

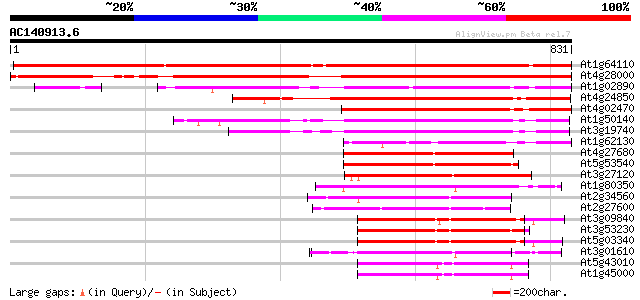
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.6 - phase: 0
(831 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g64110 unknown protein 1107 0.0
At4g28000 putative protein 933 0.0
At1g02890 hypothetical protein 469 e-132
At4g24850 unknown protein (At4g24850) 410 e-114
At4g02470 410 e-114
At1g50140 hypothetical protein, 5' partial 379 e-105
At3g19740 ATPase, putative 379 e-105
At1g62130 hypothetical protein 283 3e-76
At4g27680 unknown protein 256 5e-68
At5g53540 26S proteasome regulatory particle chain RPT6-like pro... 253 3e-67
At3g27120 spastin protein, putative 226 5e-59
At1g80350 CAD ATPase (AAA1) 216 5e-56
At2g34560 katanin like protein 199 7e-51
At2g27600 putative AAA-type ATPase 190 2e-48
At3g09840 putative transitional endoplasmic reticulum ATPase 183 3e-46
At3g53230 CDC48 - like protein 182 7e-46
At5g03340 transitional endoplasmic reticulum ATPase 182 9e-46
At3g01610 putative cell division control protein 166 6e-41
At5g43010 26S proteasome AAA-ATPase subunit RPT4a (gb|AAF22524.1) 162 9e-40
At1g45000 26S proteasome AAA-ATPase subunit RPT4a like protein 159 6e-39
>At1g64110 unknown protein
Length = 824
Score = 1107 bits (2863), Expect = 0.0
Identities = 587/834 (70%), Positives = 675/834 (80%), Gaps = 18/834 (2%)
Query: 6 MLISA-ALSVGVGVGLGLASGQTMFK-PNTYSSSSNALTPDKIENEMLRLVVDGRESNVT 63
ML+SA + VGVGVGLGLASGQ + K SSS+NA+T DK+E E+LR VVDGRES +T
Sbjct: 1 MLLSALGVGVGVGVGLGLASGQAVGKWAGGNSSSNNAVTADKMEKEILRQVVDGRESKIT 60
Query: 64 FDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVLAKAL 123
FD FPYYLSEQTRVLLTSAAYVHLKH + SKYTRNL+PASR ILLSGPAELYQQ+LAKAL
Sbjct: 61 FDEFPYYLSEQTRVLLTSAAYVHLKHFDASKYTRNLSPASRAILLSGPAELYQQMLAKAL 120
Query: 124 THYFEAKLLLFDVTDFSLKIQSRYGSSNCETS-FTRSTSETALARLSDLFGSFALFPQRE 182
H+F+AKLLL DV DF+LKIQS+YGS N E+S F RS SE+AL +LS LF SF++ PQRE
Sbjct: 121 AHFFDAKLLLLDVNDFALKIQSKYGSGNTESSSFKRSPSESALEQLSGLFSSFSILPQRE 180
Query: 183 ENQ--GKIHRQSSGSDLRQMEAEGSYS--KLRRNASASANISSIGLQSNPTNSAPGKHIT 238
E++ G + RQSSG D++ EGS + KLRRN+SA+ANIS++ SN SAP K +
Sbjct: 181 ESKAGGTLRRQSSGVDIKSSSMEGSSNPPKLRRNSSAAANISNLASSSNQV-SAPLKRSS 239
Query: 239 GWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILI 298
W FDEK+L+Q+LYKVL YVSK PIVLY+RD + L RSQR Y LFQ +L KLSGP+LI
Sbjct: 240 SWSFDEKLLVQSLYKVLAYVSKANPIVLYLRDVENFLFRSQRTYNLFQKLLQKLSGPVLI 299
Query: 299 IGSRILD-SGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKN 357
+GSRI+D S + + +DE L+++FPYNI+I+PPEDE+ LVSWKSQ E DM IQ QDN+N
Sbjct: 300 LGSRIVDLSSEDAQEIDEKLSAVFPYNIDIRPPEDETHLVSWKSQLERDMNMIQTQDNRN 359
Query: 358 HIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPC 417
HIMEVL+ NDL C DL+SI DT VLSNYIEEI+VSA+SYH+M NK+PEYRNGKL+I
Sbjct: 360 HIMEVLSENDLICDDLESISFEDTKVLSNYIEEIVVSALSYHLMNNKDPEYRNGKLVISS 419
Query: 418 NSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPT 477
SLSH +F+ GK G R+ LK + + +S K+ + ++KPE KTES V T +
Sbjct: 420 ISLSHGFSLFREGKAGGREKLKQKTKEESS-KEVKAESIKPETKTES----VTTVSSKEE 474
Query: 478 SVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVML 537
++ E P + PDNEFEKRIRPEVIPA EI VTF DIGALDE K+SLQELVML
Sbjct: 475 PEKEAKAEKVTPKAPEVAPDNEFEKRIRPEVIPAEEINVTFKDIGALDEIKESLQELVML 534
Query: 538 PLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGED 597
PLRRPDLF GGLLKPCRGILLFGPPGTGKTMLAKAIA EAGASFINVSMSTITSKWFGED
Sbjct: 535 PLRRPDLFTGGLLKPCRGILLFGPPGTGKTMLAKAIAKEAGASFINVSMSTITSKWFGED 594
Query: 598 EKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSED 657
EKNVRALFTLA+KVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMS+WDGL +K +
Sbjct: 595 EKNVRALFTLASKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSHWDGLMTKPGE 654
Query: 658 RILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELAT 717
RILVLAATNRPFDLDEAIIRRFERRIMVGLP+ ENRE ILRTLLAKEKV LD+KELA
Sbjct: 655 RILVLAATNRPFDLDEAIIRRFERRIMVGLPAVENREKILRTLLAKEKVDENLDYKELAM 714
Query: 718 MTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERV 777
MTEGY+GSDLKNLCTTAAYRPVRELIQQE KD++KKK E + + + KE ERV
Sbjct: 715 MTEGYTGSDLKNLCTTAAYRPVRELIQQERIKDTEKKKQREPTKAGEEDEGKE----ERV 770
Query: 778 ITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
ITLRPLN QDFK AK+QVAASFAAEGAGM EL+QWN+LYGEGGSRKKEQL+YFL
Sbjct: 771 ITLRPLNRQDFKEAKNQVAASFAAEGAGMGELKQWNELYGEGGSRKKEQLTYFL 824
>At4g28000 putative protein
Length = 726
Score = 933 bits (2412), Expect = 0.0
Identities = 511/833 (61%), Positives = 612/833 (73%), Gaps = 109/833 (13%)
Query: 1 MEQKGMLISAALSVGVGVGLGLASGQTMFK-PNTYSSSSNALTPDKIENEMLRLVVDGRE 59
MEQK +L SA L VGVG+G+GLASGQ++ K N S+ + LT +KIE E++R +VDGRE
Sbjct: 1 MEQKSVLFSA-LGVGVGLGIGLASGQSLGKWANGSISAEDGLTGEKIEQELVRQIVDGRE 59
Query: 60 SNVTFDNFPYYLSEQTRVLLTSAAYVHLKHAEVSKYTRNLAPASRTILLSGPAELYQQVL 119
S+VTFD FPYYLSE+TR+LLTSAAYVHLK +++SK+TRNLAP S+ ILLSGPA+ Q +
Sbjct: 60 SSVTFDEFPYYLSEKTRLLLTSAAYVHLKQSDISKHTRNLAPGSKAILLSGPADTEQVWM 119
Query: 120 AKALTHYFEAKLLLFDVTDFSLKIQSRYGSSNCETSFTRSTSETALARLSDLFGSFALFP 179
+ T + R TS L S F + P
Sbjct: 120 RQERT-----------------------------WTLRRHTSGNDLH--SRGFDVTSQPP 148
Query: 180 QREENQGKIHRQSSGSDLRQMEAEGSYSKLRRNASASANISSIGLQSNPTNSAPGKHITG 239
+ + N S+ SD+ + + S + + ++ SAN+
Sbjct: 149 RLKRN------ASAASDMSSISSR-SATSVSASSKRSANLC------------------- 182
Query: 240 WPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQRIYKLFQTMLTKLSGPILII 299
FDE++ +Q+LYKVL+ +S+T PI++Y+RD +K LC+S+R YKLFQ +LTKLSGP+L++
Sbjct: 183 --FDERLFLQSLYKVLVSISETNPIIIYLRDVEK-LCQSERFYKLFQRLLTKLSGPVLVL 239
Query: 300 GSRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHI 359
GSR+L+ ++C+ V E +++LFPYNIEI+PPEDE++L+SWK++FE DMK IQ QDNKNHI
Sbjct: 240 GSRLLEPEDDCQEVGEGISALFPYNIEIRPPEDENQLMSWKTRFEDDMKVIQFQDNKNHI 299
Query: 360 MEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNS 419
EVLAANDL+C DL SIC ADTM LS++IEEI+VSAISYH+M NKEPEY+NG+L+I NS
Sbjct: 300 AEVLAANDLECDDLGSICHADTMFLSSHIEEIVVSAISYHLMNNKEPEYKNGRLVISSNS 359
Query: 420 LSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSV 479
LSH L I Q G+ DSLKL+
Sbjct: 360 LSHGLNILQEGQGCFEDSLKLDTNI----------------------------------- 384
Query: 480 RKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPL 539
SK PDNEFEKRIRPEVIPANEIGVTF+DIG+LDETK+SLQELVMLPL
Sbjct: 385 ----------DSKEVAPDNEFEKRIRPEVIPANEIGVTFADIGSLDETKESLQELVMLPL 434
Query: 540 RRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEK 599
RRPDLF+GGLLKPCRGILLFGPPGTGKTM+AKAIANEAGASFINVSMSTITSKWFGEDEK
Sbjct: 435 RRPDLFKGGLLKPCRGILLFGPPGTGKTMMAKAIANEAGASFINVSMSTITSKWFGEDEK 494
Query: 600 NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRI 659
NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFM++WDGL S + DRI
Sbjct: 495 NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMTHWDGLMSNAGDRI 554
Query: 660 LVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMT 719
LVLAATNRPFDLDEAIIRRFERRIMVGLPS E+RE ILRTLL+KEK LDF+ELA MT
Sbjct: 555 LVLAATNRPFDLDEAIIRRFERRIMVGLPSVESREKILRTLLSKEKTE-NLDFQELAQMT 613
Query: 720 EGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQ-NSQDAQDAKEEVEQERVI 778
+GYSGSDLKN CTTAAYRPVRELI+QE KD +++K E + NS++ +AKEEV +ER I
Sbjct: 614 DGYSGSDLKNFCTTAAYRPVRELIKQECLKDQERRKREEAEKNSEEGSEAKEEVSEERGI 673
Query: 779 TLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
TLRPL+M+D K+AKSQVAASFAAEGAGMNEL+QWNDLYGEGGSRKKEQLSYFL
Sbjct: 674 TLRPLSMEDMKVAKSQVAASFAAEGAGMNELKQWNDLYGEGGSRKKEQLSYFL 726
>At1g02890 hypothetical protein
Length = 1240
Score = 469 bits (1207), Expect = e-132
Identities = 267/652 (40%), Positives = 385/652 (58%), Gaps = 116/652 (17%)
Query: 220 SSIGLQSNPTNSAPGKHITGWPFDEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRSQ 279
SS+ L+S+ ++ A +K+ I +++V S+ ++L+++D +K + +
Sbjct: 665 SSLRLESSSSDDA-----------DKLAINEIFEVAFNESERGSLILFLKDIEKSVSGNT 713
Query: 280 RIYKLFQTMLTKLSGPILII---------------------------------------G 300
+Y ++ L L I++I G
Sbjct: 714 DVYITLKSKLENLPENIVVIASQTQLDNRKEKSHPGGFLFTKFGSNQTALLDLAFPDTFG 773
Query: 301 SRILDSGNECKRVDEMLTSLFPYNIEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIM 360
R+ D E + + +T LFP + I+ PEDE+ LV WK + E D + ++ Q N I
Sbjct: 774 GRLQDRNTEMPKAVKQITRLFPNKVTIQLPEDEASLVDWKDKLERDTEILKAQANITSIR 833
Query: 361 EVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSL 420
VL+ N L C D++ +C+ D + S+ +E+++ A ++H+M EP ++ KLII S+
Sbjct: 834 AVLSKNQLVCPDIEILCIKDQTLPSDSVEKVVGFAFNHHLMNCSEPTVKDNKLIISAESI 893
Query: 421 SHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVR 480
++ L + + +E K ++K
Sbjct: 894 TYGLQLLHE---------------IQNENKSTKKSLKDV--------------------- 917
Query: 481 KTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLR 540
V +NEFEK++ +VIP ++IGV+FSDIGAL+ KD+L+ELVMLPL+
Sbjct: 918 --------------VTENEFEKKLLSDVIPPSDIGVSFSDIGALENVKDTLKELVMLPLQ 963
Query: 541 RPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEK 599
RP+LF +G L KP +GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSK EK
Sbjct: 964 RPELFGKGQLTKPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSK----GEK 1019
Query: 600 NVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRI 659
V+A+F+LA+K++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDGL +K ++R+
Sbjct: 1020 YVKAVFSLASKIAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMINWDGLRTKDKERV 1079
Query: 660 LVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMT 719
LVLAATNRPFDLDEA+IRR RR+MV LP + NR IL +LAKE++ +D + +A MT
Sbjct: 1080 LVLAATNRPFDLDEAVIRRLPRRLMVNLPDSANRSKILSVILAKEEMAEDVDLEAIANMT 1139
Query: 720 EGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVIT 779
+GYSGSDLKNLC TAA+ P+RE+ ++K+ K++ A+ +N Q
Sbjct: 1140 DGYSGSDLKNLCVTAAHLPIREI----LEKEKKERSVAQAENRAMPQLYSS-------TD 1188
Query: 780 LRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
+RPLNM DFK A QV AS A++ + MNEL+QWN+LYGEGGSRKK LSYF+
Sbjct: 1189 VRPLNMNDFKTAHDQVCASVASDSSNMNELQQWNELYGEGGSRKKTSLSYFM 1240
Score = 55.8 bits (133), Expect = 9e-08
Identities = 34/101 (33%), Positives = 60/101 (58%), Gaps = 8/101 (7%)
Query: 37 SSNALTPDKIENEMLRL-VVDGRESNVTFDNFPYYLSEQTRVLLTSAAYVHLKHA-EVSK 94
S++ LT + + LR +++ ++ V+F+NFPY+LS T+ +L + Y H+K+ E ++
Sbjct: 421 SASVLTRRQAHKDSLRGGILNPQDIEVSFENFPYFLSGTTKDVLMISTYAHIKYGKEYAE 480
Query: 95 YTRNLAPASRTILLSGPAELYQQVLAKALTHYFEAKLLLFD 135
Y +L A +E+YQ++LAKAL AKL++ D
Sbjct: 481 YASDLPTACPR------SEIYQEMLAKALAKQCGAKLMIVD 515
>At4g24850 unknown protein (At4g24850)
Length = 442
Score = 410 bits (1055), Expect = e-114
Identities = 231/506 (45%), Positives = 313/506 (61%), Gaps = 71/506 (14%)
Query: 330 PEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDS----ICVADTMVLS 385
P+DE RL WK Q + D + +++ N NH+ VL L C L++ +C+ D +
Sbjct: 2 PQDEKRLTLWKHQMDRDAETSKVKSNFNHLRMVLRRRGLGCEGLETTWSRMCLKDLTLQR 61
Query: 386 NYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAV 445
+ +E+II A HI +K P+ K+ + S+
Sbjct: 62 DSVEKIIGWAFGNHI--SKNPDTDPAKVTLSRESI------------------------- 94
Query: 446 TSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIR 505
E + + D + S + K V +N FEKR+
Sbjct: 95 ----------------------------EFGIGLLQNDLKGSTSSKKDIVVENVFEKRLL 126
Query: 506 PEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGT 564
+VI ++I VTF DIGAL++ KD L+ELVMLPL+RP+LF +G L KPC+GILLFGPPGT
Sbjct: 127 SDVILPSDIDVTFDDIGALEKVKDILKELVMLPLQRPELFCKGELTKPCKGILLFGPPGT 186
Query: 565 GKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSM 624
GKTMLAKA+A EA A+FIN+SMS+ITSKWFGE EK V+A+F+LA+K+SP++IFVDEVDSM
Sbjct: 187 GKTMLAKAVAKEADANFINISMSSITSKWFGEGEKYVKAVFSLASKMSPSVIFVDEVDSM 246
Query: 625 LGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIM 684
LG+R EHEA RKIKNEFM +WDGLT++ +R+LVLAATNRPFDLDEA+IRR RR+M
Sbjct: 247 LGRREHPREHEASRKIKNEFMMHWDGLTTQERERVLVLAATNRPFDLDEAVIRRLPRRLM 306
Query: 685 VGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQ 744
VGLP NR IL+ +LAKE + LD E+A+MT GYSGSDLKNLC TAA+RP++E+++
Sbjct: 307 VGLPDTSNRAFILKVILAKEDLSPDLDIGEIASMTNGYSGSDLKNLCVTAAHRPIKEILE 366
Query: 745 QEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGA 804
+E K+++DA K LR LN++DF+ A V+AS ++E A
Sbjct: 367 KE-----KRERDAA------LAQGKVPPPLSGSSDLRALNVEDFRDAHKWVSASVSSESA 415
Query: 805 GMNELRQWNDLYGEGGSRKKEQLSYF 830
M L+QWN L+GEGGS K++ S++
Sbjct: 416 TMTALQQWNKLHGEGGSGKQQSFSFY 441
>At4g02470
Length = 371
Score = 410 bits (1053), Expect = e-114
Identities = 205/341 (60%), Positives = 264/341 (77%), Gaps = 12/341 (3%)
Query: 492 KAEVPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGG-LL 550
K V +NEFEK++ +VIP ++IGV+F DIGAL+ K++L+ELVMLPL+RP+LF+ G L
Sbjct: 42 KDVVTENEFEKKLLSDVIPPSDIGVSFDDIGALENVKETLKELVMLPLQRPELFDKGQLT 101
Query: 551 KPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAK 610
KP +GILLFGPPGTGKTMLAKA+A EAGA+FIN+SMS+ITSKWFGE EK V+A+F+LA+K
Sbjct: 102 KPTKGILLFGPPGTGKTMLAKAVATEAGANFINISMSSITSKWFGEGEKYVKAVFSLASK 161
Query: 611 VSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFD 670
++P++IFVDEVDSMLG+R GEHEAMRK+KNEFM NWDGL +K +R+LVLAATNRPFD
Sbjct: 162 IAPSVIFVDEVDSMLGRRENPGEHEAMRKMKNEFMVNWDGLRTKDRERVLVLAATNRPFD 221
Query: 671 LDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNL 730
LDEA+IRR RR+MV LP A NR IL +LAKE++ +D + +A MT+GYSGSDLKNL
Sbjct: 222 LDEAVIRRLPRRLMVNLPDATNRSKILSVILAKEEIAPDVDLEAIANMTDGYSGSDLKNL 281
Query: 731 CTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKM 790
C TAA+ P+RE+ ++K+ K+K A+ +N + +R L M DFK
Sbjct: 282 CVTAAHFPIREI----LEKEKKEKTAAQAEN-------RPTPPLYSCTDVRSLTMNDFKA 330
Query: 791 AKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
A QV AS +++ + MNEL+QWN+LYGEGGSRKK LSYF+
Sbjct: 331 AHDQVCASVSSDSSNMNELQQWNELYGEGGSRKKTSLSYFM 371
>At1g50140 hypothetical protein, 5' partial
Length = 601
Score = 379 bits (973), Expect = e-105
Identities = 231/614 (37%), Positives = 339/614 (54%), Gaps = 103/614 (16%)
Query: 243 DEKILIQTLYKVLLYVSKTYPIVLYMRDADKLLCRS------QRIYKLFQTMLTKLSGPI 296
D I ++ L +VL + P+++Y D+ + L R+ + + M KLSGPI
Sbjct: 64 DGYIAMEALNEVLQSIQ---PLIVYFPDSTQWLSRAVPKTRRKEFVDKVKEMFDKLSGPI 120
Query: 297 LII-GSRILDSGNE-------------------CKRVDEMLTSLFPYNIEIKPPEDESRL 336
++I G +++G++ K + + LF + + PP++E L
Sbjct: 121 VMICGQNKIETGSKEREKFPLPLKGLTEGFTGRGKSEENEIYKLFTNVMRLHPPKEEDTL 180
Query: 337 VSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVLSNYIEEIIVSAI 396
+K Q D + + + N N +++ L ++L C DL + ++ E+ I A
Sbjct: 181 RLFKKQLGEDRRIVISRSNINELLKALEEHELLCTDLYQVNTDGVILTKQKAEKAIGWAK 240
Query: 397 SYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQAVTSEKKEEGAAV 456
++++ P + G+L +P R+SL++ ++ +K E ++
Sbjct: 241 NHYLASCPVPLVKGGRLSLP------------------RESLEI---SIARLRKLEDNSL 279
Query: 457 KPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEVIPANEIGV 516
KP ++ + +E+E+ V+ EIGV
Sbjct: 280 KPSQNLKN------------------------------IAKDEYERNFVSAVVAPGEIGV 309
Query: 517 TFSDIGALDETKDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
F DIGAL++ K +L ELV+LP+RRP+LF G LL+PC+GILLFGPPGTGKT+LAKA+A
Sbjct: 310 KFEDIGALEDVKKALNELVILPMRRPELFARGNLLRPCKGILLFGPPGTGKTLLAKALAT 369
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
EAGA+FI+++ ST+TSKWFG+ EK +ALF+ A K++P IIFVDE+DS+LG R EHE
Sbjct: 370 EAGANFISITGSTLTSKWFGDAEKLTKALFSFATKLAPVIIFVDEIDSLLGARGGSSEHE 429
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENREN 695
A R+++NEFM+ WDGL SK RIL+L ATNRPFDLD+A+IRR RRI V LP AENR
Sbjct: 430 ATRRMRNEFMAAWDGLRSKDSQRILILGATNRPFDLDDAVIRRLPRRIYVDLPDAENRLK 489
Query: 696 ILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKK 755
IL+ L E + F++LA TEGYSGSDLKNLC AAYRPV+EL+Q+E +K
Sbjct: 490 ILKIFLTPENLESDFQFEKLAKETEGYSGSDLKNLCIAAAYRPVQELLQEE-------QK 542
Query: 756 DAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDL 815
A + S LR L++ DF +K++V+ S A + MNELR+WN+
Sbjct: 543 GARAEASPG---------------LRSLSLDDFIQSKAKVSPSVAYDATTMNELRKWNEQ 587
Query: 816 YGEGGSRKKEQLSY 829
YGEGGSR K +
Sbjct: 588 YGEGGSRTKSPFGF 601
>At3g19740 ATPase, putative
Length = 439
Score = 379 bits (972), Expect = e-105
Identities = 211/506 (41%), Positives = 297/506 (57%), Gaps = 68/506 (13%)
Query: 325 IEIKPPEDESRLVSWKSQFEADMKKIQIQDNKNHIMEVLAANDLDCHDLDSICVADTMVL 384
+ + PP++E L+ + Q D + + + N N +++ L N+L C DL + ++
Sbjct: 1 MNLVPPKEEENLIVFNKQLGEDRRIVMSRSNLNELLKALEENELLCTDLYQVNTDGVILT 60
Query: 385 SNYIEEIIVSAISYHIMKNKEPEYRNGKLIIPCNSLSHALGIFQAGKFGDRDSLKLEAQA 444
E++I A ++++ P + G+LI+P R+S+++ +
Sbjct: 61 KQRAEKVIGWARNHYLSSCPSPSIKEGRLILP------------------RESIEISVKR 102
Query: 445 VTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRI 504
+ ++ E + P +K + + +EFE
Sbjct: 103 LKAQ----------EDISRKPTQNLKRFLFLQN-----------------IAKDEFETNF 135
Query: 505 RPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLF-EGGLLKPCRGILLFGPPG 563
V+ EIGV F DIGAL+ K +L ELV+LP+RRP+LF G LL+PC+GILLFGPPG
Sbjct: 136 VSAVVAPGEIGVKFDDIGALEHVKKTLNELVILPMRRPELFTRGNLLRPCKGILLFGPPG 195
Query: 564 TGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDS 623
TGKT+LAKA+A EAGA+FI+++ ST+TSKWFG+ EK +ALF+ A+K++P IIFVDEVDS
Sbjct: 196 TGKTLLAKALATEAGANFISITGSTLTSKWFGDAEKLTKALFSFASKLAPVIIFVDEVDS 255
Query: 624 MLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRI 683
+LG R EHEA R+++NEFM+ WDGL SK RIL+L ATNRPFDLD+A+IRR RRI
Sbjct: 256 LLGARGGAFEHEATRRMRNEFMAAWDGLRSKDSQRILILGATNRPFDLDDAVIRRLPRRI 315
Query: 684 MVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELI 743
V LP AENR IL+ L E + G +F +LA TEGYSGSDLKNLC AAYRPV+EL+
Sbjct: 316 YVDLPDAENRLKILKIFLTPENLETGFEFDKLAKETEGYSGSDLKNLCIAAAYRPVQELL 375
Query: 744 QQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEG 803
Q+E KD+ S D LRPL++ DF +K++V+ S A +
Sbjct: 376 QEE-------NKDSVTNASPD---------------LRPLSLDDFIQSKAKVSPSVAYDA 413
Query: 804 AGMNELRQWNDLYGEGGSRKKEQLSY 829
MNELR+WN+ YGEGG+R K +
Sbjct: 414 TTMNELRKWNEQYGEGGTRTKSPFGF 439
>At1g62130 hypothetical protein
Length = 372
Score = 283 bits (724), Expect = 3e-76
Identities = 164/358 (45%), Positives = 213/358 (58%), Gaps = 67/358 (18%)
Query: 495 VPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLL---- 550
V +N FE ++IP +EIGVTF DIGAL+ KD+L+ELVMLP + P+LF G L
Sbjct: 61 VTENTFEIS---DIIPPSEIGVTFDDIGALENVKDTLKELVMLPFQWPELFCKGQLTKML 117
Query: 551 -----------------KPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKW 593
+PC GILLFGP GTGKTMLAKA+A EAGA+ IN+SMS +W
Sbjct: 118 TLWIGGFLISLLLYFSTQPCNGILLFGPSGTGKTMLAKAVATEAGANLINMSMS----RW 173
Query: 594 FGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTS 653
F E EK V+A+F+LA+K+SP+IIF+DEV+SML H K KNEF+ NWDGL +
Sbjct: 174 FSEGEKYVKAVFSLASKISPSIIFLDEVESML--------HRYRLKTKNEFIINWDGLRT 225
Query: 654 KSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFK 713
++R+LVLAATNRPFDLDEA+IRR R+MVGLP A +R IL+ +L+KE + D
Sbjct: 226 NEKERVLVLAATNRPFDLDEAVIRRLPHRLMVGLPDARSRSKILKVILSKEDLSPDFDID 285
Query: 714 ELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVE 773
E+A+MT GYSG+DLK A + + G D
Sbjct: 286 EVASMTNGYSGNDLKERDAAVA----------------EGRVPPAGSGGSD--------- 320
Query: 774 QERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNELRQWNDLYGEGGSRKKEQLSYFL 831
LR L M+DF+ A V+ S +++ M LRQWN+ YGEGGSR+ E S ++
Sbjct: 321 ------LRVLKMEDFRNALELVSMSISSKSVNMTALRQWNEDYGEGGSRRNESFSQYV 372
>At4g27680 unknown protein
Length = 398
Score = 256 bits (653), Expect = 5e-68
Identities = 132/253 (52%), Positives = 175/253 (68%), Gaps = 2/253 (0%)
Query: 495 VPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFE-GGLLKPC 553
V N +E I +VI + I V F IG L+ K +L ELV+LPL+RP+LF G LL P
Sbjct: 60 VQTNPYEDVIACDVINPDHIDVEFGSIGGLETIKQALYELVILPLKRPELFAYGKLLGPQ 119
Query: 554 RGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSP 613
+G+LL+GPPGTGKTMLAKAIA E+GA FINV +S + SKWFG+ +K V A+F+LA K+ P
Sbjct: 120 KGVLLYGPPGTGKTMLAKAIAKESGAVFINVRVSNLMSKWFGDAQKLVSAVFSLAYKLQP 179
Query: 614 TIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDE 673
IIF+DEV+S LGQR R +HEAM +K EFM+ WDG ++ R++VLAATNRP +LDE
Sbjct: 180 AIIFIDEVESFLGQR-RSTDHEAMANMKTEFMALWDGFSTDPHARVMVLAATNRPSELDE 238
Query: 674 AIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTT 733
AI+RR + +G+P R IL+ L E+V +DF +A + EGY+GSD+ LC
Sbjct: 239 AILRRLPQAFEIGIPDRRERAEILKVTLKGERVEPDIDFDHIARLCEGYTGSDIFELCKK 298
Query: 734 AAYRPVRELIQQE 746
AAY P+RE++ E
Sbjct: 299 AAYFPIREILDAE 311
>At5g53540 26S proteasome regulatory particle chain RPT6-like
protein
Length = 403
Score = 253 bits (646), Expect = 3e-67
Identities = 131/260 (50%), Positives = 180/260 (68%), Gaps = 4/260 (1%)
Query: 495 VPDNEFEKRIRPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFE-GGLLKPC 553
+ N++E I +VI I V F IG L+ K +L ELV+LPL+RP+LF G LL P
Sbjct: 63 IQTNQYEDVIACDVINPLHIDVEFGSIGGLESIKQALYELVILPLKRPELFAYGKLLGPQ 122
Query: 554 RGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSP 613
+G+LL+GPPGTGKTMLAKAIA E+ A FINV +S + SKWFG+ +K V A+F+LA K+ P
Sbjct: 123 KGVLLYGPPGTGKTMLAKAIARESEAVFINVKVSNLMSKWFGDAQKLVSAVFSLAYKLQP 182
Query: 614 TIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDE 673
IIF+DEVDS LGQR R ++EAM +K EFM+ WDG T+ R++VLAATNRP +LDE
Sbjct: 183 AIIFIDEVDSFLGQR-RSTDNEAMSNMKTEFMALWDGFTTDQNARVMVLAATNRPSELDE 241
Query: 674 AIIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTT 733
AI+RRF + +G+P + R IL+ +L E V +++ +A + E Y+GSD+ LC
Sbjct: 242 AILRRFPQSFEIGMPDCQERAQILKVVLKGESVESDINYDRIARLCEDYTGSDIFELCKK 301
Query: 734 AAYRPVRELIQQEIQKDSKK 753
AAY P+RE++ E +K+ K+
Sbjct: 302 AAYFPIREIL--EAEKEGKR 319
>At3g27120 spastin protein, putative
Length = 327
Score = 226 bits (575), Expect = 5e-59
Identities = 131/289 (45%), Positives = 183/289 (62%), Gaps = 14/289 (4%)
Query: 496 PDNEFEKRIR---PEVIP--ANEI-----GVTFSDIGALDETKDSLQELVMLPLRRPDLF 545
PD E +++R P +I +NEI V + DI L+ K + E+V+ PL RPD+F
Sbjct: 17 PDGELPEKLRNLEPRLIEHVSNEIMDRDPNVRWDDIAGLEHAKKCVTEMVIWPLLRPDIF 76
Query: 546 EGGLLKPCRGILLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALF 605
+G P +G+LLFGPPGTGKTM+ KAIA EA A+F +S S++TSKW GE EK VRALF
Sbjct: 77 KG-CRSPGKGLLLFGPPGTGKTMIGKAIAGEAKATFFYISASSLTSKWIGEGEKLVRALF 135
Query: 606 TLAAKVSPTIIFVDEVDSMLGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAAT 665
+A+ P +IFVDE+DS+L QR GEHE+ R++K +F+ +G S SE +IL++ AT
Sbjct: 136 GVASCRQPAVIFVDEIDSLLSQRKSDGEHESSRRLKTQFLIEMEGFDSGSE-QILLIGAT 194
Query: 666 NRPFDLDEAIIRRFERRIMVGLPSAENRENILRTLLAKEKVH--GGLDFKELATMTEGYS 723
NRP +LDEA RR +R+ + LPS+E R I++ LL K+ + D + +TEGYS
Sbjct: 195 NRPQELDEAARRRLTKRLYIPLPSSEARAWIIQNLLKKDGLFTLSDDDMNIICNLTEGYS 254
Query: 724 GSDLKNLCTTAAYRPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEV 772
GSD+KNL A P+RE +++ I + K D QD +DA +EV
Sbjct: 255 GSDMKNLVKDATMGPLREALKRGIDITNLTKDDMRLVTLQDFKDALQEV 303
>At1g80350 CAD ATPase (AAA1)
Length = 523
Score = 216 bits (549), Expect = 5e-56
Identities = 132/374 (35%), Positives = 205/374 (54%), Gaps = 32/374 (8%)
Query: 453 GAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKA---EVPDNEFEKRIRPEVI 509
GA K S K A +G+ SK E PD + + +V+
Sbjct: 171 GATSKSTAGARSSTAGKKGAASKSNKAESMNGDAEDGKSKRGLYEGPDEDLAAMLERDVL 230
Query: 510 PANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTML 569
+ GV + D+ L E K L+E V+LPL P+ F+G + +P +G+L+FGPPGTGKT+L
Sbjct: 231 DSTP-GVRWDDVAGLSEAKRLLEEAVVLPLWMPEYFQG-IRRPWKGVLMFGPPGTGKTLL 288
Query: 570 AKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRT 629
AKA+A E G +F NVS +T+ SKW GE E+ VR LF LA +P+ IF+DE+DS+ R
Sbjct: 289 AKAVATECGTTFFNVSSATLASKWRGESERMVRCLFDLARAYAPSTIFIDEIDSLCNSRG 348
Query: 630 RVGEHEAMRKIKNEFMSNWDGL--TSKSEDR----ILVLAATNRPFDLDEAIIRRFERRI 683
GEHE+ R++K+E + DG+ T+ +ED ++VLAATN P+D+DEA+ RR E+RI
Sbjct: 349 GSGEHESSRRVKSELLVQVDGVSNTATNEDGSRKIVMVLAATNFPWDIDEALRRRLEKRI 408
Query: 684 MVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELI 743
+ LP E+R+ ++ L +V ++ +++A TEGYSG DL N+C A+ +R I
Sbjct: 409 YIPLPDFESRKALININLRTVEVASDVNIEDVARRTEGYSGDDLTNVCRDASMNGMRRKI 468
Query: 744 QQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVAASFAAEG 803
+ + + K +K+++ + P+ M DF+ A +V S ++
Sbjct: 469 AGKTRDEIKNM-------------SKDDISND------PVAMCDFEEAIRKVQPSVSSSD 509
Query: 804 AGMNELRQWNDLYG 817
+E +W +G
Sbjct: 510 IEKHE--KWLSEFG 521
>At2g34560 katanin like protein
Length = 384
Score = 199 bits (505), Expect = 7e-51
Identities = 117/309 (37%), Positives = 174/309 (55%), Gaps = 9/309 (2%)
Query: 441 EAQAVTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEF 500
E ++ E+G++ G + + V T + T++ + P P
Sbjct: 22 EEDVSNKDQPEDGSSNGNNGDVNNNSSPV-TNQDGNTALANGNVIREKPKKSMFPPFESA 80
Query: 501 EKRIRPEVIPANEI----GVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGI 556
E R E + + I + + I L+ K L+E V++P++ P F G LL P +GI
Sbjct: 81 ETRTLAESLSRDIIRGNPNIKWESIKGLENAKKLLKEAVVMPIKYPTYFNG-LLTPWKGI 139
Query: 557 LLFGPPGTGKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTII 616
LLFGPPGTGKTMLAKA+A E +F N+S S++ SKW G+ EK +R LF LA +P+ I
Sbjct: 140 LLFGPPGTGKTMLAKAVATECNTTFFNISASSVVSKWRGDSEKLIRVLFDLARHHAPSTI 199
Query: 617 FVDEVDSMLGQRTRVG--EHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEA 674
F+DE+D+++ QR G EHEA R++K E + DGL K+ + + VLAATN P++LD A
Sbjct: 200 FLDEIDAIISQRGGEGRSEHEASRRLKTELLIQMDGL-QKTNELVFVLAATNLPWELDAA 258
Query: 675 IIRRFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTA 734
++RR E+RI+V LP E R + L+ + L L +EGYSGSD++ LC A
Sbjct: 259 MLRRLEKRILVPLPDPEARRGMFEMLIPSQPGDEPLPHDVLVEKSEGYSGSDIRILCKEA 318
Query: 735 AYRPVRELI 743
A +P+R +
Sbjct: 319 AMQPLRRTL 327
>At2g27600 putative AAA-type ATPase
Length = 435
Score = 190 bits (483), Expect = 2e-48
Identities = 113/295 (38%), Positives = 168/295 (56%), Gaps = 8/295 (2%)
Query: 449 KKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRPEV 508
+ EE AV EG + P +A + T + + E P+ +
Sbjct: 65 RAEEIRAVLDEGGS---GPGSNGDAAVATRPKTKPKDGEGGGKDGEDPEQSKLRAGLNSA 121
Query: 509 IPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTM 568
I + + +SD+ L+ K +LQE V+LP++ P F G +P R LL+GPPGTGK+
Sbjct: 122 IVREKPNIKWSDVAGLESAKQALQEAVILPVKFPQFFTGKR-RPWRAFLLYGPPGTGKSY 180
Query: 569 LAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR 628
LAKA+A EA ++F +VS S + SKW GE EK V LF +A + +P+IIFVDE+DS+ G R
Sbjct: 181 LAKAVATEADSTFFSVSSSDLVSKWMGESEKLVSNLFEMARESAPSIIFVDEIDSLCGTR 240
Query: 629 TRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIRRFERRIMVGLP 688
E EA R+IK E + G+ +++++LVLAATN P+ LD+AI RRF++RI + LP
Sbjct: 241 GEGNESEASRRIKTELLVQMQGV-GHNDEKVLVLAATNTPYALDQAIRRRFDKRIYIPLP 299
Query: 689 SAENRENILRTLLAKEKVHG--GLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE 741
A+ R+++ + L + H DF+ L TEG+SGSD+ + PVR+
Sbjct: 300 EAKARQHMFKVHLG-DTPHNLTEPDFEYLGQKTEGFSGSDVSVCVKDVLFEPVRK 353
>At3g09840 putative transitional endoplasmic reticulum ATPase
Length = 809
Score = 183 bits (465), Expect = 3e-46
Identities = 103/314 (32%), Positives = 174/314 (54%), Gaps = 11/314 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V+++DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 477 VSWNDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 536
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E A+FI+V + + WFGE E NVR +F A + +P ++F DE+DS+ QR +
Sbjct: 537 ECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGGGSGGD 596
Query: 636 ---AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSA 690
A ++ N+ ++ DG+ +K + ++ ATNRP +D A++R R ++ I + LP
Sbjct: 597 GGGAADRVLNQLLTEMDGMNAKK--TVFIIGATNRPDIIDSALLRPGRLDQLIYIPLPDE 654
Query: 691 ENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKD 750
++R NI + L K + +D LA T+G+SG+D+ +C A +RE I+++I+K+
Sbjct: 655 DSRLNIFKAALRKSPIAKDVDIGALAKYTQGFSGADITEICQRACKYAIRENIEKDIEKE 714
Query: 751 SKKKKDAEGQNSQDAQDAKEEVEQ---ERVITLRPLNMQDFKMAKSQVAASFAAEGAGMN 807
++ ++ E +D D E++ E + ++ D + K Q A + G
Sbjct: 715 KRRSENPEAM-EEDGVDEVSEIKAAHFEESMKYARRSVSDADIRKYQAFAQTLQQSRGFG 773
Query: 808 ELRQWNDLYGEGGS 821
++ + G G +
Sbjct: 774 SEFRFENSAGSGAT 787
Score = 171 bits (434), Expect = 1e-42
Identities = 102/250 (40%), Positives = 153/250 (60%), Gaps = 9/250 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G + + ++ELV LPLR P LF+ +KP +GILL+GPPG+GKT++A+A+AN
Sbjct: 204 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVAN 263
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E GA F ++ I SK GE E N+R F A K +P+IIF+DE+DS+ +R + E
Sbjct: 264 ETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-NGE 322
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
R+I ++ ++ DGL KS ++V+ ATNRP +D A+ R RF+R I +G+P R
Sbjct: 323 VERRIVSQLLTLMDGL--KSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIGR 380
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQ-EIQKDSK 752
+LR K+ +D + ++ T GY G+DL LCT AA + +RE + +++ DS
Sbjct: 381 LEVLRIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVIDLEDDS- 439
Query: 753 KKKDAEGQNS 762
DAE NS
Sbjct: 440 --IDAEILNS 447
>At3g53230 CDC48 - like protein
Length = 815
Score = 182 bits (462), Expect = 7e-46
Identities = 95/258 (36%), Positives = 156/258 (59%), Gaps = 6/258 (2%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V++ DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 478 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 537
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE- 633
E A+FI++ + + WFGE E NVR +F A + +P ++F DE+DS+ QR VG+
Sbjct: 538 ECQANFISIKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSVGDA 597
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
A ++ N+ ++ DG+ +K + ++ ATNRP +D A++R R ++ I + LP E
Sbjct: 598 GGAADRVLNQLLTEMDGMNAKK--TVFIIGATNRPDIIDPALLRPGRLDQLIYIPLPDEE 655
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
+R I ++ L K V +D + LA T+G+SG+D+ +C + +RE I+++I+K+
Sbjct: 656 SRYQIFKSCLRKSPVAKDVDLRALAKYTQGFSGADITEICQRSCKYAIRENIEKDIEKER 715
Query: 752 KKKKDAEGQNSQDAQDAK 769
K+ + E + + A+
Sbjct: 716 KRAESPEAMEEDEEEIAE 733
Score = 171 bits (433), Expect = 2e-42
Identities = 100/249 (40%), Positives = 152/249 (60%), Gaps = 7/249 (2%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G + + ++ELV LPLR P LF+ +KP +GILL+GPPG+GKT++A+A+AN
Sbjct: 205 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVAN 264
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E GA F ++ I SK GE E N+R F A K +P+IIF+DE+DS+ +R + E
Sbjct: 265 ETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-HGE 323
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
R+I ++ ++ DGL KS ++V+ ATNRP +D A+ R RF+R I +G+P R
Sbjct: 324 VERRIVSQLLTLMDGL--KSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIGR 381
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDSKK 753
+LR K+ +D + ++ T GY G+DL LCT AA + +RE + ++ +
Sbjct: 382 LEVLRIHTKNMKLAEDVDLERVSKDTHGYVGADLAALCTEAALQCIRE--KMDVIDLDDE 439
Query: 754 KKDAEGQNS 762
+ DAE NS
Sbjct: 440 EIDAEILNS 448
>At5g03340 transitional endoplasmic reticulum ATPase
Length = 810
Score = 182 bits (461), Expect = 9e-46
Identities = 103/311 (33%), Positives = 169/311 (54%), Gaps = 9/311 (2%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V++ DIG L+ K LQE V P+ P+ FE + P +G+L +GPPG GKT+LAKAIAN
Sbjct: 477 VSWEDIGGLENVKRELQETVQYPVEHPEKFEKFGMSPSKGVLFYGPPGCGKTLLAKAIAN 536
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQR-TRVGE- 633
E A+FI+V + + WFGE E NVR +F A + +P ++F DE+DS+ QR G+
Sbjct: 537 ECQANFISVKGPELLTMWFGESEANVREIFDKARQSAPCVLFFDELDSIATQRGNSAGDA 596
Query: 634 HEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAE 691
A ++ N+ ++ DG+ +K + ++ ATNRP +D A++R R ++ I + LP +
Sbjct: 597 GGAADRVLNQLLTEMDGMNAKK--TVFIIGATNRPDIIDSALLRPGRLDQLIYIPLPDED 654
Query: 692 NRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQEIQKDS 751
+R NI + L K V +D LA T+G+SG+D+ +C A +RE I+++I+ +
Sbjct: 655 SRLNIFKACLRKSPVAKDVDVTALAKYTQGFSGADITEICQRACKYAIRENIEKDIENER 714
Query: 752 KKKKDAEGQNSQDAQDAKEEVEQ---ERVITLRPLNMQDFKMAKSQVAASFAAEGAGMNE 808
++ ++ E D E+ E + ++ D + K Q A + G
Sbjct: 715 RRSQNPEAMEEDMVDDEVSEIRAAHFEESMKYARRSVSDADIRKYQAFAQTLQQSRGFGS 774
Query: 809 LRQWNDLYGEG 819
+++ G G
Sbjct: 775 EFRFDSTAGVG 785
Score = 171 bits (434), Expect = 1e-42
Identities = 102/250 (40%), Positives = 153/250 (60%), Gaps = 9/250 (3%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
V + D+G + + ++ELV LPLR P LF+ +KP +GILL+GPPG+GKT++A+A+AN
Sbjct: 204 VGYDDVGGVRKQMAQIRELVELPLRHPQLFKSIGVKPPKGILLYGPPGSGKTLIARAVAN 263
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVGEHE 635
E GA F ++ I SK GE E N+R F A K +P+IIF+DE+DS+ +R + E
Sbjct: 264 ETGAFFFCINGPEIMSKLAGESESNLRKAFEEAEKNAPSIIFIDEIDSIAPKREKT-NGE 322
Query: 636 AMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSAENR 693
R+I ++ ++ DGL KS ++V+ ATNRP +D A+ R RF+R I +G+P R
Sbjct: 323 VERRIVSQLLTLMDGL--KSRAHVIVMGATNRPNSIDPALRRFGRFDREIDIGVPDEIGR 380
Query: 694 ENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRELIQQ-EIQKDSK 752
+LR K+ +D + ++ T GY G+DL LCT AA + +RE + +++ DS
Sbjct: 381 LEVLRIHTKNMKLAEDVDLERISKDTHGYVGADLAALCTEAALQCIREKMDVIDLEDDS- 439
Query: 753 KKKDAEGQNS 762
DAE NS
Sbjct: 440 --IDAEILNS 447
>At3g01610 putative cell division control protein
Length = 703
Score = 166 bits (419), Expect = 6e-41
Identities = 113/381 (29%), Positives = 187/381 (48%), Gaps = 39/381 (10%)
Query: 447 SEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRIRP 506
+EK E V +G+++ + EA++ S+ G + + P
Sbjct: 64 AEKNVEVETVSNKGRSKLATMGARKEAKVSLSLSGATGNGDLEVEGTKGP---------- 113
Query: 507 EVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGK 566
TF D G + + D L+ V+ P+ P+ F+ +KP GIL GPPG GK
Sbjct: 114 ----------TFKDFGGIKKILDELEMNVLFPILNPEPFKKIGVKPPSGILFHGPPGCGK 163
Query: 567 TMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLG 626
T LA AIANEAG F +S + + S G E+N+R LF+ A + +P+I+F+DE+D+ +G
Sbjct: 164 TKLANAIANEAGVPFYKISATEVISGVSGASEENIRELFSKAYRTAPSIVFIDEIDA-IG 222
Query: 627 QRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDR--------ILVLAATNRPFDLDEAIIR- 677
+ + E ++I + ++ DG +K + +LV+ ATNRP LD A+ R
Sbjct: 223 SKRENQQREMEKRIVTQLLTCMDGPGNKGDKNAPDSSAGFVLVIGATNRPDALDPALRRS 282
Query: 678 -RFERRIMVGLPSAENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAY 736
RFE I + P + R IL + K ++ G D K +A +T G+ G+DL+++ A
Sbjct: 283 GRFETEIALTAPDEDARAEILSVVAQKLRLEGPFDKKRIARLTPGFVGADLESVAYLAGR 342
Query: 737 RPVRELIQQEIQKDSKKKKDAEGQNSQDAQDAKEEVEQERVITLRPLNMQDFKMAKSQVA 796
+ ++ ++ + S++ D E S EE E E++ + M DF+ A + V
Sbjct: 343 KAIKRILD---SRKSEQSGDGEDDKSWLRMPWPEE-ELEKLF----VKMSDFEEAVNLVQ 394
Query: 797 ASFAAEGAGMNELRQWNDLYG 817
AS EG + +W+D+ G
Sbjct: 395 ASLTREGFSIVPDVKWDDVGG 415
Score = 147 bits (372), Expect = 2e-35
Identities = 94/302 (31%), Positives = 163/302 (53%), Gaps = 18/302 (5%)
Query: 445 VTSEKKEEGAAVKPEGKTESPAPAVKTEAEIPTSVRKTDGENSVPASKAEVPDNEFEKRI 504
+ +K E + + K+ P + E E V+ +D E +V +A + F
Sbjct: 348 ILDSRKSEQSGDGEDDKSWLRMPWPEEELE-KLFVKMSDFEEAVNLVQASLTREGFS--- 403
Query: 505 RPEVIPANEIGVTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGT 564
++P V + D+G LD + ++ P+++PD+++ + G LL+GPPG
Sbjct: 404 ---IVP----DVKWDDVGGLDHLRLQFNRYIVRPIKKPDIYKAFGVDLETGFLLYGPPGC 456
Query: 565 GKTMLAKAIANEAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSM 624
GKT++AKA ANEAGA+F+++ + + +K+ GE E +R LF A +P +IF DEVD++
Sbjct: 457 GKTLIAKAAANEAGANFMHIKGAELLNKYVGESELAIRTLFQRARTCAPCVIFFDEVDAL 516
Query: 625 LGQRTRVGEHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERR 682
R + G + ++ N+F+ DG ++ + V+ ATNRP +D A +R RF
Sbjct: 517 TTSRGKEGAW-VVERLLNQFLVELDGGERRN---VYVIGATNRPDVVDPAFLRPGRFGNL 572
Query: 683 IMVGLPSAENRENILRTLLAKEKVHGGLDFKELA-TMTEGYSGSDLKNLCTTAAYRPVRE 741
+ V LP+A+ R +IL+ + K+ + +D +A EG+SG+DL +L A ++ V E
Sbjct: 573 LYVPLPNADERASILKAIARKKPIDPSVDLDGIAKNNCEGFSGADLAHLVQKATFQAVEE 632
Query: 742 LI 743
+I
Sbjct: 633 MI 634
>At5g43010 26S proteasome AAA-ATPase subunit RPT4a (gb|AAF22524.1)
Length = 399
Score = 162 bits (409), Expect = 9e-40
Identities = 92/262 (35%), Positives = 156/262 (59%), Gaps = 12/262 (4%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
+++S +G L + L+E + LPL P+LF +KP +G+LL+GPPGTGKT+LA+AIA+
Sbjct: 136 ISYSAVGGLGDQIRELRESIELPLMNPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIAS 195
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVG--- 632
A+F+ V S I K+ GE + +R +F A + P IIF+DE+D++ G+R G
Sbjct: 196 NIDANFLKVVSSAIIDKYIGESARLIREMFNYAREHQPCIIFMDEIDAIGGRRFSEGTSA 255
Query: 633 EHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSA 690
+ E R + E ++ DG + ++ ++ ATNRP LD A++R R +R+I + LP+
Sbjct: 256 DREIQRTLM-ELLNQLDGFDNLG--KVKMIMATNRPDVLDPALLRPGRLDRKIEIPLPNE 312
Query: 691 ENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE----LIQQE 746
++R +IL+ A HG +D++ + + EG++G+DL+N+CT A +R +I ++
Sbjct: 313 QSRMDILKIHAAGIAKHGEIDYEAIVKLAEGFNGADLRNICTEAGMFAIRAERDYVIHED 372
Query: 747 IQKDSKKKKDAEGQNSQDAQDA 768
K +K +A+ S +A
Sbjct: 373 FMKAVRKLSEAKKLESSSHYNA 394
>At1g45000 26S proteasome AAA-ATPase subunit RPT4a like protein
Length = 399
Score = 159 bits (402), Expect = 6e-39
Identities = 91/262 (34%), Positives = 154/262 (58%), Gaps = 12/262 (4%)
Query: 516 VTFSDIGALDETKDSLQELVMLPLRRPDLFEGGLLKPCRGILLFGPPGTGKTMLAKAIAN 575
+++S +G L + L+E + LPL P+LF +KP +G+LL+GPPGTGKT+LA+AIA+
Sbjct: 136 ISYSAVGGLGDQIRELRESIELPLMNPELFLRVGIKPPKGVLLYGPPGTGKTLLARAIAS 195
Query: 576 EAGASFINVSMSTITSKWFGEDEKNVRALFTLAAKVSPTIIFVDEVDSMLGQRTRVG--- 632
A+F+ V S I K+ GE + +R +F A + P IIF+DE+D++ G+R G
Sbjct: 196 NIDANFLKVVSSAIIDKYIGESARLIREMFNYAREHQPCIIFMDEIDAIGGRRFSEGTSA 255
Query: 633 EHEAMRKIKNEFMSNWDGLTSKSEDRILVLAATNRPFDLDEAIIR--RFERRIMVGLPSA 690
+ E R + E ++ DG ++ ++ ATNRP LD A++R R +R+I + LP+
Sbjct: 256 DREIQRTLM-ELLNQLDGFDQLG--KVKMIMATNRPDVLDPALLRPGRLDRKIEIPLPNE 312
Query: 691 ENRENILRTLLAKEKVHGGLDFKELATMTEGYSGSDLKNLCTTAAYRPVRE----LIQQE 746
++R IL+ + HG +D++ + + EG++G+DL+N+CT A +R +I ++
Sbjct: 313 QSRMEILKIHASGIAKHGEIDYEAIVKLGEGFNGADLRNICTEAGMFAIRAERDYVIHED 372
Query: 747 IQKDSKKKKDAEGQNSQDAQDA 768
K +K +A+ S +A
Sbjct: 373 FMKAVRKLSEAKKLESSSHYNA 394
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,595,140
Number of Sequences: 26719
Number of extensions: 751342
Number of successful extensions: 3124
Number of sequences better than 10.0: 171
Number of HSP's better than 10.0 without gapping: 139
Number of HSP's successfully gapped in prelim test: 35
Number of HSP's that attempted gapping in prelim test: 2696
Number of HSP's gapped (non-prelim): 276
length of query: 831
length of database: 11,318,596
effective HSP length: 108
effective length of query: 723
effective length of database: 8,432,944
effective search space: 6097018512
effective search space used: 6097018512
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140913.6


