 YSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALI 385
+ GI +LL K + + HL+L+ +FL+D V +L +F L LNL C LT FF++I
Sbjct: 282 FDGILYLLDKYQSLVHLNLKGANFLSDEMVMKLGMFFRRLTFLNLSFCSKLTGLAFFSII 341
Query: 386 TNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNL 445
C SL + M TN D K L+L+ L D+ + + P +
Sbjct: 342 ERCVSLRCMIMVGTNFGVEEYTKDTKD--FKSGVKFLYLSRNHNLLDECLEKISRHCPFI 399
Query: 446 QQLHLSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKSLG-TNFDLPDLEVLNLTNTEV 503
+ L ++ IT +GI + +C K+R L+++ C +KSLG +F+LP LE L T +
Sbjct: 400 ESLDVAQCPGITRDGILEVWRNCGKLRSLDISRCTGIKSLGVVDFELPKLESLRACGTWI 459
Query: 504 DDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASM 563
DDEAL +IS +C LL L L C ++ +GV VV +C +LREINL C K+ M
Sbjct: 460 DDEALDMISKKCRGLLHLDLQGCLNVSSRGVKEVVQSCIRLREINLKYC-EADKKMYTWM 518
Query: 564 VVSRPSLRKIHVPPNFPLSDRNRKLFSRHGCLL 596
V + PSLRKI P F + + F RHGC++
Sbjct: 519 VFANPSLRKIVPPCGFSPTKALKNFFLRHGCVV 551
>At5g01720 putative F-box protein family, AtFBL3
Length = 665
Score = 110 bits (276), Expect = 2e-24
Identities = 133/527 (25%), Positives = 213/527 (40%), Gaps = 73/527 (13%)
Query: 53 LKSLSVASKHFLSVTNRHKFCLTILYPALPVLPGLLQRFTKLTSLDLSYYYGDLDALLTQ 112
LKS S+ K F + ++H+ L L LP +L R+ T LDL++ D L+
Sbjct: 35 LKSFSLTCKSFYQLESKHRGSLKPLRS--DYLPRILTRYRNTTDLDLTFCPRVTDYALSV 92
Query: 113 ISSFPMLKLTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPL 172
+ L SL+LS A GL + L + SN + D ++A
Sbjct: 93 VGCLSGPTLRSLDLSRSGSFSAAGLLRLALKCVNLVEIDLSNATEMRDADAAVVA----- 147
Query: 173 LEELDLAYPSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFL 232
EA SL +KL + + L + + CK L
Sbjct: 148 ------------------------EARSLERLKLGRCKM-----LTDMGIGCIAVGCKKL 178
Query: 233 QEVILLRCEQLTIAGVDLALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLL 292
V L C + GV L ++ + +L ++ TG + +L L+ L LL
Sbjct: 179 NTVSLKWCVGVGDLGVGLLAVKCKDIRTLDLSYLPITG------KCLHDILKLQHLEELL 232
Query: 293 LTG-FRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLN 351
L G F + D L S+ + L++L S C T+ G++ LLS + +Q LDL
Sbjct: 233 LEGCFGVDDDSLKSLRHDCKSLKKLDASSCQNLTHRGLTSLLSGAGYLQRLDLS------ 286
Query: 352 DHCVAELSLFLGDLL-------SLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGT 404
HC + +SL L S+ L C + T A+ T C SL E+++++
Sbjct: 287 -HCSSVISLDFASSLKKVSALQSIRLDGCSV-TPDGLKAIGTLCNSLKEVSLSKCVSVTD 344
Query: 405 TIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPL 464
+SL+ +L + + L + L +I A P L L + ++ E +
Sbjct: 345 EGLSSLVMKLKD--LRKLDITCCRKLSRVSITQIANSCPLLVSLKMESCSLVSREAFWLI 402
Query: 465 LESCRKIRHLNLTCLS-----LKSLGTNFDLPDLEV---LNLTNTEVDDEALYIISNRCP 516
+ CR + L+LT LKS+ + L L++ LN+T D+ L I C
Sbjct: 403 GQKCRLLEELDLTDNEIDDEGLKSISSCLSLSSLKLGICLNIT-----DKGLSYIGMGCS 457
Query: 517 ALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASM 563
L +L L R ITD G+ + C L IN+ C ++ K + S+
Sbjct: 458 NLRELDLYRSVGITDVGISTIAQGCIHLETINISYCQDITDKSLVSL 504
Score = 43.9 bits (102), Expect = 3e-04
Identities = 48/248 (19%), Positives = 106/248 (42%), Gaps = 37/248 (14%)
Query: 332 LLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLL-SLNLGNCRLLTVSTFFALITNCPS 390
+L++ + LDL + + D+ ++ + G L SL+L + + L C +
Sbjct: 67 ILTRYRNTTDLDLTFCPRVTDYALSVVGCLSGPTLRSLDLSRSGSFSAAGLLRLALKCVN 126
Query: 391 LTEINM-NRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLH 449
L EI++ N T ++ A AA + + +L++L
Sbjct: 127 LVEIDLSNATEMRD---------------------ADAAVVAEAR---------SLERLK 156
Query: 450 LSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKSLGTNF---DLPDLEVLNLTNTEVDD 505
L +T+ GI + C+K+ ++L C+ + LG D+ L+L+ +
Sbjct: 157 LGRCKMLTDMGIGCIAVGCKKLNTVSLKWCVGVGDLGVGLLAVKCKDIRTLDLSYLPITG 216
Query: 506 EALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVV 565
+ L+ I + L +L+L C + D + + ++C L++++ C N+ + + S++
Sbjct: 217 KCLHDIL-KLQHLEELLLEGCFGVDDDSLKSLRHDCKSLKKLDASSCQNLTHRGLTSLLS 275
Query: 566 SRPSLRKI 573
L+++
Sbjct: 276 GAGYLQRL 283
Score = 38.5 bits (88), Expect = 0.011
Identities = 51/233 (21%), Positives = 93/233 (39%), Gaps = 52/233 (22%)
Query: 195 GLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRC-----EQLTIAGVD 249
GL +L + L LRK++++ L+ ++ + +C L + + C E + G
Sbjct: 346 GLSSLVMKLKDLRKLDITCCRKLSRVSITQIANSCPLLVSLKMESCSLVSREAFWLIGQK 405
Query: 250 LALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSL--------KGLTSL---------- 291
LL++ LT I GL+ ++S S L L KGL+ +
Sbjct: 406 CRLLEELDLTDNEID---DEGLKSISSCLSLSSLKLGICLNITDKGLSYIGMGCSNLREL 462
Query: 292 -LLTGFRISDQFLSSIAMESLPLRRLVLSYC-------------------------PGYT 325
L I+D +S+IA + L + +SYC P T
Sbjct: 463 DLYRSVGITDVGISTIAQGCIHLETINISYCQDITDKSLVSLSKCSLLQTFESRGCPNIT 522
Query: 326 YSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTV 378
G++ + + KR+ +DL+ +ND + L+ F +L +N+ + + V
Sbjct: 523 SQGLAAIAVRCKRLAKVDLKKCPSINDAGLLALAHFSQNLKQINVSDTAVTEV 575
>At4g15475 unknown protein
Length = 610
Score = 99.0 bits (245), Expect = 7e-21
Identities = 142/604 (23%), Positives = 252/604 (41%), Gaps = 88/604 (14%)
Query: 28 LPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTILYPALP--VLP 85
LP+++ +FR L++ + D + S+ K +LS+ + L I P +
Sbjct: 11 LPEELILEIFRRLESKPNRD-------ACSLVCKRWLSLERFSRTTLRIGASFSPDDFIS 63
Query: 86 GLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLKLTSLNLSNQLILPANGLRAFSQNIT 145
L +RF +TS+ + + L +S P K + S P++ R + +T
Sbjct: 64 LLSRRFLYITSIHVDER---ISVSLPSLSPSPKRKRGRDSSS-----PSSSKR---KKLT 112
Query: 146 TLTSLICSNLISLNSTDIHL--IADTFPLLEELDLAYPSKIINHTHATFSTGLEALSLAL 203
T N+ S + TD L +A+ FP +E L L + + S GL +L+
Sbjct: 113 DKTHSGAENVESSSLTDTGLTALANGFPRIENLSLIWCPNVS-------SVGLCSLAQKC 165
Query: 204 IKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGV-DLALLQKPTLTSLS 262
L+ ++L Y+ L+ + K CK L+E+ L CE LT GV DL + +L S+
Sbjct: 166 TSLKSLDLQ-GCYVGDQGLAAVGKFCKQLEELNLRFCEGLTDVGVIDLVVGCSKSLKSIG 224
Query: 263 ITCTVTT---GLEHLTSH-------FIDS-LLSLKGLTS----------LLLTGFRISDQ 301
+ + LE + SH ++DS + KGL + L L ++D
Sbjct: 225 VAASAKITDLSLEAVGSHCKLLEVLYLDSEYIHDKGLIAVAQGCHRLKNLKLQCVSVTDV 284
Query: 302 FLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLF 361
+++ L RL L +T G+ + SK+++ L L F++ + ++
Sbjct: 285 AFAAVGELCTSLERLALYSFQHFTDKGMRAIGKGSKKLKDLTLSDCYFVSCKGLEAIAHG 344
Query: 362 LGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVN--PQF 419
+L + + C + A+ +CP L E+ + + I NS + +
Sbjct: 345 CKELERVEINGCHNIGTRGIEAIGKSCPRLKELAL----LYCQRIGNSALQEIGKGCKSL 400
Query: 420 KSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCL 479
+ L L + + D + A NL++LH+ Y I +GI + + C+ + L+L
Sbjct: 401 EILHLVDCSGIGDIAMCSIAKGCRNLKKLHIRRCYEIGNKGIISIGKHCKSLTELSLRFC 460
Query: 480 ------SLKSLGTNFDLPDLEVLN------------------LTNTEVD------DEALY 509
+L ++G L L V LT+ ++ D L
Sbjct: 461 DKVGNKALIAIGKGCSLQQLNVSGCNQISDAGITAIARGCPQLTHLDISVLQNIGDMPLA 520
Query: 510 IISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPS 569
+ CP L LVL C +ITD G+ H+V C L ++ CP + + VA++V S P
Sbjct: 521 ELGEGCPMLKDLVLSHCHHITDNGLNHLVQKCKLLETCHMVYCPGITSAGVATVVSSCPH 580
Query: 570 LRKI 573
++K+
Sbjct: 581 IKKV 584
Score = 67.0 bits (162), Expect = 3e-11
Identities = 71/325 (21%), Positives = 141/325 (42%), Gaps = 33/325 (10%)
Query: 298 ISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAE 357
++D L+++A + L L +CP + G+ L K ++ LDLQ ++ D +A
Sbjct: 127 LTDTGLTALANGFPRIENLSLIWCPNVSSVGLCSLAQKCTSLKSLDLQGC-YVGDQGLAA 185
Query: 358 LSLFLGDLLSLNLGNCRLLTVSTFFALITNCP-SLTEINMNRT------NIQGTTIPNSL 410
+ F L LNL C LT L+ C SL I + + +++ L
Sbjct: 186 VGKFCKQLEELNLRFCEGLTDVGVIDLVVGCSKSLKSIGVAASAKITDLSLEAVGSHCKL 245
Query: 411 MDRL-VNPQF---KSLFLASAACLEDQNI---------IMFAA---LFPNLQQLHLSCSY 454
++ L ++ ++ K L + C +N+ + FAA L +L++L L
Sbjct: 246 LEVLYLDSEYIHDKGLIAVAQGCHRLKNLKLQCVSVTDVAFAAVGELCTSLERLALYSFQ 305
Query: 455 NITEEGIRPLLESCRKIRHLNLT------CLSLKSLGTNF-DLPDLEVLNLTNTEVDDEA 507
+ T++G+R + + +K++ L L+ C L+++ +L +E+ N +
Sbjct: 306 HFTDKGMRAIGKGSKKLKDLTLSDCYFVSCKGLEAIAHGCKELERVEINGCHN--IGTRG 363
Query: 508 LYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSR 567
+ I CP L +L LL C I + + + C L ++L C + + S+
Sbjct: 364 IEAIGKSCPRLKELALLYCQRIGNSALQEIGKGCKSLEILHLVDCSGIGDIAMCSIAKGC 423
Query: 568 PSLRKIHVPPNFPLSDRNRKLFSRH 592
+L+K+H+ + + ++ +H
Sbjct: 424 RNLKKLHIRRCYEIGNKGIISIGKH 448
Score = 33.5 bits (75), Expect = 0.35
Identities = 29/118 (24%), Positives = 49/118 (40%), Gaps = 7/118 (5%)
Query: 146 TLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTGLEALSLALIK 205
+L L S ++ I IA P L LD++ I + A G L
Sbjct: 476 SLQQLNVSGCNQISDAGITAIARGCPQLTHLDISVLQNIGDMPLAELGEGCPML------ 529
Query: 206 LRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSI 263
+ + LS+ ++ L+HL + CK L+ ++ C +T AGV + P + + I
Sbjct: 530 -KDLVLSHCHHITDNGLNHLVQKCKLLETCHMVYCPGITSAGVATVVSSCPHIKKVLI 586
>At5g27920 unknown protein
Length = 642
Score = 87.8 bits (216), Expect = 2e-17
Identities = 135/586 (23%), Positives = 234/586 (39%), Gaps = 73/586 (12%)
Query: 20 SSSSSELD-LPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTILY 78
S+S S L L +D+ RV+ L D RK + + SK FL V + + + IL
Sbjct: 2 STSPSILSVLSEDLLVRVYECL----DPPCRKTW----RLISKDFLRVDSLTRTTIRIL- 52
Query: 79 PALPVLPGLLQRFTKLTSLDLSY---YYGDLDALLTQISSFPMLKLTSLNLSNQLILPAN 135
+ LP LL ++ L+SLDLS D+ L + L + SLNLS + A
Sbjct: 53 -RVEFLPTLLFKYPNLSSLDLSVCPKLDDDVVLRLALDGAISTLGIKSLNLSRSTAVRAR 111
Query: 136 GLRAFSQ-------------------------NITTLTSLICSNLISLNSTDIHLIADTF 170
GL ++ + T L L +SL+ + I
Sbjct: 112 GLETLARMCHALERVDVSHCWGFGDREAAALSSATGLRELKMDKCLSLSDVGLARIVVGC 171
Query: 171 PLLEELDLAYPSKIIN---HTHATFSTGLEALSLA--------------LIKLRKVNLSY 213
L ++ L + +I + GL++L ++ L+KL +++
Sbjct: 172 SNLNKISLKWCMEISDLGIDLLCKICKGLKSLDVSYLKITNDSIRSIALLVKLEVLDMVS 231
Query: 214 HGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSITCTVTTGLEH 273
++ L L LQEV + RC++++++G+ + P + L + V+
Sbjct: 232 CPLIDDGGLQFLENGSPSLQEVDVTRCDRVSLSGLISIVRGHPDIQLLKASHCVS----E 287
Query: 274 LTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLL 333
++ F+ + LK L ++ + G +SD L S++ L + LS C T G+ L
Sbjct: 288 VSGSFLKYIKGLKHLKTIWIDGAHVSDSSLVSLSSSCRSLMEIGLSRCVDVTDIGMISLA 347
Query: 334 SKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTE 393
++ L+L F+ D ++ ++ +L +L L +C L+T +L C S+
Sbjct: 348 RNCLNLKTLNLACCGFVTDVAISAVAQSCRNLGTLKLESCHLITEKGLQSL--GCYSMLV 405
Query: 394 INMNRTNIQGTTIPN-SLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSC 452
++ T+ G + + N Q L L + D+ I + L +L L
Sbjct: 406 QELDLTDCYGVNDRGLEYISKCSNLQ--RLKLGLCTNISDKGIFHIGSKCSKLLELDLYR 463
Query: 453 SYNITEEGIRPLLESCRKIRHLNLT-CLSLKSLGTNFD-----LPDLEVLNLTNTEVDDE 506
++G+ L C+ + L L+ C L G L LE+ L N +
Sbjct: 464 CAGFGDDGLAALSRGCKSLNRLILSYCCELTDTGVEQIRQLELLSHLELRGLKN--ITGV 521
Query: 507 ALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGC 552
L I++ C L L + C+ I D G + LR+INL C
Sbjct: 522 GLAAIASGCKKLGYLDVKLCENIDDSGFWALAYFSKNLRQINLCNC 567
Score = 62.0 bits (149), Expect = 9e-10
Identities = 69/320 (21%), Positives = 147/320 (45%), Gaps = 10/320 (3%)
Query: 252 LLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLT-GFRISDQFLSSIAMES 310
L + P L+SL ++ + + +D +S G+ SL L+ + + L ++A
Sbjct: 61 LFKYPNLSSLDLSVCPKLDDDVVLRLALDGAISTLGIKSLNLSRSTAVRARGLETLARMC 120
Query: 311 LPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNL 370
L R+ +S+C G+ + LS + ++ L + L+D +A + + +L ++L
Sbjct: 121 HALERVDVSHCWGFGDREAA-ALSSATGLRELKMDKCLSLSDVGLARIVVGCSNLNKISL 179
Query: 371 GNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACL 430
C ++ L C L ++++ I +I + + LV + + L + S +
Sbjct: 180 KWCMEISDLGIDLLCKICKGLKSLDVSYLKITNDSIRSIAL--LV--KLEVLDMVSCPLI 235
Query: 431 EDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKS---LGT 486
+D + P+LQ++ ++ ++ G+ ++ I+ L + C+S S L
Sbjct: 236 DDGGLQFLENGSPSLQEVDVTRCDRVSLSGLISIVRGHPDIQLLKASHCVSEVSGSFLKY 295
Query: 487 NFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLRE 546
L L+ + + V D +L +S+ C +L+++ L RC +TD G++ + NC L+
Sbjct: 296 IKGLKHLKTIWIDGAHVSDSSLVSLSSSCRSLMEIGLSRCVDVTDIGMISLARNCLNLKT 355
Query: 547 INLDGCPNVQAKVVASMVVS 566
+NL C V ++++ S
Sbjct: 356 LNLACCGFVTDVAISAVAQS 375
Score = 55.5 bits (132), Expect = 9e-08
Identities = 87/457 (19%), Positives = 181/457 (39%), Gaps = 65/457 (14%)
Query: 138 RAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTGLE 197
R S++ + SL + + L + + +P L LDL+ K+ + L+
Sbjct: 32 RLISKDFLRVDSLTRTTIRILRVEFLPTLLFKYPNLSSLDLSVCPKLDDDV--VLRLALD 89
Query: 198 ALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPT 257
+++ + ++ +NLS + L L + C L+ V + C + A L
Sbjct: 90 G-AISTLGIKSLNLSRSTAVRARGLETLARMCHALERVDVSHC--WGFGDREAAALSS-- 144
Query: 258 LTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLV 317
TGL L +D LSL SD L+ I + L ++
Sbjct: 145 ----------ATGLRELK---MDKCLSL-------------SDVGLARIVVGCSNLNKIS 178
Query: 318 LSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLT 377
L +C + GI L K ++ LD+ Y ND + ++L + L L++ +C L+
Sbjct: 179 LKWCMEISDLGIDLLCKICKGLKSLDVSYLKITNDS-IRSIALLV-KLEVLDMVSCPLID 236
Query: 378 VSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQN--I 435
L PSL E+++ R + + S + +V L ++ C+ + +
Sbjct: 237 DGGLQFLENGSPSLQEVDVTRCD----RVSLSGLISIVRGHPDIQLLKASHCVSEVSGSF 292
Query: 436 IMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLT-CLSLKSLGTNFDLPDLE 494
+ + +L+ + + ++ +++ + L SCR + + L+ C+ + +G
Sbjct: 293 LKYIKGLKHLKTIWIDGAH-VSDSSLVSLSSSCRSLMEIGLSRCVDVTDIG--------- 342
Query: 495 VLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPN 554
+ ++ C L L L C ++TD + V +C L + L+ C
Sbjct: 343 -------------MISLARNCLNLKTLNLACCGFVTDVAISAVAQSCRNLGTLKLESCHL 389
Query: 555 VQAKVVASMVVSRPSLRKIHVPPNFPLSDRNRKLFSR 591
+ K + S+ ++++ + + ++DR + S+
Sbjct: 390 ITEKGLQSLGCYSMLVQELDLTDCYGVNDRGLEYISK 426
>At2g25490 putative glucose regulated repressor protein
Length = 628
Score = 72.8 bits (177), Expect = 5e-13
Identities = 101/463 (21%), Positives = 191/463 (40%), Gaps = 46/463 (9%)
Query: 123 SLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPS 182
S+ SN + GLR+ ++ +L SL N+ ++ + IA+ LE+L+L
Sbjct: 155 SIRGSNSAKVSDLGLRSIGRSCPSLGSLSLWNVSTITDNGLLEIAEGCAQLEKLEL---- 210
Query: 183 KIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQ 242
N GL A++ + L ++ L + L + ++C L+ V + C
Sbjct: 211 ---NRCSTITDKGLVAIAKSCPNLTELTLEACSRIGDEGLLAIARSCSKLKSVSIKNCPL 267
Query: 243 LTIAGVDLAL------LQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGF 296
+ G+ L L K L L++T + H D L L GL+ + GF
Sbjct: 268 VRDQGIASLLSNTTCSLAKLKLQMLNVTDVSLAVVGHYGLSITD--LVLAGLSHVSEKGF 325
Query: 297 RISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVA 356
+ + + ++ L L ++ C G T G+ + ++ + + L+D+ +
Sbjct: 326 WVMG---NGVGLQKL--NSLTITACQGVTDMGLESVGKGCPNMKKAIISKSPLLSDNGLV 380
Query: 357 ELSLFLGDLLSLNLGNCRLLTVSTFFALITNC----PSLTEINMNRTNIQGTTIPNSLMD 412
+ L SL L C +T FF + NC + + +N T +P S
Sbjct: 381 SFAKASLSLESLQLEECHRVTQFGFFGSLLNCGEKLKAFSLVNCLSIRDLTTGLPASSHC 440
Query: 413 RLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLES----- 467
+ +SL + + D N+ L P L+ + L ITE G L++S
Sbjct: 441 SAL----RSLSIRNCPGFGDANLAAIGKLCPQLEDIDLCGLKGITESGFLHLIQSSLVKI 496
Query: 468 ----CRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTN-TEVDDEALYIISNRCPALLQLV 522
C + ++ ++ ++ T LEVLN+ + + D +L I+ C L L
Sbjct: 497 NFSGCSNLTDRVISAITARNGWT------LEVLNIDGCSNITDASLVSIAANCQILSDLD 550
Query: 523 LLRCDYITDKGVMHVV-NNCTQLREINLDGCPNVQAKVVASMV 564
+ +C I+D G+ + ++ +L+ +++ GC V K + ++V
Sbjct: 551 ISKC-AISDSGIQALASSDKLKLQILSVAGCSMVTDKSLPAIV 592
Score = 69.3 bits (168), Expect = 6e-12
Identities = 101/464 (21%), Positives = 197/464 (41%), Gaps = 44/464 (9%)
Query: 121 LTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAY 180
L SL+L N + NGL ++ L L + ++ + IA + P L EL L
Sbjct: 179 LGSLSLWNVSTITDNGLLEIAEGCAQLEKLELNRCSTITDKGLVAIAKSCPNLTELTLEA 238
Query: 181 PSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRC 240
S+I + GL A++ + KL+ V++ + ++ L N + L+
Sbjct: 239 CSRIGDE-------GLLAIARSCSKLKSVSIKNCPLVRDQGIASLLSNTTC--SLAKLKL 289
Query: 241 EQLTIAGVDLALLQKPTLTSLSITCTVTTGLEHLTSH---FIDSLLSLKGLTSLLLTGFR 297
+ L + V LA++ LSIT V GL H++ + + + L+ L SL +T +
Sbjct: 290 QMLNVTDVSLAVVGH---YGLSITDLVLAGLSHVSEKGFWVMGNGVGLQKLNSLTITACQ 346
Query: 298 -ISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVA 356
++D L S+ +++ ++S P + +G+ S ++ L L+ H V
Sbjct: 347 GVTDMGLESVGKGCPNMKKAIISKSPLLSDNGLVSFAKASLSLESLQLEEC-----HRVT 401
Query: 357 ELSLFLGDLLS-------LNLGNC-RLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPN 408
+ F G LL+ +L NC + ++T ++C +L +++ N G N
Sbjct: 402 QFGFF-GSLLNCGEKLKAFSLVNCLSIRDLTTGLPASSHCSALRSLSIR--NCPGFGDAN 458
Query: 409 SLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPL---- 464
+ PQ + + L + + + + +L +++ S N+T+ I +
Sbjct: 459 LAAIGKLCPQLEDIDLCGLKGITESGFLHL--IQSSLVKINFSGCSNLTDRVISAITARN 516
Query: 465 ---LESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQL 521
LE N+T SL S+ N + L L+++ + D + +++ LQ+
Sbjct: 517 GWTLEVLNIDGCSNITDASLVSIAANCQI--LSDLDISKCAISDSGIQALASSDKLKLQI 574
Query: 522 VLLR-CDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMV 564
+ + C +TDK + +V + L +NL C ++ V +V
Sbjct: 575 LSVAGCSMVTDKSLPAIVGLGSTLLGLNLQQCRSISNSTVDFLV 618
Score = 60.8 bits (146), Expect = 2e-09
Identities = 90/411 (21%), Positives = 159/411 (37%), Gaps = 75/411 (18%)
Query: 195 GLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQ 254
GL ++ + L ++L + L + + C L+++ L RC +T G+
Sbjct: 168 GLRSIGRSCPSLGSLSLWNVSTITDNGLLEIAEGCAQLEKLELNRCSTITDKGLVAIAKS 227
Query: 255 KPTLTSLSI-TCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMESLPL 313
P LT L++ C+ RI D+ L +IA L
Sbjct: 228 CPNLTELTLEACS------------------------------RIGDEGLLAIARSCSKL 257
Query: 314 RRLVLSYCPGYTYSGISFLLSKSK-RIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGN 372
+ + + CP GI+ LLS + + L LQ + V ++SL + L++ +
Sbjct: 258 KSVSIKNCPLVRDQGIASLLSNTTCSLAKLKLQMLN------VTDVSLAVVGHYGLSITD 311
Query: 373 CRLLTVSTF----FALITNCPSLTEIN-MNRTNIQGTTIPNSLMDRLVNPQFKSLFLASA 427
L +S F ++ N L ++N + T QG T P K ++ +
Sbjct: 312 LVLAGLSHVSEKGFWVMGNGVGLQKLNSLTITACQGVTDMGLESVGKGCPNMKKAIISKS 371
Query: 428 ACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESC-RKIRHLNLT-CLSLKSLG 485
L D ++ FA +L+ L L + +T+ G L +C K++ +L CLS++ L
Sbjct: 372 PLLSDNGLVSFAKASLSLESLQLEECHRVTQFGFFGSLLNCGEKLKAFSLVNCLSIRDLT 431
Query: 486 TNFDLPD----LEVLNLTNTE-VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNN 540
T L L++ N D L I CP L + L IT+ G +H++ +
Sbjct: 432 TGLPASSHCSALRSLSIRNCPGFGDANLAAIGKLCPQLEDIDLCGLKGITESGFLHLIQS 491
Query: 541 ---------CTQLRE----------------INLDGCPNVQAKVVASMVVS 566
C+ L + +N+DGC N+ + S+ +
Sbjct: 492 SLVKINFSGCSNLTDRVISAITARNGWTLEVLNIDGCSNITDASLVSIAAN 542
Score = 54.3 bits (129), Expect = 2e-07
Identities = 50/214 (23%), Positives = 99/214 (45%), Gaps = 13/214 (6%)
Query: 387 NCPSLTEINMNRTNIQGTTIPNSLMDRLVN-PQFKSLFLASAACLEDQNIIMFAALFPNL 445
+CPSL +++ + T N L++ Q + L L + + D+ ++ A PNL
Sbjct: 175 SCPSLGSLSLWNVS---TITDNGLLEIAEGCAQLEKLELNRCSTITDKGLVAIAKSCPNL 231
Query: 446 QQLHLSCSYNITEEGIRPLLESCRKIRHLNL-TCLSLKSLGTNFDLPD----LEVLNLTN 500
+L L I +EG+ + SC K++ +++ C ++ G L + L L L
Sbjct: 232 TELTLEACSRIGDEGLLAIARSCSKLKSVSIKNCPLVRDQGIASLLSNTTCSLAKLKLQM 291
Query: 501 TEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREIN---LDGCPNVQA 557
V D +L ++ + ++ LVL ++++KG V+ N L+++N + C V
Sbjct: 292 LNVTDVSLAVVGHYGLSITDLVLAGLSHVSEKG-FWVMGNGVGLQKLNSLTITACQGVTD 350
Query: 558 KVVASMVVSRPSLRKIHVPPNFPLSDRNRKLFSR 591
+ S+ P+++K + + LSD F++
Sbjct: 351 MGLESVGKGCPNMKKAIISKSPLLSDNGLVSFAK 384
Score = 52.0 bits (123), Expect = 9e-07
Identities = 35/116 (30%), Positives = 58/116 (49%), Gaps = 4/116 (3%)
Query: 467 SCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTE-VDDEALYIISNRCPALLQLVLLR 525
S R ++ L L+S+G + P L L+L N + D L I+ C L +L L R
Sbjct: 155 SIRGSNSAKVSDLGLRSIGRS--CPSLGSLSLWNVSTITDNGLLEIAEGCAQLEKLELNR 212
Query: 526 CDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHVPPNFPL 581
C ITDKG++ + +C L E+ L+ C + + + ++ S L+ + + N PL
Sbjct: 213 CSTITDKGLVAIAKSCPNLTELTLEACSRIGDEGLLAIARSCSKLKSVSI-KNCPL 267
Score = 50.8 bits (120), Expect = 2e-06
Identities = 35/140 (25%), Positives = 63/140 (45%), Gaps = 10/140 (7%)
Query: 441 LFPNLQQLHLSCSYNITEEGIRPLLESC-------RKIRHLNLTCLSLKSLGTNFDLPDL 493
L +++Q + ITE+G E C +K + L +++ + G L L
Sbjct: 98 LVSSIRQKEIDVPSKITEDGDD--CEGCLSRSLDGKKATDVRLAAIAVGTAGRG-GLGKL 154
Query: 494 EVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCP 553
+ + +V D L I CP+L L L ITD G++ + C QL ++ L+ C
Sbjct: 155 SIRGSNSAKVSDLGLRSIGRSCPSLGSLSLWNVSTITDNGLLEIAEGCAQLEKLELNRCS 214
Query: 554 NVQAKVVASMVVSRPSLRKI 573
+ K + ++ S P+L ++
Sbjct: 215 TITDKGLVAIAKSCPNLTEL 234
Score = 46.2 bits (108), Expect = 5e-05
Identities = 65/286 (22%), Positives = 114/286 (39%), Gaps = 22/286 (7%)
Query: 286 KGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKS-----KRIQ 340
+G S L G + +D L++IA+ + L G + +S L +S +
Sbjct: 121 EGCLSRSLDGKKATDVRLAAIAVGTAGRGGLGKLSIRGSNSAKVSDLGLRSIGRSCPSLG 180
Query: 341 HLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTN 400
L L + D+ + E++ L L L C +T A+ +CP+LTE+ + +
Sbjct: 181 SLSLWNVSTITDNGLLEIAEGCAQLEKLELNRCSTITDKGLVAIAKSCPNLTELTLEACS 240
Query: 401 IQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPN----LQQLHLSCSYNI 456
G L + KS+ + + + DQ I A+L N L +L L N+
Sbjct: 241 RIGD--EGLLAIARSCSKLKSVSIKNCPLVRDQGI---ASLLSNTTCSLAKLKLQ-MLNV 294
Query: 457 TEEGIRPLLESCRKIRHLNLTCLSLKS------LGTNFDLPDLEVLNLTNTE-VDDEALY 509
T+ + + I L L LS S +G L L L +T + V D L
Sbjct: 295 TDVSLAVVGHYGLSITDLVLAGLSHVSEKGFWVMGNGVGLQKLNSLTITACQGVTDMGLE 354
Query: 510 IISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNV 555
+ CP + + ++ + ++D G++ L + L+ C V
Sbjct: 355 SVGKGCPNMKKAIISKSPLLSDNGLVSFAKASLSLESLQLEECHRV 400
>At3g58530 unknown protein
Length = 353
Score = 67.0 bits (162), Expect = 3e-11
Identities = 59/240 (24%), Positives = 116/240 (47%), Gaps = 13/240 (5%)
Query: 333 LSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLS----LNLGNCRLLTVSTFFALITNC 388
L + ++++H++L++ + D + + D L LNL C+ ++ + A+ + C
Sbjct: 77 LPRYRQVKHINLEFAQGVVDSHLKLVKTECPDALLSLEWLNLNVCQKISDNGIEAITSIC 136
Query: 389 PSLTEINMN-RTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQ 447
P L ++ + I N + + L L+ L D+++ + A +P+L+
Sbjct: 137 PKLKVFSIYWNVRVTDAGIRNLVKNCR---HITDLNLSGCKSLTDKSMQLVAESYPDLES 193
Query: 448 LHLSCSYNITEEGIRPLLESCRKIRHLNLTCLS---LKSLGTNFDLPDLEVLNLTNTE-V 503
L+++ IT++G+ +L+ C ++ LNL LS K+ L DL L++ + +
Sbjct: 194 LNITRCVKITDDGLLQVLQKCFSLQTLNLYALSGFTDKAYMKISLLADLRFLDICGAQNI 253
Query: 504 DDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASM 563
DE + I+ +C L L L C ITD GV + N+CT L ++L G V + + ++
Sbjct: 254 SDEGIGHIA-KCNKLESLNLTWCVRITDAGVNTIANSCTSLEFLSLFGIVGVTDRCLETL 312
Score = 51.2 bits (121), Expect = 2e-06
Identities = 69/299 (23%), Positives = 119/299 (39%), Gaps = 58/299 (19%)
Query: 280 DSLLSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRI 339
D+LLSL+ L L +ISD + +I L+ + + T +GI L+ + I
Sbjct: 108 DALLSLEWLN--LNVCQKISDNGIEAITSICPKLKVFSIYWNVRVTDAGIRNLVKNCRHI 165
Query: 340 QHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRT 399
L+L L D + ++ DL SLN+ C +T ++ C SL +N+
Sbjct: 166 TDLNLSGCKSLTDKSMQLVAESYPDLESLNITRCVKITDDGLLQVLQKCFSLQTLNLYA- 224
Query: 400 NIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEE 459
+ G T M +L +L+ L + + NI++E
Sbjct: 225 -LSGFT---------------------------DKAYMKISLLADLRFLDICGAQNISDE 256
Query: 460 GIRPLLESCRKIRHLNLT-CLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPAL 518
GI + + C K+ LNLT C+ + G N I+N C +L
Sbjct: 257 GIGHIAK-CNKLESLNLTWCVRITDAGVN----------------------TIANSCTSL 293
Query: 519 LQLVLLRCDYITDKGVMHVVNNC-TQLREINLDGCPNVQAKVVASMVVSRPSLR--KIH 574
L L +TD+ + + C T L ++++GC ++ + ++ P L K+H
Sbjct: 294 EFLSLFGIVGVTDRCLETLSQTCSTTLTTLDVNGCTGIKRRSREELLQMFPRLTCFKVH 352
Score = 49.3 bits (116), Expect = 6e-06
Identities = 34/126 (26%), Positives = 62/126 (48%), Gaps = 13/126 (10%)
Query: 469 RKIRHLNLTCLS------LKSLGTNFDLPD----LEVLNLTNTE-VDDEALYIISNRCPA 517
R+++H+NL LK + T + PD LE LNL + + D + I++ CP
Sbjct: 81 RQVKHINLEFAQGVVDSHLKLVKT--ECPDALLSLEWLNLNVCQKISDNGIEAITSICPK 138
Query: 518 LLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHVPP 577
L + +TD G+ ++V NC + ++NL GC ++ K + + S P L +++
Sbjct: 139 LKVFSIYWNVRVTDAGIRNLVKNCRHITDLNLSGCKSLTDKSMQLVAESYPDLESLNITR 198
Query: 578 NFPLSD 583
++D
Sbjct: 199 CVKITD 204
Score = 48.1 bits (113), Expect = 1e-05
Identities = 59/249 (23%), Positives = 104/249 (41%), Gaps = 33/249 (13%)
Query: 168 DTFPLLEELDLAYPSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFK 227
D LE L+L KI ++ G+EA++ KL+ ++ ++ + + +L K
Sbjct: 108 DALLSLEWLNLNVCQKISDN-------GIEAITSICPKLKVFSIYWNVRVTDAGIRNLVK 160
Query: 228 NCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKG 287
NC+ + ++ L C+ LT + L P L SL+IT V + L + SL+
Sbjct: 161 NCRHITDLNLSGCKSLTDKSMQLVAESYPDLESLNITRCVKITDDGLL-QVLQKCFSLQT 219
Query: 288 LTSLLLTGF-----------------------RISDQFLSSIAMESLPLRRLVLSYCPGY 324
L L+GF ISD+ + IA + L L L++C
Sbjct: 220 LNLYALSGFTDKAYMKISLLADLRFLDICGAQNISDEGIGHIA-KCNKLESLNLTWCVRI 278
Query: 325 TYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLG-DLLSLNLGNCRLLTVSTFFA 383
T +G++ + + ++ L L + D C+ LS L +L++ C + +
Sbjct: 279 TDAGVNTIANSCTSLEFLSLFGIVGVTDRCLETLSQTCSTTLTTLDVNGCTGIKRRSREE 338
Query: 384 LITNCPSLT 392
L+ P LT
Sbjct: 339 LLQMFPRLT 347
Score = 32.7 bits (73), Expect = 0.59
Identities = 36/146 (24%), Positives = 63/146 (42%), Gaps = 19/146 (13%)
Query: 136 GLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKI--------INH 187
G+R +N +T L S SL + L+A+++P LE L++ KI +
Sbjct: 154 GIRNLVKNCRHITDLNLSGCKSLTDKSMQLVAESYPDLESLNITRCVKITDDGLLQVLQK 213
Query: 188 THATFSTGLEALS----------LALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVIL 237
+ + L ALS L LR +++ ++ + H+ K C L+ + L
Sbjct: 214 CFSLQTLNLYALSGFTDKAYMKISLLADLRFLDICGAQNISDEGIGHIAK-CNKLESLNL 272
Query: 238 LRCEQLTIAGVDLALLQKPTLTSLSI 263
C ++T AGV+ +L LS+
Sbjct: 273 TWCVRITDAGVNTIANSCTSLEFLSL 298
>At5g23340 unknown protein
Length = 405
Score = 65.1 bits (157), Expect = 1e-10
Identities = 85/326 (26%), Positives = 147/326 (45%), Gaps = 26/326 (7%)
Query: 83 VLPGLLQRFTKLTSLDL------SYYYGDLDALLTQIS-SFPMLKLTSLNLSNQLILPAN 135
+L L RFT++ LDL S+Y G D+ L IS F L++ LNL N +
Sbjct: 57 MLRRLASRFTQIVELDLSQSISRSFYPGVTDSDLAVISEGFKFLRV--LNLHNCKGITDT 114
Query: 136 GLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTG 195
GL + + ++ L L S L+ + +A+ L L LA I + +
Sbjct: 115 GLASIGRCLSLLQFLDVSYCRKLSDKGLSAVAEGCHDLRALHLAGCRFITDES------- 167
Query: 196 LEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGV-DLALLQ 254
L++LS L + L + + L+ L K C+ ++ + + +C + AGV +A
Sbjct: 168 LKSLSERCRDLEALGLQGCTNITDSGLADLVKGCRKIKSLDINKCSNVGDAGVSSVAKAC 227
Query: 255 KPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFR-ISDQFLSSIAMESL-P 312
+L +L + G E ++S K L +L++ G R ISD+ + +A
Sbjct: 228 ASSLKTLKLLDCYKVGNESISS----LAQFCKNLETLIIGGCRDISDESIMLLADSCKDS 283
Query: 313 LRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSL--FLGDLLSLNL 370
L+ L + +C + S +S +L + K ++ LD+ + + D +L LG L L +
Sbjct: 284 LKNLRMDWCLNISDSSLSCILKQCKNLEALDIGCCEEVTDTAFRDLGSDDVLG-LKVLKV 342
Query: 371 GNCRLLTVSTFFALITNCPSLTEINM 396
NC +TV+ L+ C SL I++
Sbjct: 343 SNCTKITVTGIGKLLDKCSSLEYIDV 368
Score = 57.8 bits (138), Expect = 2e-08
Identities = 60/285 (21%), Positives = 119/285 (41%), Gaps = 61/285 (21%)
Query: 298 ISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAE 357
++D L+ I+ LR L L C G T +G++ + +Q LD+ Y L+D ++
Sbjct: 85 VTDSDLAVISEGFKFLRVLNLHNCKGITDTGLASIGRCLSLLQFLDVSYCRKLSDKGLSA 144
Query: 358 LSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNP 417
++ DL +L+L CR +T + +L C L + +QG T
Sbjct: 145 VAEGCHDLRALHLAGCRFITDESLKSLSERCRDLEAL-----GLQGCT------------ 187
Query: 418 QFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLT 477
NIT+ G+ L++ CRKI+ L++
Sbjct: 188 -------------------------------------NITDSGLADLVKGCRKIKSLDIN 210
Query: 478 -CLSLKSLGTN----FDLPDLEVLNLTNT-EVDDEALYIISNRCPALLQLVLLRCDYITD 531
C ++ G + L+ L L + +V +E++ ++ C L L++ C I+D
Sbjct: 211 KCSNVGDAGVSSVAKACASSLKTLKLLDCYKVGNESISSLAQFCKNLETLIIGGCRDISD 270
Query: 532 KGVMHVVNNC-TQLREINLDGCPNVQAKVVASMVVSRPSLRKIHV 575
+ +M + ++C L+ + +D C N+ ++ ++ +L + +
Sbjct: 271 ESIMLLADSCKDSLKNLRMDWCLNISDSSLSCILKQCKNLEALDI 315
Score = 52.0 bits (123), Expect = 9e-07
Identities = 80/356 (22%), Positives = 148/356 (41%), Gaps = 55/356 (15%)
Query: 137 LRAFSQNITTLTSLICSNLIS------LNSTDIHLIADTFPLLEELDLAYPSKIINHTHA 190
LR + T + L S IS + +D+ +I++ F L L+L ++
Sbjct: 58 LRRLASRFTQIVELDLSQSISRSFYPGVTDSDLAVISEGFKFLRVLNL-------HNCKG 110
Query: 191 TFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDL 250
TGL ++ L L+ +++SY L+ LS + + C L+ + L C +T
Sbjct: 111 ITDTGLASIGRCLSLLQFLDVSYCRKLSDKGLSAVAEGCHDLRALHLAGCRFIT------ 164
Query: 251 ALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMES 310
+L SLS C LE L L+G T+ I+D L+ +
Sbjct: 165 ----DESLKSLSERC---RDLE---------ALGLQGCTN-------ITDSGLADLVKGC 201
Query: 311 LPLRRLVLSYCPGYTYSGISFLLSK-SKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLN 369
++ L ++ C +G+S + + ++ L L + + ++ L+ F +L +L
Sbjct: 202 RKIKSLDINKCSNVGDAGVSSVAKACASSLKTLKLLDCYKVGNESISSLAQFCKNLETLI 261
Query: 370 LGNCRLLTVSTFFALITNC-PSLTEINMN-RTNIQGTTIPNSLMDRLVNPQFKSLFLASA 427
+G CR ++ + L +C SL + M+ NI +++ + Q K+L
Sbjct: 262 IGGCRDISDESIMLLADSCKDSLKNLRMDWCLNISDSSL------SCILKQCKNLEALDI 315
Query: 428 ACLEDQNIIMFAALFPN----LQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCL 479
C E+ F L + L+ L +S IT GI LL+ C + ++++ L
Sbjct: 316 GCCEEVTDTAFRDLGSDDVLGLKVLKVSNCTKITVTGIGKLLDKCSSLEYIDVRSL 371
Score = 45.4 bits (106), Expect = 9e-05
Identities = 37/150 (24%), Positives = 69/150 (45%), Gaps = 15/150 (10%)
Query: 439 AALFPNLQQLHLSCSYN------ITEEGIRPLLESCRKIRHLNL------TCLSLKSLGT 486
A+ F + +L LS S + +T+ + + E + +R LNL T L S+G
Sbjct: 62 ASRFTQIVELDLSQSISRSFYPGVTDSDLAVISEGFKFLRVLNLHNCKGITDTGLASIGR 121
Query: 487 NFDLPDLEVLNLTNT-EVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLR 545
L L+ L+++ ++ D+ L ++ C L L L C +ITD+ + + C L
Sbjct: 122 CLSL--LQFLDVSYCRKLSDKGLSAVAEGCHDLRALHLAGCRFITDESLKSLSERCRDLE 179
Query: 546 EINLDGCPNVQAKVVASMVVSRPSLRKIHV 575
+ L GC N+ +A +V ++ + +
Sbjct: 180 ALGLQGCTNITDSGLADLVKGCRKIKSLDI 209
Score = 28.9 bits (63), Expect = 8.5
Identities = 22/88 (25%), Positives = 40/88 (45%)
Query: 503 VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVAS 562
V D L +IS L L L C ITD G+ + + L+ +++ C + K +++
Sbjct: 85 VTDSDLAVISEGFKFLRVLNLHNCKGITDTGLASIGRCLSLLQFLDVSYCRKLSDKGLSA 144
Query: 563 MVVSRPSLRKIHVPPNFPLSDRNRKLFS 590
+ LR +H+ ++D + K S
Sbjct: 145 VAEGCHDLRALHLAGCRFITDESLKSLS 172
>At4g33210 unknown protein
Length = 990
Score = 65.1 bits (157), Expect = 1e-10
Identities = 130/561 (23%), Positives = 216/561 (38%), Gaps = 94/561 (16%)
Query: 94 LTSLDLSYYYGDLDALLTQISSFPMLKLTSLNLSNQLILPANGLRAFSQNITTLTSLICS 153
LT L L G A +T I++ P L++ L+ N L + L + +++ + C
Sbjct: 444 LTVLKLHSCEGITSASMTWIANSPALEVLELDNCNLLTTVSLHL----SRLQSISLVHCR 499
Query: 154 NLISLNSTDIHLIADTF---PLLEELDLAYPSKIINHTHATFSTGLEALSLALIKLRKVN 210
LN I L + T P L + + S + L L L L++V+
Sbjct: 500 KFTDLNLQSIMLSSITVSNCPALRRITIT--SNALRRLALQKQENLTTLVLQCHSLQEVD 557
Query: 211 LSYHGYLNGTLLSHLFKN---CKFLQEVILLRCEQLTIAG------VDLALLQKPTLTSL 261
LS L+ ++ +F + C L+ +IL CE LT L+L+ +TSL
Sbjct: 558 LSDCESLSNSVCK-IFSDDGGCPMLKSLILDNCESLTAVRFCNSSLASLSLVGCRAVTSL 616
Query: 262 SITCT-----VTTGLEHLTSHFIDSL-------------------------LSLKGLTSL 291
+ C G +HL + F + L LKG L
Sbjct: 617 ELKCPRIEQICLDGCDHLETAFFQPVALRSLNLGICPKLSVLNIEAPYMVSLELKGCGVL 676
Query: 292 --------LLTGF------RISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSK 337
LLT ++ D LS+ + LVL CP G+S L +
Sbjct: 677 SEASIMCPLLTSLDASFCSQLRDDCLSATTASCPLIESLVLMSCPSIGSDGLSSL-NGLP 735
Query: 338 RIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITN--CPSLTEIN 395
+ LDL YT +N V + + L L L C+ LT S+ L P+L E++
Sbjct: 736 NLTVLDLSYTFLMNLEPVFKSCI---QLKVLKLQACKYLTDSSLEPLYKEGALPALEELD 792
Query: 396 MNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYN 455
++ + T I D L+ L+ C+ ++ + + S +
Sbjct: 793 LSYGTLCQTAI-----DDLLACCTHLTHLSLNGCVNMHDLDWGSTSVHLFDYFGVYSSSD 847
Query: 456 ITEEGIRP---LLES-----CRKIRHL---------NLTCLSLKSLGTNFDLPDLEVLNL 498
T+E LL++ C IR + +L+ L+L SL N DL NL
Sbjct: 848 NTQEPAETANRLLQNLNCVGCPNIRKVLIPPAARFYHLSTLNL-SLSVNLKEVDLTCSNL 906
Query: 499 TNTEVDDE-ALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQA 557
+ + +L ++ CP L L L C+ + + GV ++ C+ L ++L CP + +
Sbjct: 907 VLLNLSNCCSLEVLKLGCPRLASLFLQSCN-MDEAGVEAAISGCSSLETLDLRFCPKISS 965
Query: 558 KVVASMVVSRPSLRKIHVPPN 578
++ PSL+++ PN
Sbjct: 966 VSMSKFRTVCPSLKRVFSSPN 986
Score = 49.3 bits (116), Expect = 6e-06
Identities = 53/228 (23%), Positives = 100/228 (43%), Gaps = 40/228 (17%)
Query: 368 LNLGNCRLLTVSTFFALITNCPSLTEINM------NRTNIQGTTIPNSLMDRLVNPQF-- 419
LN N R+ ++ F + + P+ TE+N+ N ++ T +L +
Sbjct: 236 LNFENIRI-SMEQFENMCSRYPNATEVNVYGAPAVNALAMKAATTLRNLEVLTIGKGHIS 294
Query: 420 KSLFLASAACLEDQNIIMFAALFPN-LQQLHLSCSYNITEEGIRPL-LESCRKIRHLNLT 477
+S F A C +++ + A+ N Q++HLS + +R L + CR +R L++
Sbjct: 295 ESFFQALGECNMLRSVTVSDAILGNGAQEIHLS------HDRLRELKITKCRVMR-LSIR 347
Query: 478 CLSLKSLGTN--------FDLPDLEVLNLTNT-EVDDEALYIISNRCPALLQLVLLRCDY 528
C L+SL + P L++L++ + ++ D A+ + CP L L + C
Sbjct: 348 CPQLRSLSLKRSNMSQAMLNCPLLQLLDIASCHKLLDAAIRSAAISCPQLESLDVSNCSC 407
Query: 529 ITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKIHVP 576
++D+ + + C L +N CPN+ SL +H+P
Sbjct: 408 VSDETLREIAQACANLHILNASYCPNI-------------SLESVHLP 442
Score = 45.8 bits (107), Expect = 7e-05
Identities = 73/336 (21%), Positives = 135/336 (39%), Gaps = 41/336 (12%)
Query: 250 LALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTG---------FRISD 300
LA+ TL +L + +T G H++ F +L L S+ ++ +S
Sbjct: 272 LAMKAATTLRNLEV---LTIGKGHISESFFQALGECNMLRSVTVSDAILGNGAQEIHLSH 328
Query: 301 QFLSSIAMESLPLRRLVLSYCP-----GYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCV 355
L + + + RL + CP S +S + +Q LD+ L D +
Sbjct: 329 DRLRELKITKCRVMRLSIR-CPQLRSLSLKRSNMSQAMLNCPLLQLLDIASCHKLLDAAI 387
Query: 356 AELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLV 415
++ L SL++ NC ++ T + C +L +N + PN ++ +
Sbjct: 388 RSAAISCPQLESLDVSNCSCVSDETLREIAQACANLHILN-------ASYCPNISLESVH 440
Query: 416 NPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHL-SCSYNITEEGIRPLLES-----CR 469
P L L S + + + + A P L+ L L +C+ T L+S CR
Sbjct: 441 LPMLTVLKLHSCEGITSAS-MTWIANSPALEVLELDNCNLLTTVSLHLSRLQSISLVHCR 499
Query: 470 KIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEV------DDEALYIISNRCPALLQLVL 523
K LNL + L S+ T + P L + +T+ + E L + +C +L ++ L
Sbjct: 500 KFTDLNLQSIMLSSI-TVSNCPALRRITITSNALRRLALQKQENLTTLVLQCHSLQEVDL 558
Query: 524 LRCDYITDK--GVMHVVNNCTQLREINLDGCPNVQA 557
C+ +++ + C L+ + LD C ++ A
Sbjct: 559 SDCESLSNSVCKIFSDDGGCPMLKSLILDNCESLTA 594
>At3g50080 unknown protein
Length = 522
Score = 65.1 bits (157), Expect = 1e-10
Identities = 113/423 (26%), Positives = 178/423 (41%), Gaps = 66/423 (15%)
Query: 27 DLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVT--NRHKFCLTILYPALPVL 84
+LPDD +F+ L D RKR S+ SK +L V NRH+ L LP L
Sbjct: 43 NLPDDCLAHIFQFLSAGD----RKR----CSLVSKRWLLVDGQNRHRLSLDAKSEILPFL 94
Query: 85 PGLLQRFTKLTSLDL-----SYYYGDLDALLTQISSFPMLKLTSLNLSNQLILPANGLRA 139
P + RF +T L L S+ D +AL I S L + L + G+ +
Sbjct: 95 PCIFNRFDSVTKLALRCDRRSFSLSD-EALF--IVSIRCSNLIRVKLRGCREITDLGMES 151
Query: 140 FSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTGLEAL 199
F++N +L L C + + + I+ + + +LEEL L K I H E +
Sbjct: 152 FARNCKSLRKLSCGS-CTFGAKGINAMLEHCKVLEELSL----KRIRGLHEL----AEPI 202
Query: 200 SLAL-IKLRKVNLS--YHGYLNGTLLSHLFKNCKFLQEVILLRC-----EQLTIAGVDLA 251
L+L LR V L +G + G+L++ + L++V ++RC + G +
Sbjct: 203 KLSLSASLRSVFLKELVNGQVFGSLVA-----TRTLKKVKIIRCLGNWDRVFEMNGNGNS 257
Query: 252 LLQKPTLTSLSITCTVTTGLEHL----TSHFID----SLLSL-------KGLTSLLLTGF 296
L + L L +T G+ T H + S L L K L L + G+
Sbjct: 258 SLTEIRLERLQVTDIGLFGISKCSNLETLHIVKTPDCSNLGLASVVERCKLLRKLHIDGW 317
Query: 297 ---RISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLND- 352
RI DQ L S+A L L+ LVL TY +S + S K+++ L L + + D
Sbjct: 318 RVKRIGDQGLMSVAKHCLNLQELVLIGVDA-TYMSLSAIASNCKKLERLALCGSGTIGDA 376
Query: 353 --HCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSL 410
C+AE + L + C + V AL CP L ++ + + ++ + L
Sbjct: 377 EIGCIAEKCV---TLRKFCIKGCLISDVGV-QALALGCPKLVKLKVKKCSLVTGEVREWL 432
Query: 411 MDR 413
+R
Sbjct: 433 RER 435
Score = 45.4 bits (106), Expect = 9e-05
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 4/75 (5%)
Query: 503 VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGC----PNVQAK 558
+ DEAL+I+S RC L+++ L C ITD G+ NC LR+++ C + A
Sbjct: 118 LSDEALFIVSIRCSNLIRVKLRGCREITDLGMESFARNCKSLRKLSCGSCTFGAKGINAM 177
Query: 559 VVASMVVSRPSLRKI 573
+ V+ SL++I
Sbjct: 178 LEHCKVLEELSLKRI 192
Score = 40.8 bits (94), Expect = 0.002
Identities = 32/138 (23%), Positives = 63/138 (45%), Gaps = 7/138 (5%)
Query: 444 NLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLP------DLEVLN 497
NL+ LH+ + + + G+ ++E C+ +R L++ +K +G + +L+ L
Sbjct: 282 NLETLHIVKTPDCSNLGLASVVERCKLLRKLHIDGWRVKRIGDQGLMSVAKHCLNLQELV 341
Query: 498 LTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQA 557
L + +L I++ C L +L L I D + + C LR+ + GC +
Sbjct: 342 LIGVDATYMSLSAIASNCKKLERLALCGSGTIGDAEIGCIAEKCVTLRKFCIKGC-LISD 400
Query: 558 KVVASMVVSRPSLRKIHV 575
V ++ + P L K+ V
Sbjct: 401 VGVQALALGCPKLVKLKV 418
Score = 34.3 bits (77), Expect = 0.20
Identities = 45/219 (20%), Positives = 89/219 (40%), Gaps = 37/219 (16%)
Query: 350 LNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNS 409
L+D + +S+ +L+ + L CR +T + NC SL +++ I
Sbjct: 118 LSDEALFIVSIRCSNLIRVKLRGCREITDLGMESFARNCKSLRKLSCGSCTFGAKGI--- 174
Query: 410 LMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPL----- 464
+ L LE+ ++ L + + LS S ++ ++ L
Sbjct: 175 -----------NAMLEHCKVLEELSLKRIRGLHELAEPIKLSLSASLRSVFLKELVNGQV 223
Query: 465 ---LESCRKIRHLNLTCLSLKSLGTNFDL---------PDLEVLNLTNTEVDDEALYIIS 512
L + R ++ + + ++ LG N+D L + L +V D L+ IS
Sbjct: 224 FGSLVATRTLKKVKI----IRCLG-NWDRVFEMNGNGNSSLTEIRLERLQVTDIGLFGIS 278
Query: 513 NRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDG 551
+C L L +++ ++ G+ VV C LR++++DG
Sbjct: 279 -KCSNLETLHIVKTPDCSNLGLASVVERCKLLRKLHIDG 316
Score = 28.9 bits (63), Expect = 8.5
Identities = 39/174 (22%), Positives = 70/174 (39%), Gaps = 22/174 (12%)
Query: 331 FLLSKSKRIQHLDLQYTDFLNDHCVAELSLFL--GDLLSLNLGNCRLLTVSTFFALITNC 388
FL +S + D +T L D C+A + FL GD +L + R L V
Sbjct: 25 FLDCESIGFEDGDYDFTANLPDDCLAHIFQFLSAGDRKRCSLVSKRWLLVDGQ------- 77
Query: 389 PSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAAC------LEDQNIIMFAALF 442
N +R ++ + + + N +F S+ + C L D+ + + +
Sbjct: 78 ------NRHRLSLDAKSEILPFLPCIFN-RFDSVTKLALRCDRRSFSLSDEALFIVSIRC 130
Query: 443 PNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVL 496
NL ++ L IT+ G+ +C+ +R L+ + + G N L +VL
Sbjct: 131 SNLIRVKLRGCREITDLGMESFARNCKSLRKLSCGSCTFGAKGINAMLEHCKVL 184
>At1g77000 unknown protein
Length = 360
Score = 63.2 bits (152), Expect = 4e-10
Identities = 45/157 (28%), Positives = 81/157 (50%), Gaps = 10/157 (6%)
Query: 418 QFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLT 477
+ + L L+ ++ + D ++ A NL +L+LS + ++ + L CRK++ LNL
Sbjct: 118 ELQDLDLSKSSKITDHSLYSLARGCTNLTKLNLSGCTSFSDTALAHLTRFCRKLKILNLC 177
Query: 478 -CL------SLKSLGTNFDLPDLEVLNLTNTE-VDDEALYIISNRCPALLQLVLLRCDYI 529
C+ +L+++G N + L+ LNL E + D+ + ++ CP L L L C I
Sbjct: 178 GCVEAVSDNTLQAIGENCN--QLQSLNLGWCENISDDGVMSLAYGCPDLRTLDLCSCVLI 235
Query: 530 TDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVS 566
TD+ V+ + N C LR + L C N+ + + S+ S
Sbjct: 236 TDESVVALANRCIHLRSLGLYYCRNITDRAMYSLAQS 272
Score = 49.3 bits (116), Expect = 6e-06
Identities = 40/145 (27%), Positives = 57/145 (38%), Gaps = 22/145 (15%)
Query: 430 LEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFD 489
LED + A LQ L LS S IT+ + L C + LNL+
Sbjct: 104 LEDNAVEAIANHCHELQDLDLSKSSKITDHSLYSLARGCTNLTKLNLSGC---------- 153
Query: 490 LPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRC-DYITDKGVMHVVNNCTQLREIN 548
T D AL ++ C L L L C + ++D + + NC QL+ +N
Sbjct: 154 -----------TSFSDTALAHLTRFCRKLKILNLCGCVEAVSDNTLQAIGENCNQLQSLN 202
Query: 549 LDGCPNVQAKVVASMVVSRPSLRKI 573
L C N+ V S+ P LR +
Sbjct: 203 LGWCENISDDGVMSLAYGCPDLRTL 227
Score = 45.1 bits (105), Expect = 1e-04
Identities = 60/241 (24%), Positives = 100/241 (40%), Gaps = 31/241 (12%)
Query: 158 LNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYL 217
L + IA+ L++LDL+ SKI +H+ L +L+ L K+NLS
Sbjct: 104 LEDNAVEAIANHCHELQDLDLSKSSKITDHS-------LYSLARGCTNLTKLNLSGCTSF 156
Query: 218 NGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSITCTVTTGL-----E 272
+ T L+HL + C+ L+ + L C + + TL ++ C L E
Sbjct: 157 SDTALAHLTRFCRKLKILNLCGC---------VEAVSDNTLQAIGENCNQLQSLNLGWCE 207
Query: 273 HLTSHFIDSL-LSLKGLTSL-LLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGIS 330
+++ + SL L +L L + I+D+ + ++A + LR L L YC T +
Sbjct: 208 NISDDGVMSLAYGCPDLRTLDLCSCVLITDESVVALANRCIHLRSLGLYYCRNITDRAMY 267
Query: 331 FLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPS 390
L + +H + V + L SLN+ C LT S A+ P+
Sbjct: 268 SLAQSGVKNKHEMWR--------AVKKGKFDEEGLRSLNISQCTYLTPSAVQAVCDTFPA 319
Query: 391 L 391
L
Sbjct: 320 L 320
Score = 44.3 bits (103), Expect = 2e-04
Identities = 30/99 (30%), Positives = 45/99 (45%), Gaps = 5/99 (5%)
Query: 465 LESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLL 524
L C+K N+ L L SL F VL +++D A+ I+N C L L L
Sbjct: 71 LSWCKK----NMNSLVL-SLAPKFVKLQTLVLRQDKPQLEDNAVEAIANHCHELQDLDLS 125
Query: 525 RCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASM 563
+ ITD + + CT L ++NL GC + +A +
Sbjct: 126 KSSKITDHSLYSLARGCTNLTKLNLSGCTSFSDTALAHL 164
Score = 36.2 bits (82), Expect = 0.053
Identities = 57/249 (22%), Positives = 96/249 (37%), Gaps = 21/249 (8%)
Query: 217 LNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLSIT-CTV--TTGLEH 273
L + + +C LQ++ L + ++T + LT L+++ CT T L H
Sbjct: 104 LEDNAVEAIANHCHELQDLDLSKSSKITDHSLYSLARGCTNLTKLNLSGCTSFSDTALAH 163
Query: 274 LTSHFID-SLLSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFL 332
LT +L+L G +SD L +I L+ L L +C + G+ L
Sbjct: 164 LTRFCRKLKILNLCGCVEA------VSDNTLQAIGENCNQLQSLNLGWCENISDDGVMSL 217
Query: 333 LSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFAL----ITNC 388
++ LDL + D V L+ L SL L CR +T ++L + N
Sbjct: 218 AYGCPDLRTLDLCSCVLITDESVVALANRCIHLRSLGLYYCRNITDRAMYSLAQSGVKNK 277
Query: 389 PSL-TEINMNRTNIQGTTIPNSLMDRLVNPQ----FKSLFLASAACLEDQNIIMFAALFP 443
+ + + + +G N + P F A C +++M L
Sbjct: 278 HEMWRAVKKGKFDEEGLRSLNISQCTYLTPSAVQAVCDTFPALHTCSGRHSLVMSGCL-- 335
Query: 444 NLQQLHLSC 452
NLQ +H +C
Sbjct: 336 NLQSVHCAC 344
>At1g21410 unknown protein
Length = 360
Score = 63.2 bits (152), Expect = 4e-10
Identities = 54/212 (25%), Positives = 102/212 (47%), Gaps = 15/212 (7%)
Query: 365 LLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNP--QFKSL 422
L L L C S +L+ L +N+ + Q + ++ ++ + N + + L
Sbjct: 66 LTRLRLSWCNNNMNSLVLSLVPKFVKLQTLNLRQDKPQ---LEDNAVEAIANHCHELQEL 122
Query: 423 FLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNL------ 476
L+ + + D+++ A P+L +L+LS + ++ I L CRK++ LNL
Sbjct: 123 DLSKSLKITDRSLYALAHGCPDLTKLNLSGCTSFSDTAIAYLTRFCRKLKVLNLCGCVKA 182
Query: 477 -TCLSLKSLGTNFDLPDLEVLNLTNTE-VDDEALYIISNRCPALLQLVLLRCDYITDKGV 534
T +L+++G N + ++ LNL E + D+ + ++ CP L L L C ITD+ V
Sbjct: 183 VTDNALEAIGNNCN--QMQSLNLGWCENISDDGVMSLAYGCPDLRTLDLCGCVLITDESV 240
Query: 535 MHVVNNCTQLREINLDGCPNVQAKVVASMVVS 566
+ + + C LR + L C N+ + + S+ S
Sbjct: 241 VALADWCVHLRSLGLYYCRNITDRAMYSLAQS 272
Score = 57.0 bits (136), Expect = 3e-08
Identities = 51/185 (27%), Positives = 76/185 (40%), Gaps = 25/185 (13%)
Query: 391 LTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHL 450
LT + ++ N ++ SL+ + V Q +L LED + A LQ+L L
Sbjct: 66 LTRLRLSWCNNNMNSLVLSLVPKFVKLQTLNL-RQDKPQLEDNAVEAIANHCHELQELDL 124
Query: 451 SCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTN-TEVDDEALY 509
S S IT+ + L C PDL LNL+ T D A+
Sbjct: 125 SKSLKITDRSLYALAHGC----------------------PDLTKLNLSGCTSFSDTAIA 162
Query: 510 IISNRCPALLQLVLLRC-DYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRP 568
++ C L L L C +TD + + NNC Q++ +NL C N+ V S+ P
Sbjct: 163 YLTRFCRKLKVLNLCGCVKAVTDNALEAIGNNCNQMQSLNLGWCENISDDGVMSLAYGCP 222
Query: 569 SLRKI 573
LR +
Sbjct: 223 DLRTL 227
Score = 38.5 bits (88), Expect = 0.011
Identities = 62/264 (23%), Positives = 106/264 (39%), Gaps = 24/264 (9%)
Query: 204 IKLRKVNLSYHG-YLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTSLS 262
+KL+ +NL L + + +C LQE+ L + ++T + P LT L+
Sbjct: 90 VKLQTLNLRQDKPQLEDNAVEAIANHCHELQELDLSKSLKITDRSLYALAHGCPDLTKLN 149
Query: 263 IT-CTV--TTGLEHLTSHFIDSL--LSLKGLTSLLLTGFRISDQFLSSIAMESLPLRRLV 317
++ CT T + +LT F L L+L G ++D L +I ++ L
Sbjct: 150 LSGCTSFSDTAIAYLT-RFCRKLKVLNLCGCVKA------VTDNALEAIGNNCNQMQSLN 202
Query: 318 LSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLT 377
L +C + G+ L ++ LDL + D V L+ + L SL L CR +T
Sbjct: 203 LGWCENISDDGVMSLAYGCPDLRTLDLCGCVLITDESVVALADWCVHLRSLGLYYCRNIT 262
Query: 378 VSTFFAL----ITNCP-SLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSL----FLASAA 428
++L + N P S + + + +G N + P F A
Sbjct: 263 DRAMYSLAQSGVKNKPGSWKSVKKGKYDEEGLRSLNISQCTALTPSAVQAVCDSFPALHT 322
Query: 429 CLEDQNIIMFAALFPNLQQLHLSC 452
C +++M L NL +H +C
Sbjct: 323 CSGRHSLVMSGCL--NLTTVHCAC 344
>At3g26810 transport inhibitor response 1-like protein
Length = 575
Score = 60.5 bits (145), Expect = 3e-09
Identities = 76/302 (25%), Positives = 126/302 (41%), Gaps = 58/302 (19%)
Query: 287 GLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQY 346
GL L L ++D+ L ++ + + LVL C G+T G++ + + + ++ LDLQ
Sbjct: 101 GLEELRLKRMVVTDESLELLSRSFVNFKSLVLVSCEGFTTDGLASIAANCRHLRDLDLQE 160
Query: 347 TDFLNDHCVAELSLF---LGDLLSLNL----GNCRLLTVSTFFALITNCPSLTEINMNRT 399
+ ++DH LS F L++LN G L+ + L+ P+L + +NR
Sbjct: 161 NE-IDDHRGQWLSCFPDTCTTLVTLNFACLEGETNLVALE---RLVARSPNLKSLKLNR- 215
Query: 400 NIQGTTIPNSLMDRLV--NPQFKSLFLAS-----------------AACLEDQNIIMFAA 440
+P + RL+ PQ L + S C +++ F
Sbjct: 216 -----AVPLDALARLMACAPQIVDLGVGSYENDPDSESYLKLMAVIKKCTSLRSLSGFLE 270
Query: 441 LFP-----------NLQQLHLSCSYNITEEGIRPLLESCRKIRHL-NLTCLSLKSL---- 484
P NL L+LS + I + L++ C+K++ L L + K L
Sbjct: 271 AAPHCLSAFHPICHNLTSLNLSYAAEIHGSHLIKLIQHCKKLQRLWILDSIGDKGLEVVA 330
Query: 485 GTNFDLPDLEV-----LNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVN 539
T +L +L V L NT V +E L IS CP L +L C +T+ ++ V
Sbjct: 331 STCKELQELRVFPSDLLGGGNTAVTEEGLVAISAGCPK-LHSILYFCQQMTNAALVTVAK 389
Query: 540 NC 541
NC
Sbjct: 390 NC 391
Score = 38.5 bits (88), Expect = 0.011
Identities = 20/57 (35%), Positives = 30/57 (52%)
Query: 493 LEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINL 549
LE L L V DE+L ++S LVL+ C+ T G+ + NC LR+++L
Sbjct: 102 LEELRLKRMVVTDESLELLSRSFVNFKSLVLVSCEGFTTDGLASIAANCRHLRDLDL 158
Score = 36.2 bits (82), Expect = 0.053
Identities = 116/557 (20%), Positives = 204/557 (35%), Gaps = 106/557 (19%)
Query: 29 PDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTILYPALPVLPGLL 88
PD++ E VF + + H+ R SL S + + +R K + Y P LL
Sbjct: 5 PDEVIEHVFDFVTS-----HKDRNAISLVCKSWYKIERYSRQKVFIGNCYAINP--ERLL 57
Query: 89 QRFTKLTSLDLSY--YYGDLDALLTQISSFPMLKLTSLNLSN---------QLILPANGL 137
+RF L SL L ++ D + + + F + + +L S ++++ L
Sbjct: 58 RRFPCLKSLTLKGKPHFADFNLVPHEWGGFVLPWIEALARSRVGLEELRLKRMVVTDESL 117
Query: 138 RAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFS---- 193
S++ SL+ + + + IA L +LDL ++I +H S
Sbjct: 118 ELLSRSFVNFKSLVLVSCEGFTTDGLASIAANCRHLRDLDLQ-ENEIDDHRGQWLSCFPD 176
Query: 194 --TGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLA 251
T L L+ A ++ G N L L L+ + L R L +A
Sbjct: 177 TCTTLVTLNFACLE---------GETNLVALERLVARSPNLKSLKLNRAVPLDALARLMA 227
Query: 252 LLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLL-LTGF-RISDQFLSSIAME 309
P + L + + + ++ + +K TSL L+GF + LS+
Sbjct: 228 CA--PQIVDLGVGSYEN---DPDSESYLKLMAVIKKCTSLRSLSGFLEAAPHCLSAFHPI 282
Query: 310 SLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDH--------C--VAELS 359
L L LSY S + L+ K++Q L + D + D C + EL
Sbjct: 283 CHNLTSLNLSYAAEIHGSHLIKLIQHCKKLQRLWI--LDSIGDKGLEVVASTCKELQELR 340
Query: 360 LFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEIN---MNRTNIQGTTIPNSLMDRLVN 416
+F DLL G +T A+ CP L I TN T+ + +
Sbjct: 341 VFPSDLLG---GGNTAVTEEGLVAISAGCPKLHSILYFCQQMTNAALVTVAKNCPN---- 393
Query: 417 PQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNL 476
F+ C+ + N P +EG ++++C+ +R L+L
Sbjct: 394 ------FIRFRLCILEPNKPDHVTSQP-------------LDEGFGAIVKACKSLRRLSL 434
Query: 477 TCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMH 536
+ L D LYI L+++ + TDKG+++
Sbjct: 435 SGLL----------------------TDQVFLYI--GMYANQLEMLSIAFAGDTDKGMLY 470
Query: 537 VVNNCTQLREINLDGCP 553
V+N C +++++ + P
Sbjct: 471 VLNGCKKMKKLEIRDSP 487
>At1g80570 unknown protein
Length = 467
Score = 60.1 bits (144), Expect = 3e-09
Identities = 110/476 (23%), Positives = 187/476 (39%), Gaps = 82/476 (17%)
Query: 27 DLPDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTI---LYPALPV 83
+LPD + + L DD + SLS++ K F S+ N ++ L I L PA
Sbjct: 3 ELPDHLVWDILSKLHTTDDRN-------SLSLSCKRFFSLDNEQRYSLRIGCGLVPASDA 55
Query: 84 LPGLLQRFTKLTSLDLSYYYGDLDALLTQISSFPMLKLTSLNLSNQLILPANGLRAFSQN 143
L L +RF L+ +++ Y G + L Q+ +L LT+ N
Sbjct: 56 LLSLCRRFPNLSKVEI-IYSGWMSKLGKQVDDQGLLVLTT-------------------N 95
Query: 144 ITTLTSLICSNLISLNSTDI-HLIADTFPLLEELDLAYPSKIINHTHATFSTGLEALSLA 202
+LT L S + I HL + P L L L + +I TG LSLA
Sbjct: 96 CHSLTDLTLSFCTFITDVGIGHL--SSCPELSSLKLNFAPRI---------TGCGVLSLA 144
Query: 203 L--IKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQKPTLTS 260
+ KLR+++L + F + L+E+ + C I DL L+
Sbjct: 145 VGCKKLRRLHLIRCLNVASVEWLEYFGKLETLEELCIKNCR--AIGEGDLIKLRNSWRKL 202
Query: 261 LSITCTVTTGLEHL-------TSHFIDSLLSLKGLTSLLLTGFRISD-QFLSSIAMESLP 312
S+ V ++ + L+ L L L I+ + L+ +
Sbjct: 203 TSLQFEVDANYRYMKVYDQLDVERWPKQLVPCDSLVELSLGNCIIAPGRGLACVLRNCKN 262
Query: 313 LRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQY-TDF-----------LNDHCVAELSL 360
L +L L C G + S I L+ K+ ++ + L+ +DF L D ++ ++
Sbjct: 263 LEKLHLDMCTGVSDSDIIALVQKASHLRSISLRVPSDFTLPLLNNITLRLTDESLSAIAQ 322
Query: 361 FLGDLLSLNLG-------NCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSL-MD 412
L S + + T+ LI CP + ++ + N + M+
Sbjct: 323 HCSKLESFKISFSDGEFPSLFSFTLQGIITLIQKCP------VRELSLDHVCVFNDMGME 376
Query: 413 RLVNPQ-FKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLES 467
L + Q + L L + D+ +I+ + FP+L L LS +T++G+RPL+ S
Sbjct: 377 ALCSAQKLEILELVHCQEVSDEGLILVSQ-FPSLNVLKLSKCLGVTDDGMRPLVGS 431
Score = 52.0 bits (123), Expect = 9e-07
Identities = 24/74 (32%), Positives = 43/74 (57%), Gaps = 1/74 (1%)
Query: 502 EVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVA 561
+VDD+ L +++ C +L L L C +ITD G+ H +++C +L + L+ P + V
Sbjct: 83 QVDDQGLLVLTTNCHSLTDLTLSFCTFITDVGIGH-LSSCPELSSLKLNFAPRITGCGVL 141
Query: 562 SMVVSRPSLRKIHV 575
S+ V LR++H+
Sbjct: 142 SLAVGCKKLRRLHL 155
Score = 44.7 bits (104), Expect = 2e-04
Identities = 56/219 (25%), Positives = 93/219 (41%), Gaps = 29/219 (13%)
Query: 384 LITNCPSLTEINMNRTNIQGTTIPNSLMDRLVN-PQFKSLFLASAACLEDQNIIMFAALF 442
L TNC SLT++ ++ T I + + L + P+ SL L A + ++ A
Sbjct: 92 LTTNCHSLTDLTLSFC----TFITDVGIGHLSSCPELSSLKLNFAPRITGCGVLSLAVGC 147
Query: 443 PNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCL-SLKSLGTNFDLPDLEVL--NLT 499
L++LHL N+ LE K+ L C+ + +++G DL L LT
Sbjct: 148 KKLRRLHLIRCLNVASV---EWLEYFGKLETLEELCIKNCRAIGEG-DLIKLRNSWRKLT 203
Query: 500 NTEVDDEALYIISN---------------RCPALLQLVLLRCDYITDKGVMHVVNNCTQL 544
+ + + +A Y C +L++L L C +G+ V+ NC L
Sbjct: 204 SLQFEVDANYRYMKVYDQLDVERWPKQLVPCDSLVELSLGNCIIAPGRGLACVLRNCKNL 263
Query: 545 REINLDGCPNVQAKVVASMVVSRPSLRKI--HVPPNFPL 581
+++LD C V + ++V LR I VP +F L
Sbjct: 264 EKLHLDMCTGVSDSDIIALVQKASHLRSISLRVPSDFTL 302
>At5g25350 leucine-rich repeats containing protein
Length = 623
Score = 58.5 bits (140), Expect = 1e-08
Identities = 91/421 (21%), Positives = 172/421 (40%), Gaps = 40/421 (9%)
Query: 136 GLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFSTG 195
GL A + +L + NL +++ + IA + P++E+LDL+ I + +G
Sbjct: 158 GLGAVAHGCPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGITD-------SG 210
Query: 196 LEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQK 255
L A++ + L + + + L + + C L+ + + C ++ GV L Q
Sbjct: 211 LVAIAENCVNLSDLTIDSCSGVGNEGLRAIARRCVNLRSISIRSCPRIGDQGVAFLLAQA 270
Query: 256 PT-LTSLSITCTVTTGLE-HLTSHFIDSL--LSLKGLTSLLLTGFRISDQFLSSIAMESL 311
+ LT + + +GL + H+ ++ L L GL + GF ++ A
Sbjct: 271 GSYLTKVKLQMLNVSGLSLAVIGHYGAAVTDLVLHGLQGVNEKGF-----WVMGNAKGLK 325
Query: 312 PLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLG 371
L+ L + C G T G+ + + ++H+ L ++ + L+ L SL L
Sbjct: 326 KLKSLSVMSCRGMTDVGLEAVGNGCPDLKHVSLNKCLLVSGKGLVALAKSALSLESLKLE 385
Query: 372 NCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAAC-- 429
C + + NC S + + N G + NS L +P SL S C
Sbjct: 386 ECHRINQFGLMGFLMNCGSKLKA-FSLANCLGISDFNS-ESSLPSPSCSSLRSLSIRCCP 443
Query: 430 -LEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHLNLTCLSLKSLGTNF 488
D ++ LQ + L +T+ G+R LL+S ++ L ++L
Sbjct: 444 GFGDASLAFLGKFCHQLQDVELCGLNGVTDAGVRELLQS----NNVGLVKVNLS------ 493
Query: 489 DLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREIN 548
E +N+++ V A+ + R L L L C IT+ ++ V NC + +++
Sbjct: 494 -----ECINVSDNTV--SAISVCHGR--TLESLNLDGCKNITNASLVAVAKNCYSVNDLD 544
Query: 549 L 549
+
Sbjct: 545 I 545
Score = 49.7 bits (117), Expect = 5e-06
Identities = 27/86 (31%), Positives = 45/86 (51%), Gaps = 1/86 (1%)
Query: 491 PDLEVLNLTNTE-VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINL 549
P L +++L N V D L I+ CP + +L L RC ITD G++ + NC L ++ +
Sbjct: 167 PSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGITDSGLVAIAENCVNLSDLTI 226
Query: 550 DGCPNVQAKVVASMVVSRPSLRKIHV 575
D C V + + ++ +LR I +
Sbjct: 227 DSCSGVGNEGLRAIARRCVNLRSISI 252
Score = 48.5 bits (114), Expect = 1e-05
Identities = 83/418 (19%), Positives = 162/418 (37%), Gaps = 94/418 (22%)
Query: 195 GLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLALLQ 254
GL A++ LR V+L ++ LS + ++C ++++ L RC +T +G
Sbjct: 158 GLGAVAHGCPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGITDSG------- 210
Query: 255 KPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFRISDQFLSSIAMESLPLR 314
L +++ C +L+ IDS + ++ L +IA + LR
Sbjct: 211 ---LVAIAENCV------NLSDLTIDSCSG-------------VGNEGLRAIARRCVNLR 248
Query: 315 RLVLSYCPGYTYSGISFLLSKSK------RIQHLDLQYTDF--LNDHCVAELSLFLGDLL 366
+ + CP G++FLL+++ ++Q L++ + + A L L L
Sbjct: 249 SISIRSCPRIGDQGVAFLLAQAGSYLTKVKLQMLNVSGLSLAVIGHYGAAVTDLVLHGLQ 308
Query: 367 SLN------LGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFK 420
+N +GN + L ++++ C +T++ + + N P K
Sbjct: 309 GVNEKGFWVMGNAKGLKKLKSLSVMS-CRGMTDVGLE-------AVGNGC------PDLK 354
Query: 421 SLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESC-RKIRHLNLT-C 478
+ L + + ++ A +L+ L L + I + G+ L +C K++ +L C
Sbjct: 355 HVSLNKCLLVSGKGLVALAKSALSLESLKLEECHRINQFGLMGFLMNCGSKLKAFSLANC 414
Query: 479 LSLKSLGTNFDLPDLEVLNLTNTEV------DDEALYIISNRCPALLQLVLLRCDYITDK 532
L + + LP +L + + D +L + C L + L + +TD
Sbjct: 415 LGISDFNSESSLPSPSCSSLRSLSIRCCPGFGDASLAFLGKFCHQLQDVELCGLNGVTDA 474
Query: 533 GVMHVVNN------------CTQ----------------LREINLDGCPNV-QAKVVA 561
GV ++ + C L +NLDGC N+ A +VA
Sbjct: 475 GVRELLQSNNVGLVKVNLSECINVSDNTVSAISVCHGRTLESLNLDGCKNITNASLVA 532
Score = 45.8 bits (107), Expect = 7e-05
Identities = 52/227 (22%), Positives = 104/227 (44%), Gaps = 27/227 (11%)
Query: 350 LNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNS 409
++D ++E++ + L+L C +T S A+ NC +L+++ ++ + G +
Sbjct: 180 VSDLGLSEIARSCPMIEKLDLSRCPGITDSGLVAIAENCVNLSDLTIDSCSGVGNEGLRA 239
Query: 410 LMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIRPLLESCR 469
+ R VN +S+ + S + DQ + A + SY +T+
Sbjct: 240 IARRCVN--LRSISIRSCPRIGDQGVAFLLAQ---------AGSY-LTKV---------- 277
Query: 470 KIRHLNLTCLSLKSLGT-NFDLPDLEVLNLTNTEVDDEALYIISNR--CPALLQLVLLRC 526
K++ LN++ LSL +G + DL + L V+++ +++ N L L ++ C
Sbjct: 278 KLQMLNVSGLSLAVIGHYGAAVTDLVLHGLQG--VNEKGFWVMGNAKGLKKLKSLSVMSC 335
Query: 527 DYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLRKI 573
+TD G+ V N C L+ ++L+ C V K + ++ S SL +
Sbjct: 336 RGMTDVGLEAVGNGCPDLKHVSLNKCLLVSGKGLVALAKSALSLESL 382
Score = 45.4 bits (106), Expect = 9e-05
Identities = 81/365 (22%), Positives = 154/365 (42%), Gaps = 21/365 (5%)
Query: 109 LLTQISSF-PMLKLTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIA 167
LL Q S+ +KL LN+S L +T L+ L +N ++
Sbjct: 266 LLAQAGSYLTKVKLQMLNVSGL------SLAVIGHYGAAVTDLVLHGLQGVNEKGFWVMG 319
Query: 168 DTFPLLEELDLAYPSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFK 227
+ L + L+ S GLEA+ L+ V+L+ ++G L L K
Sbjct: 320 NAKGLKKLKSLSVMS-----CRGMTDVGLEAVGNGCPDLKHVSLNKCLLVSGKGLVALAK 374
Query: 228 NCKFLQEVILLRCEQLTIAGVDLALLQKPT-LTSLSIT-CTVTTGLEHLTSHFIDSLLSL 285
+ L+ + L C ++ G+ L+ + L + S+ C + +S S SL
Sbjct: 375 SALSLESLKLEECHRINQFGLMGFLMNCGSKLKAFSLANCLGISDFNSESSLPSPSCSSL 434
Query: 286 KGLTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQ 345
+ L+ GF D L+ + L+ + L G T +G+ LL +S + + +
Sbjct: 435 RSLSIRCCPGF--GDASLAFLGKFCHQLQDVELCGLNGVTDAGVRELL-QSNNVGLVKVN 491
Query: 346 YTDFLN--DHCVAELSLFLGDLL-SLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQ 402
++ +N D+ V+ +S+ G L SLNL C+ +T ++ A+ NC S+ +++++ T +
Sbjct: 492 LSECINVSDNTVSAISVCHGRTLESLNLDGCKNITNASLVAVAKNCYSVNDLDISNTLVS 551
Query: 403 GTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGIR 462
I +L + + L + + + D++ L L L++ I+ +
Sbjct: 552 DHGI-KALASSPNHLNLQVLSIGGCSSITDKSKACIQKLGRTLLGLNIQRCGRISSSTVD 610
Query: 463 PLLES 467
LLE+
Sbjct: 611 TLLEN 615
Score = 34.7 bits (78), Expect = 0.16
Identities = 13/55 (23%), Positives = 30/55 (53%)
Query: 501 TEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNV 555
++V D L +++ CP+L + L ++D G+ + +C + +++L CP +
Sbjct: 152 SKVTDVGLGAVAHGCPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSRCPGI 206
>At3g62980 transport inhibitor response 1 (TIR1)
Length = 594
Score = 57.0 bits (136), Expect = 3e-08
Identities = 67/275 (24%), Positives = 116/275 (41%), Gaps = 49/275 (17%)
Query: 288 LTSLLLTGFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYT 347
L + L ++D L IA + LVLS C G++ G++ + + + ++ LDL+ +
Sbjct: 107 LEEIRLKRMVVTDDCLELIAKSFKNFKVLVLSSCEGFSTDGLAAIAATCRNLKELDLRES 166
Query: 348 DF--LNDHCVAELSLFLGDLLSLNLGNCRL--LTVSTFFALITNCPSLTEINMNRTNIQG 403
D ++ H ++ L+SLN+ +C ++ S L+T CP+L + +NR
Sbjct: 167 DVDDVSGHWLSHFPDTYTSLVSLNI-SCLASEVSFSALERLVTRCPNLKSLKLNR----- 220
Query: 404 TTIPNSLMDRLVN--PQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNITEEGI 461
+P + L+ PQ + L A + P++ G+
Sbjct: 221 -AVPLEKLATLLQRAPQLEELGTGGYT----------AEVRPDVY------------SGL 257
Query: 462 RPLLESCRKIRHLNLTCLSLKSLGTNFDLP-------DLEVLNLTNTEVDDEALYIISNR 514
L C+++R CLS LP L LNL+ V L + +
Sbjct: 258 SVALSGCKELR-----CLSGFWDAVPAYLPAVYSVCSRLTTLNLSYATVQSYDLVKLLCQ 312
Query: 515 CPALLQLVLLRCDYITDKGVMHVVNNCTQLREINL 549
CP L +L +L DYI D G+ + + C LRE+ +
Sbjct: 313 CPKLQRLWVL--DYIEDAGLEVLASTCKDLRELRV 345
>At1g12820 transport inhibitor response 1, putative
Length = 577
Score = 56.2 bits (134), Expect = 5e-08
Identities = 94/413 (22%), Positives = 165/413 (39%), Gaps = 74/413 (17%)
Query: 180 YPSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLR 239
+P ++I H ++ + S++L V S+H + NC + L+R
Sbjct: 4 FPDEVIEHVFDFVASHKDRNSISL-----VCKSWHKIERFSRKEVFIGNCYAINPERLIR 58
Query: 240 ----CEQLTIAGVDLALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLL-SLKGLTSLLLT 294
+ LT+ G KP ++ G H +I++L S GL L L
Sbjct: 59 RFPCLKSLTLKG-------KPHFADFNLVPHEWGGFVH---PWIEALARSRVGLEELRLK 108
Query: 295 GFRISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHC 354
++D+ L ++ + LVL C G+T G++ + + + ++ LDLQ + ++DH
Sbjct: 109 RMVVTDESLDLLSRSFANFKSLVLVSCEGFTTDGLASIAANCRHLRELDLQENE-IDDHR 167
Query: 355 VAELSLF---LGDLLSLNLGNCRLLT-VSTFFALITNCPSLTEINMNRTNIQGTTIPNSL 410
L+ F L+SLN + T V+ L+ P+L + +NR +P
Sbjct: 168 GQWLNCFPDSCTTLMSLNFACLKGETNVAALERLVARSPNLKSLKLNR------AVPLDA 221
Query: 411 MDRLVN--PQFKSLFLAS--------------------------AACLEDQNIIM--FAA 440
+ RL++ PQ L + S + LE + + F
Sbjct: 222 LARLMSCAPQLVDLGVGSYENEPDPESFAKLMTAIKKYTSLRSLSGFLEVAPLCLPAFYP 281
Query: 441 LFPNLQQLHLSCSYNITEEGIRPLLESCRKIRHL-NLTCLSLKSL----GTNFDLPDLEV 495
+ NL L+LS + I + L++ C++++ L L + K L T +L +L V
Sbjct: 282 ICQNLISLNLSYAAEIQGNHLIKLIQLCKRLQRLWILDSIGDKGLAVVAATCKELQELRV 341
Query: 496 L-------NLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNC 541
N V + L IS CP L +L C +T+ ++ V NC
Sbjct: 342 FPSDVHGEEDNNASVTEVGLVAISAGCPK-LHSILYFCKQMTNAALIAVAKNC 393
Score = 39.7 bits (91), Expect = 0.005
Identities = 21/57 (36%), Positives = 30/57 (51%)
Query: 493 LEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINL 549
LE L L V DE+L ++S LVL+ C+ T G+ + NC LRE++L
Sbjct: 102 LEELRLKRMVVTDESLDLLSRSFANFKSLVLVSCEGFTTDGLASIAANCRHLRELDL 158
Score = 35.0 bits (79), Expect = 0.12
Identities = 119/577 (20%), Positives = 213/577 (36%), Gaps = 124/577 (21%)
Query: 29 PDDIWERVFRLLKNNDDDDHRKRYLKSLSVASKHFLSVTNRHKFCLTILYPALPVLPGLL 88
PD++ E VF + + H+ R SL S H + +R + + Y P L+
Sbjct: 5 PDEVIEHVFDFVAS-----HKDRNSISLVCKSWHKIERFSRKEVFIGNCYAINP--ERLI 57
Query: 89 QRFTKLTSLDLSY--YYGDLDALLTQISSFPMLKLTSLNLSN---------QLILPANGL 137
+RF L SL L ++ D + + + F + +L S ++++ L
Sbjct: 58 RRFPCLKSLTLKGKPHFADFNLVPHEWGGFVHPWIEALARSRVGLEELRLKRMVVTDESL 117
Query: 138 RAFSQNITTLTSLICSNLISLNSTDIHLIADTFPLLEELDLAYPSKIINHTHATFS---- 193
S++ SL+ + + + IA L ELDL ++I +H +
Sbjct: 118 DLLSRSFANFKSLVLVSCEGFTTDGLASIAANCRHLRELDLQ-ENEIDDHRGQWLNCFPD 176
Query: 194 --TGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLTIAGVDLA 251
T L +L+ A +K G N L L L+ + L R L LA
Sbjct: 177 SCTTLMSLNFACLK---------GETNVAALERLVARSPNLKSLKLNRAVPLDA----LA 223
Query: 252 LLQK--PTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLL-LTGF-RISDQFLSSIA 307
L P L L + E F + ++K TSL L+GF ++ L +
Sbjct: 224 RLMSCAPQLVDLGVGSYEN---EPDPESFAKLMTAIKKYTSLRSLSGFLEVAPLCLPAFY 280
Query: 308 MESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLS 367
L L LSY + + L+ KR+Q L + D + D +A ++
Sbjct: 281 PICQNLISLNLSYAAEIQGNHLIKLIQLCKRLQRLWI--LDSIGDKGLAVVA-------- 330
Query: 368 LNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASA 427
C L E+ + +++ G N+ + + L SA
Sbjct: 331 ------------------ATCKELQELRVFPSDVHGEEDNNASVTEV------GLVAISA 366
Query: 428 ACLEDQNIIMF------AALF------PNLQQLHLSC---------SYNITEEGIRPLLE 466
C + +I+ F AAL PN + L ++ +EG +++
Sbjct: 367 GCPKLHSILYFCKQMTNAALIAVAKNCPNFIRFRLCILEPHKPDHITFQSLDEGFGAIVQ 426
Query: 467 SCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRC 526
+C+ +R L+++ L D LYI L+++ +
Sbjct: 427 ACKGLRRLSVSGLL----------------------TDQVFLYI--GMYAEQLEMLSIAF 462
Query: 527 DYITDKGVMHVVNNCTQLREINLDGCPNVQAKVVASM 563
TDKG+++V+N C ++R++ + P A ++A +
Sbjct: 463 AGDTDKGMLYVLNGCKKMRKLEIRDSPFGNAALLADV 499
>At5g67140 unknown protein
Length = 228
Score = 52.4 bits (124), Expect = 7e-07
Identities = 38/132 (28%), Positives = 69/132 (51%), Gaps = 11/132 (8%)
Query: 430 LEDQNIIMFAALFPNLQQLHLSCSY---NITEEGIRPLLESCRKIRHLNLTCLSLKSLGT 486
++D + L NL++L +S S +IT+ G+ + S R + +LN L + T
Sbjct: 59 MDDDSTSRLVHLAFNLKELDISRSRWGCHITDNGLYQIA-SARCVSNLNSVSLWGMTAIT 117
Query: 487 NFDL-------PDLEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVN 539
+ + L+ LN+ T + DE+L+ I+ RC L + + C ++T++G++ +VN
Sbjct: 118 DSGVVQLISRTSSLQHLNIGGTFITDESLFAIAERCHQLKTIGMWCCRHVTERGLLVLVN 177
Query: 540 NCTQLREINLDG 551
C +L INL G
Sbjct: 178 KCRKLESINLWG 189
Score = 37.4 bits (85), Expect = 0.024
Identities = 35/130 (26%), Positives = 57/130 (42%), Gaps = 4/130 (3%)
Query: 275 TSHFIDSLLSLKGLT-SLLLTGFRISDQFLSSIAMESLP--LRRLVLSYCPGYTYSGISF 331
TS + +LK L S G I+D L IA L + L T SG+
Sbjct: 64 TSRLVHLAFNLKELDISRSRWGCHITDNGLYQIASARCVSNLNSVSLWGMTAITDSGVVQ 123
Query: 332 LLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSL 391
L+S++ +QHL++ T F+ D + ++ L ++ + CR +T L+ C L
Sbjct: 124 LISRTSSLQHLNIGGT-FITDESLFAIAERCHQLKTIGMWCCRHVTERGLLVLVNKCRKL 182
Query: 392 TEINMNRTNI 401
IN+ T +
Sbjct: 183 ESINLWGTRV 192
>At4g03190 putative homolog of transport inhibitor response 1
Length = 585
Score = 52.4 bits (124), Expect = 7e-07
Identities = 65/266 (24%), Positives = 115/266 (42%), Gaps = 51/266 (19%)
Query: 298 ISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSKRIQHLDLQYT-------DFL 350
++D+ L IA + LVL+ C G++ GI+ + + + ++ L+L+ D+L
Sbjct: 113 VTDECLEKIAASFKDFKVLVLTSCEGFSTDGIAAIAATCRNLRVLELRECIVEDLGGDWL 172
Query: 351 NDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMNRTNIQGTTIPNSL 410
+ + SL D L+ + +S L++ P+L + +N P
Sbjct: 173 SYFPESSTSLVSLDFSCLD----SEVKISDLERLVSRSPNLKSLKLN---------PAVT 219
Query: 411 MDRLVN-----PQFKSLFLAS-AACLEDQNIIMFAALFPNLQQLH-LSCSYNITEEGIRP 463
+D LV+ PQ L S AA L+ + + F N +QL LS +++ E +
Sbjct: 220 LDGLVSLLRCAPQLTELGTGSFAAQLKPEAFSKLSEAFSNCKQLQSLSGLWDVLPEYLPA 279
Query: 464 LLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVL 523
L C + LNL+ +++ +PDL L RC L +L +
Sbjct: 280 LYSVCPGLTSLNLSYATVR-------MPDLVEL---------------LRRCSKLQKLWV 317
Query: 524 LRCDYITDKGVMHVVNNCTQLREINL 549
+ D I DKG+ V + C +LRE+ +
Sbjct: 318 M--DLIEDKGLEAVASYCKELRELRV 341
Score = 28.9 bits (63), Expect = 8.5
Identities = 17/60 (28%), Positives = 25/60 (41%)
Query: 493 LEVLNLTNTEVDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGC 552
LE + + V DE L I+ LVL C+ + G+ + C LR + L C
Sbjct: 103 LEEIRMKRMVVTDECLEKIAASFKDFKVLVLTSCEGFSTDGIAAIAATCRNLRVLELREC 162
>At1g55590 unknown protein
Length = 607
Score = 52.0 bits (123), Expect = 9e-07
Identities = 66/273 (24%), Positives = 120/273 (43%), Gaps = 38/273 (13%)
Query: 132 LPANGLRA--FSQNITTLTSL-ICSN----LISLNSTDIHLIADTFPLLEELDLAYPSKI 184
L GL+A F Q +T+L+ + C N +N I L+++ LE + L K+
Sbjct: 265 LTYTGLQALGFCQQLTSLSLVRTCYNRKISFKRINDMGIFLLSEACKGLESVRLGGFPKV 324
Query: 185 INHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLRCEQLT 244
+ G +L + L+K + L+ + + LQEV L C +T
Sbjct: 325 SD-------AGFASLLHSCRNLKKFEVRGAFLLSDLAFHDVTGSSCSLQEVRLSTCPLIT 377
Query: 245 IAGV-------DLALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLSLKGLTSLLLTGFR 297
V +L +L + S+S +C ++S+ +L+ LTSL L G
Sbjct: 378 SEAVKKLGLCGNLEVLDLGSCKSISDSC-------------LNSVSALRKLTSLNLAGAD 424
Query: 298 ISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSK----SKRIQHLDLQYTDFLNDH 353
++D + ++ +P+ +L L C + GIS+LL+ SK + LDL + ++D
Sbjct: 425 VTDSGMLALGKSDVPITQLSLRGCRRVSDRGISYLLNNEGTISKTLSTLDLGHMPGISDR 484
Query: 354 CVAELSLFLGDLLSLNLGNCRLLTVSTFFALIT 386
+ ++ L L++ +C +T S+ +L T
Sbjct: 485 AIHTITHCCKALTELSIRSCFHVTDSSIESLAT 517
Score = 45.4 bits (106), Expect = 9e-05
Identities = 124/544 (22%), Positives = 207/544 (37%), Gaps = 93/544 (17%)
Query: 94 LTSLDLSYYYGD---LDALLTQISSFPMLKLTSLNLSNQLILPANG--------LRA--F 140
LTSLDLS + D L+ +L L L L L+ + G LR
Sbjct: 44 LTSLDLSVFSPDDETLNHVLRGCIGLSSLTLNCLRLNAASVRGVLGPHLRELHLLRCSLL 103
Query: 141 SQNITTLTSLICSNL-------ISLNSTDIHL-----IADTFPLLEELDLAYPSKIINHT 188
S + T +C NL L+S D+ + + P LE L L +++ T
Sbjct: 104 SSTVLTYIGTLCPNLRVLTLEMADLDSPDVFQSNLTQMLNGCPYLESLQLNIRGILVDAT 163
Query: 189 -----HATFSTGLEALSLA-LIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQE-------- 234
+ L+AL L L++ + L + GT LS N L
Sbjct: 164 AFQSVRFSLPETLKALRLQPLLESEAILLMNRFKVTGTYLSQPDYNSALLSPSPSFTLQS 223
Query: 235 ---VILLRCEQLTIAG-------VDLALLQKPTLTSLSITCTVTTGLEHLTSHFIDSLLS 284
V+ L ++L IA V L L +P TGL+ L F L S
Sbjct: 224 LSLVLDLISDRLIIAITGSLPQLVKLDLEDRPEKEPFPDNDLTYTGLQALG--FCQQLTS 281
Query: 285 LKGLTSLLLTGF-------RISDQFLSSIAMESLPLRRLVLSYCPGYTYSGISFLLSKSK 337
L SL+ T + RI+D + ++ L + L P + +G + LL +
Sbjct: 282 L----SLVRTCYNRKISFKRINDMGIFLLSEACKGLESVRLGGFPKVSDAGFASLLHSCR 337
Query: 338 RIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNCPSLTEINMN 397
++ +++ L+D +++ L + L C L+T S + C +L +++
Sbjct: 338 NLKKFEVRGAFLLSDLAFHDVTGSSCSLQEVRLSTCPLIT-SEAVKKLGLCGNLEVLDLG 396
Query: 398 RTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQLHLSCSYNIT 457
+ NS+ + SL LA A + + + + P + QL L ++
Sbjct: 397 SCKSISDSCLNSVS---ALRKLTSLNLAGADVTDSGMLALGKSDVP-ITQLSLRGCRRVS 452
Query: 458 EEGIRPLLESCRKIRHLNLTCLSLKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPA 517
+ GI LL + I K+L T DL + + D A++ I++ C A
Sbjct: 453 DRGISYLLNNEGTIS---------KTLST-LDLGHMP-------GISDRAIHTITHCCKA 495
Query: 518 LLQLVLLRCDYITDKGVMHVV-------NNCTQLREINLDGCPNVQAKVVASMVVSRPSL 570
L +L + C ++TD + + QLR++N+ C V A +S+PS
Sbjct: 496 LTELSIRSCFHVTDSSIESLATWERQAEGGSKQLRKLNVHNC--VSLTTGALRWLSKPSF 553
Query: 571 RKIH 574
+H
Sbjct: 554 AGLH 557
Score = 31.6 bits (70), Expect = 1.3
Identities = 30/101 (29%), Positives = 49/101 (47%), Gaps = 9/101 (8%)
Query: 475 NLTCLS--LKSLGTNFDLPDLEVLNLTNTEVDDEALYIISNRCPALLQLVL--LRCDYIT 530
++ C+S L+S + LP L L+L+ DDE L + C L L L LR + +
Sbjct: 24 SVACVSTTLRSAVVSGVLPSLTSLDLSVFSPDDETLNHVLRGCIGLSSLTLNCLRLNAAS 83
Query: 531 DKGVMHVVNNCTQLREINLDGCPNVQAKVVASMVVSRPSLR 571
+GV+ LRE++L C + + V+ + P+LR
Sbjct: 84 VRGVLG-----PHLRELHLLRCSLLSSTVLTYIGTLCPNLR 119
>At5g40470 unknown protein
Length = 496
Score = 50.1 bits (118), Expect = 4e-06
Identities = 56/261 (21%), Positives = 115/261 (43%), Gaps = 19/261 (7%)
Query: 329 ISFLLSKSKRIQHLDLQYTDFLNDHCVAELSLFLGDLLSLNLGNCRLLTVSTFFALITNC 388
+++L ++++ L L+ + + E+ L L +L + L CR + + C
Sbjct: 219 VTWLWKSCRKLKKLSLRSCGSIGE----EIGLCLKNLEEIELRTCRSIVDVVLLKVSEIC 274
Query: 389 PSLTEINMNRTNIQGTTIPNSLMDRLVNPQFKSLFLASAACLEDQNIIMFAALFPNLQQL 448
SL + ++ + + R + + L L L D +++ AA F L L
Sbjct: 275 ESLKSLLIHDGGSKDGLVQFMSNARCYDT-LERLDLRLPMDLTDDHLVSLAANFKCLSTL 333
Query: 449 HLSCSYNITEEGIRPL-LESCRKIRHLNLTCLS--------LKSLGTNFDLPDLEVLNLT 499
L+ +T ++ L L + L+L + L ++G + L L L+LT
Sbjct: 334 RLTSCIFVTGFSLKALALSFSSSLEELSLLSCNAIERERGLLATIGQH--LGRLRKLDLT 391
Query: 500 NTE--VDDEALYIISNRCPALLQLVLLRCDYITDKGVMHVVNNCTQLREINLDGCPNVQA 557
E D E + ++++ C L+++VL C ++T ++ + NC +L+ +++ GC ++
Sbjct: 392 RNEWLFDKEVVSMLAS-CNGLVEVVLRDCKHLTGAVLVALNKNCVKLKTLDILGCRLIEP 450
Query: 558 KVVASMVVSRPSLRKIHVPPN 578
V V+ L+K+ V N
Sbjct: 451 DDVEGFVMKTQCLKKLVVEEN 471
Score = 31.6 bits (70), Expect = 1.3
Identities = 36/136 (26%), Positives = 62/136 (45%), Gaps = 8/136 (5%)
Query: 121 LTSLNLSNQLILPANGLRAFSQNITTLTSLICSNLISLNSTDIHLIADTFPL-LEELDLA 179
L L+L + L + L + + N L++L ++ I + + +A +F LEEL L
Sbjct: 304 LERLDLRLPMDLTDDHLVSLAANFKCLSTLRLTSCIFVTGFSLKALALSFSSSLEELSLL 363
Query: 180 YPSKIINHTHATFSTGLEALSLALIKLRKVNLSYHGYLNGTLLSHLFKNCKFLQEVILLR 239
+ I + G L +LRK++L+ + +L + + +C L EV+L
Sbjct: 364 SCNAIERERGLLATIGQH-----LGRLRKLDLTRNEWLFDKEVVSMLASCNGLVEVVLRD 418
Query: 240 CEQLTIAGVDLALLQK 255
C+ LT G L L K
Sbjct: 419 CKHLT--GAVLVALNK 432
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,557,580
Number of Sequences: 26719
Number of extensions: 511988
Number of successful extensions: 2325
Number of sequences better than 10.0: 136
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 95
Number of HSP's that attempted gapping in prelim test: 1822
Number of HSP's gapped (non-prelim): 346
length of query: 598
length of database: 11,318,596
effective HSP length: 105
effective length of query: 493
effective length of database: 8,513,101
effective search space: 4196958793
effective search space used: 4196958793
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140915.5


