

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.5 + phase: 0 /pseudo
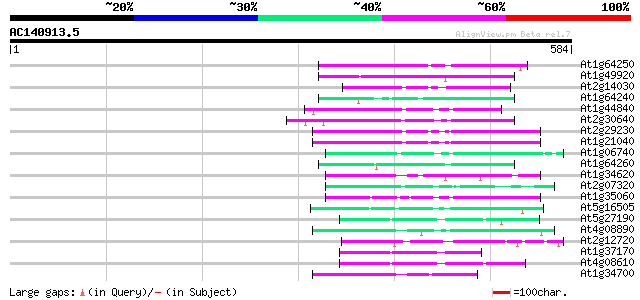
(584 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g64250 hypothetical protein 87 3e-17
At1g49920 hypothetical protein 76 5e-14
At2g14030 Mutator-like transposase 73 5e-13
At1g64240 hypothetical protein 72 7e-13
At1g44840 hypothetical protein 72 7e-13
At2g30640 Mutator-like transposase 72 9e-13
At2g29230 Mutator-like transposase 71 1e-12
At1g21040 hypothetical protein 70 2e-12
At1g06740 mudrA-like protein 69 6e-12
At1g64260 hypothetical protein 68 1e-11
At1g34620 Mutator-like protein 67 2e-11
At2g07320 Mutator-like transposase 64 2e-10
At1g35060 hypothetical protein 62 9e-10
At5g16505 mutator-like transposase-like protein (MQK4.25) 61 2e-09
At5g27190 putative protein 56 5e-08
At4g08890 putative transposon protein 56 6e-08
At2g12720 putative protein 55 8e-08
At1g37170 hypothetical protein 55 8e-08
At4g08610 predicted transposon protein 55 1e-07
At1g34700 hypothetical protein 55 1e-07
>At1g64250 hypothetical protein
Length = 790
Score = 86.7 bits (213), Expect = 3e-17
Identities = 63/225 (28%), Positives = 94/225 (41%), Gaps = 15/225 (6%)
Query: 322 FDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFK-GTRNLP 380
FD Y I+K N A W+D P+ W QA+D GRR+ MT + + F + LP
Sbjct: 531 FDSYMNEIEKKNSEARKWLDQFPQYQWAQAHDSGRRYRVMTIDAENLFAFCESFQSLGLP 590
Query: 381 VTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRER 440
VT + + FF RV GD++T+ + ++ + S H V E+
Sbjct: 591 VTATALLLFDEMRFFFYSGLCDSSGRVNRGDMYTKPVMDQLEKLMTDSIPHVVMPL--EK 648
Query: 441 YTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIP 500
F V E + E + V L C CG+FQ PC HV+A C + + +
Sbjct: 649 GLFQVTEPLQEDE------WIVQLSEWSCTCGEFQLKKFPCLHVLAVCEKLKINPLQYVD 702
Query: 501 GVLKNESVYSIYNTTFKVVQDKSYWLPYDG------PVLCHNPNM 539
+ +Y Y TF V + + W G PV+ PN+
Sbjct: 703 DCYSLDRLYKTYAATFSPVPEVAAWPEASGVPTLFPPVILPPPNV 747
>At1g49920 hypothetical protein
Length = 785
Score = 76.3 bits (186), Expect = 5e-14
Identities = 54/209 (25%), Positives = 88/209 (41%), Gaps = 6/209 (2%)
Query: 322 FDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPV 381
FD Y IK+ N A W+D P W A+D+GRR+G M + E+ V K R + +
Sbjct: 508 FDSYMKEIKERNPEAWKWLDQFPPHQWALAHDDGRRYGIMRID-TEALFAVCKRFRKVAM 566
Query: 382 TPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTH--HVEQFNRE 439
V + +L FA+ + + GD++TE+ +++ S+T + R+
Sbjct: 567 AGGVMLLFGQLKDAFAESFKLSRGSLKHGDVYTEHVMEKLEEFETDSDTWVITITPLERD 626
Query: 440 RYTFSVREIVNYR---EGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYT 496
Y S+ R + + V L C CG+FQ PC H +A C + +
Sbjct: 627 AYQVSMAPKKKTRLMGQSNDSTSGIVQLNDTTCTCGEFQKNKFPCLHALAVCDELKINPL 686
Query: 497 TLIPGVLKNESVYSIYNTTFKVVQDKSYW 525
+ E + Y+ F V + S W
Sbjct: 687 QYVDDCYTVERYHKTYSAKFSPVPELSAW 715
>At2g14030 Mutator-like transposase
Length = 874
Score = 72.8 bits (177), Expect = 5e-13
Identities = 55/175 (31%), Positives = 75/175 (42%), Gaps = 12/175 (6%)
Query: 347 NWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADRAQKGFVR 406
+WT+AY G R+ MTSN+VES N V K R LP+ +++ L +FA R + +
Sbjct: 672 HWTRAYFLGERYNVMTSNVVESLNPVLKEARELPIFSLLEFIRTTLISWFAMRRKAARSK 731
Query: 407 VGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKVDLRA 466
+ + + + KS T V + +RY + VR G F V L
Sbjct: 732 TSA---LLPKMREVVHQNFEKSVTFAVHRI--DRYDYKVR-------GEGSSVFHVKLME 779
Query: 467 GWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESVYSIYNTTFKVVQD 521
C C F LHLPC H IAA + L+ ES Y T K V D
Sbjct: 780 RTCSCRAFDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSIESWRMSYQRTIKPVHD 834
>At1g64240 hypothetical protein
Length = 938
Score = 72.4 bits (176), Expect = 7e-13
Identities = 55/211 (26%), Positives = 84/211 (39%), Gaps = 21/211 (9%)
Query: 322 FDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHM---TSNIVESWNFVFKGTRN 378
FD Y I+K N A W+D P+ W A+D GRR+G M T+ + E +N
Sbjct: 700 FDSYMKDIEKKNSEARKWLDQFPQNQWALAHDNGRRYGIMEIETTTLFEDFNVSHLDNHV 759
Query: 379 LPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNR 438
L T Y L F D + F + C N + + T + + +
Sbjct: 760 L--------TGYVLRLF--DELRHSFDEFFCFSRGSRKCGNVYTEPV----TEKLAESRK 805
Query: 439 ERYTFSVREIVN--YREGRPMRT--FKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHD 494
+ T+ V + N ++ P + V L C CG+FQ+ PC H +A C + +
Sbjct: 806 DSVTYDVMPLDNNAFQVTAPQENDEWTVQLSDCSCTCGEFQSCKFPCLHALAVCKLLKIN 865
Query: 495 YTTLIPGVLKNESVYSIYNTTFKVVQDKSYW 525
+ E +Y Y TF V + S W
Sbjct: 866 PLEYVDDCYTLERLYKTYTATFSPVPELSAW 896
>At1g44840 hypothetical protein
Length = 926
Score = 72.4 bits (176), Expect = 7e-13
Identities = 57/209 (27%), Positives = 87/209 (41%), Gaps = 16/209 (7%)
Query: 308 KKLVQLI----YALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTS 363
K L QL+ YA F + G I+ NQ ++ +I NWT+ Y G+R+ MTS
Sbjct: 676 KALTQLVKNAGYAFTGTKFKEFYGQIETTNQNCGKYLHDIGMANWTRHYFRGQRFNLMTS 735
Query: 364 NIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKD 423
NI E+ N R+ + +++ L +F R +K G I
Sbjct: 736 NIAETLNKALNKGRSSHIVELIRFIRSMLTRWFNARRKKSLKHKGP---VPPEVDKQITK 792
Query: 424 DIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSH 483
++ +N V + Y EI GR + VDL C C ++ L +PCSH
Sbjct: 793 TMLTTNGSKVGRITNWSY-----EINGMLGGRNV----VDLEKKQCTCKRYDKLKIPCSH 843
Query: 484 VIAACSSFRHDYTTLIPGVLKNESVYSIY 512
+ A +SF+ Y L+ K + S Y
Sbjct: 844 ALVAANSFKISYKALVDDCFKPHAWVSSY 872
>At2g30640 Mutator-like transposase
Length = 754
Score = 72.0 bits (175), Expect = 9e-13
Identities = 59/245 (24%), Positives = 100/245 (40%), Gaps = 21/245 (8%)
Query: 289 FQTNMNKLKAHSMTQGMV----GKKLVQLIYALNVPLFDYY----RGVIKKANQRALDWV 340
F T + L H +T+ + LV L++ L D+ G I + + A W+
Sbjct: 457 FPTAFHGLCVHCLTESVRTQFNNSILVNLVWEAAKCLTDFEFEGKMGEIAQISPEAASWI 516
Query: 341 DNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADRA 400
NI W EG R+GH+T+N+ ES N + LP+ +++S +L F +R
Sbjct: 517 RNIQHSQWATYCFEGTRFGHLTANVSESLNSWVQDASGLPIIQMLESIRRQLMTLFNERR 576
Query: 401 QKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTF 460
+ G + + + + I + + V + N ++ V EG+ +
Sbjct: 577 ETSMQWSG---MLVPSAERHVLEAIEECRLYPVHKANEAQFE------VMTSEGK----W 623
Query: 461 KVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESVYSIYNTTFKVVQ 520
VD+R C C ++ LPCSH +AA + R + + Y T V
Sbjct: 624 IVDIRCRTCYCRGWELYGLPCSHAVAALLACRQNVYRFTESYFTVANYRRTYAETIHPVP 683
Query: 521 DKSYW 525
DK+ W
Sbjct: 684 DKTEW 688
>At2g29230 Mutator-like transposase
Length = 915
Score = 71.2 bits (173), Expect = 1e-12
Identities = 65/239 (27%), Positives = 98/239 (40%), Gaps = 14/239 (5%)
Query: 316 ALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKG 375
A + F Y + + + +++++ +WT+AY G+R+ MTSN+ ES N V K
Sbjct: 656 AFRICEFHTYFNEVIRLDPACARYLESVGFCHWTRAYFLGKRYNVMTSNVAESLNAVLKE 715
Query: 376 TRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQ 435
R LP+ +++ L +FA R + + + + KS V +
Sbjct: 716 ARELPIISLLEFIRTTLISWFAMRREAARTEASP---LPPKMREVVHRNFEKSVRFAVHR 772
Query: 436 FNRERYTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDY 495
+RY + +RE EG + + V L C C F LHLPC H IAA +
Sbjct: 773 L--DRYDYEIRE-----EGASV--YHVKLMERTCSCRAFDLLHLPCPHAIAAAVAEGVPI 823
Query: 496 TTLIPGVLKNESVYSIYNTTFKVVQDKS--YWLPYDGPVLCHNPNMRRLKKGRPNSTRI 552
L+ ES Y T K V + + LP L P R GRP RI
Sbjct: 824 QGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALPEPIASLRLFPPATRRPSGRPKKKRI 882
>At1g21040 hypothetical protein
Length = 904
Score = 70.5 bits (171), Expect = 2e-12
Identities = 65/239 (27%), Positives = 97/239 (40%), Gaps = 14/239 (5%)
Query: 316 ALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKG 375
A + F Y + + + +++++ +WT+AY G R+ MTSN+ ES N V K
Sbjct: 656 AFRICEFHTYFNEVIRLDPACARYLESVGFCHWTRAYFLGERYNVMTSNVAESLNAVLKE 715
Query: 376 TRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQ 435
R LP+ +++ L +FA R + + + + KS V +
Sbjct: 716 ARELPIISLLEFIRTTLISWFAMRREAARTEASP---LPPKMREVVHRNFEKSVRFAVHR 772
Query: 436 FNRERYTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDY 495
+RY + +RE EG + + V L C C F LHLPC H IAA +
Sbjct: 773 L--DRYDYEIRE-----EGASV--YHVKLMERTCSCRAFDLLHLPCPHAIAAAVAEGVPI 823
Query: 496 TTLIPGVLKNESVYSIYNTTFKVVQDKS--YWLPYDGPVLCHNPNMRRLKKGRPNSTRI 552
L+ ES Y T K V + + LP L P R GRP RI
Sbjct: 824 QGLMAPEYSVESWRMSYLGTIKPVPEVGDVFALPEPIASLRLFPPATRRPSGRPKKKRI 882
>At1g06740 mudrA-like protein
Length = 726
Score = 69.3 bits (168), Expect = 6e-12
Identities = 60/250 (24%), Positives = 98/250 (39%), Gaps = 19/250 (7%)
Query: 329 IKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIV-ESWNFVFKGTRNLPVTPIVQS 387
I++ + A W+ N W +Y EG R+G +T+N++ ES + + T LP+ ++
Sbjct: 483 IEQISPEASLWIQNKSPARWASSYFEGTRFGQLTANVITESLSNWVEDTSGLPIIQTMEC 542
Query: 388 TYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVRE 447
+ L +R + + ++ + + I +S H V + N +
Sbjct: 543 IHRHLINMLKERRETS---LHWSNVLVPSAEKQMLAAIEQSRAHRVYRANEAEFE----- 594
Query: 448 IVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNES 507
V EG + V++ C CG++Q LPCSH + A S D E+
Sbjct: 595 -VMTCEGNVV----VNIENCSCLCGRWQVYGLPCSHAVGALLSCEEDVYRYTESCFTVEN 649
Query: 508 VYSIYNTTFKVVQDKSYWLPYDGPVLCHNP-NMRRLKKGRPNSTRIRTEMDEEVVERTPI 566
Y T + + DK W D N + KG P R+R E D + V R
Sbjct: 650 YRRAYAETLEPISDKVQWKENDSERDSENVIKTPKAMKGAPRKRRVRAE-DRDRVRRVV- 707
Query: 567 PRQCWLCRHT 576
C C T
Sbjct: 708 --HCGRCNQT 715
>At1g64260 hypothetical protein
Length = 1490
Score = 68.2 bits (165), Expect = 1e-11
Identities = 52/210 (24%), Positives = 81/210 (37%), Gaps = 19/210 (9%)
Query: 322 FDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVE-SWNFVFKGTRNLP 380
FD Y IK+ N A W+D IP+ W A+D G R+G I+E +F R P
Sbjct: 1274 FDSYMNDIKEKNPEAWKWLDQIPRHKWALAHDSGLRYG-----IIEIDREALFAVCRGFP 1328
Query: 381 -----VTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQ 435
+T V + L F + + G ++TE + +++ + S + + Q
Sbjct: 1329 YCTVAMTGGVMLMFDELRSSFDKSLSSIYSSLNRGVVYTEPFMDKLEEFMTDSIPYVITQ 1388
Query: 436 FNRERYTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDY 495
R+ + S E + V L C C KFQ+ PC H +A + +
Sbjct: 1389 LERDSFKVS--------ESSEKEEWIVQLNVSTCTCRKFQSYKFPCLHALAVFEKLKINP 1440
Query: 496 TTLIPGVLKNESVYSIYNTTFKVVQDKSYW 525
+ E Y TF V D + W
Sbjct: 1441 LQYVDECYTVEQYCKTYAATFSPVPDVAAW 1470
Score = 60.5 bits (145), Expect = 3e-09
Identities = 51/206 (24%), Positives = 78/206 (37%), Gaps = 9/206 (4%)
Query: 322 FDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNF--VFKGTRNL 379
F Y IK+ N A W+D P+ W A+D GRR+G M N + F+ ++
Sbjct: 510 FVSYMNDIKEKNPEARKWLDQFPQNRWALAHDNGRRYGIMEINTKALFAVCNAFEQAGHV 569
Query: 380 PVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRE 439
VT V + L F + GD++TE + +++ T+ +
Sbjct: 570 -VTGSVLLLFDELRSKFDKSFSCSRSSLNCGDVYTEPVMDKLEEFRTTFVTYSYIVTPLD 628
Query: 440 RYTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLI 499
F V ++ E V L C CG FQ PC H +A C + + +
Sbjct: 629 NNAFQVATALDKGE------CIVQLSDCSCTCGDFQRYKFPCLHALAVCKKLKFNPLQYV 682
Query: 500 PGVLKNESVYSIYNTTFKVVQDKSYW 525
E + Y T F V + S W
Sbjct: 683 DDCYTLERLKRTYATIFSHVPEMSAW 708
>At1g34620 Mutator-like protein
Length = 1034
Score = 67.4 bits (163), Expect = 2e-11
Identities = 60/235 (25%), Positives = 96/235 (40%), Gaps = 31/235 (13%)
Query: 329 IKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQST 388
I+K N ++ ++ WT+ + +GRR+ M SNI ESWN V + R P+ +++
Sbjct: 690 IQKLNPGCAAYLVDLGFSEWTRVHSKGRRFNIMDSNICESWNNVIREAREYPLICMLEYI 749
Query: 389 YYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREI 448
L +FA R + E C + + VE+ + + SV+ I
Sbjct: 750 RTTLMDWFATRRAQ-----------AEDCPTTLAPRV----QERVEENYQSAMSMSVKPI 794
Query: 449 VNYR---EGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAAC--------SSFRHDYTT 497
N+ + R F V L C C +FQ L +PC+H IAA S + Y
Sbjct: 795 CNFEFQVQERTGECFIVKLDESTCSCLEFQGLGIPCAHAIAAAARIGVPTDSLAANGYFN 854
Query: 498 LIPGVLKNESVYSIYNTTFKVVQDKSYWLPYDGPVLCHNPNMRRLKKGRPNSTRI 552
+ + E +Y I++ +V + + G P R GRP RI
Sbjct: 855 ELVKLSYEEKIYPIHSVGGEVAPEIA-----AGTTGELQPPFVRRPPGRPRKIRI 904
Score = 30.4 bits (67), Expect = 2.8
Identities = 20/67 (29%), Positives = 29/67 (42%), Gaps = 7/67 (10%)
Query: 31 GMTFHSKEDCLHVIKSFYIRQSMDYDVIH*DPRRYVIQCTLETCQFNLRA-------SFR 83
G F SK DC I I + + P+ V++C +TC + + A SF+
Sbjct: 368 GRVFKSKSDCKIKIAIHAINRKFHFRTARSTPKFMVLKCISKTCPWRVYASKVDTSDSFQ 427
Query: 84 KRVANGR 90
R AN R
Sbjct: 428 VRQANQR 434
>At2g07320 Mutator-like transposase
Length = 534
Score = 63.9 bits (154), Expect = 2e-10
Identities = 57/239 (23%), Positives = 92/239 (37%), Gaps = 41/239 (17%)
Query: 329 IKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQST 388
IK N +++ I W++ Y +G R+ MTSNI E+ N + R P+ +V+
Sbjct: 21 IKSRNYECDKYLEKIGPARWSRVYFQGERYNLMTSNIAETLNNAIRLGRCAPILELVKFI 80
Query: 389 YYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREI 448
L + + R +K G + Y + ++K ++ Y EI
Sbjct: 81 RAMLTRWCSARRKKSIKHTG---MVQPYVDKVMAQTMMKMEGSKIQNVTNWTY-----EI 132
Query: 449 VNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESV 508
V G M +F D++ C C F L +PC H + A +S D+ TL+ K
Sbjct: 133 VGRFGGNHMVSFD-DMK---CSCKVFDKLKIPCGHAMLAANSIGLDFGTLVGECFKT--- 185
Query: 509 YSIYNTTFKVVQDKSYWLPYDGPVLCHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPIP 567
K++ L Y G V P + + + EEV ER +P
Sbjct: 186 -------------KTWVLTYKGIV-------------NPEANALELDQSEEVKERVILP 218
>At1g35060 hypothetical protein
Length = 873
Score = 62.0 bits (149), Expect = 9e-10
Identities = 59/226 (26%), Positives = 93/226 (41%), Gaps = 15/226 (6%)
Query: 329 IKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWN-FVFKGTRNLPVTPIVQS 387
I N + ++ ++ +WT+ Y G+R+ MTSNI E+ N +FKG R+ + +++
Sbjct: 652 INAINPLCIKYLHDVGMAHWTRLYFPGQRFNLMTSNIAETLNKALFKG-RSSHIVELLRF 710
Query: 388 TYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVRE 447
L +F +K + SG + E I ++ S+ V + Y E
Sbjct: 711 IRSMLTRWFNAHRKKS--QAHSGPVPPEV-DKQISKNLTTSSGSKVGRVTSWSY-----E 762
Query: 448 IVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNES 507
+V G + VDL C C ++ L +PC H + A +S Y L+ K S
Sbjct: 763 VVGKLGGSNV----VDLEKKQCTCKRYDKLKIPCGHALVAANSINLSYKALVDDYFKPHS 818
Query: 508 -VYSIYNTTFKVVQDKSYWLPYDGPVLCHNPNMRRLKKGRPNSTRI 552
V S F K +P + P P R GRP RI
Sbjct: 819 WVASYKGAVFPEANGKEEDIPEELPHRSMLPPYTRRPSGRPKVARI 864
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 61.2 bits (147), Expect = 2e-09
Identities = 55/249 (22%), Positives = 98/249 (39%), Gaps = 21/249 (8%)
Query: 314 IYALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVES-WNFV 372
+YAL F+ + + +Q + W + W AY +G R+GH I E +N+
Sbjct: 335 VYALTPAEFETKSNEMIEISQDVVQWFELYLPHLWAVAYFQGVRYGHFGLGITEVLYNWA 394
Query: 373 FKGTRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHH 432
+ LP+ +++ ++++ +F +R + + +G + + I + + + +
Sbjct: 395 LE-CHELPIIQMMEHIRHQISSWFDNRRE---LSMGWNSILVPSAERRITEAVADARCYQ 450
Query: 433 VEQFNRERYTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAACSSFR 492
V + N + EIV+ RT VD+R C C ++Q LPC+H AA S
Sbjct: 451 VLRANEVEF-----EIVSTE-----RTNIVDIRTRDCSCRRWQIYGLPCAHAAAALISCG 500
Query: 493 HDYTTLIPGVLKNESVYSIYNTTFKVVQDKSYWLPYDGPV------LCHNPNMRRLKKGR 546
+ S Y+ + D+S W L P R GR
Sbjct: 501 RNVHLFAEPCFTVSSYQQTYSQMIGEIPDRSLWKDEGEEAGGGVESLRIRPPKTRRPPGR 560
Query: 547 PNSTRIRTE 555
P +R E
Sbjct: 561 PKKKVVRVE 569
Score = 32.3 bits (72), Expect = 0.75
Identities = 20/80 (25%), Positives = 34/80 (42%), Gaps = 4/80 (5%)
Query: 2 CNVDVDAYDALPTDDHLNTNVPVDGAIEQGMTFHSKEDCLHVIKSFYIRQSMDYDVIH*D 61
C + A L +DD L T + + G F E C +K I D ++ D
Sbjct: 3 CLMADGALITLDSDDILAT----EHVLAVGKEFPDVETCRRTLKDLAIALHFDLRIVKSD 58
Query: 62 PRRYVIQCTLETCQFNLRAS 81
R++ +C+ E C + + A+
Sbjct: 59 RSRFIAKCSKEGCPWRIHAA 78
>At5g27190 putative protein
Length = 779
Score = 56.2 bits (134), Expect = 5e-08
Identities = 59/219 (26%), Positives = 86/219 (38%), Gaps = 21/219 (9%)
Query: 344 PKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADRAQKG 403
P ENW Y EG R+ TSN ES N F+ R + P++ + +++ +F ++ +K
Sbjct: 538 PVENWAMCYFEGDRYNIDTSNACESLNSTFERARKYFLLPLLDAIIEKISEWF-NKHRKE 596
Query: 404 FVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKVD 463
V+ +N I + T V + N +SV G ++ VD
Sbjct: 597 SVKYSKTRGLVPVVENEIHSLCPIAKTMPVTELNSFERQYSV-------IGEGGISYYVD 649
Query: 464 LRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESVYS----------IYN 513
L C C +F PC H IAA +F T I L+ E + S Y+
Sbjct: 650 LLRKTCSCRRFDVDKYPCVHAIAAARNFVG--TEWILDGLEMEDLTSHYYHMNTWILAYH 707
Query: 514 TTFKVVQDKSYW-LPYDGPVLCHNPNMRRLKKGRPNSTR 551
T V S W +P + L P K GR TR
Sbjct: 708 RTIYPVPHSSVWIIPDEIRQLIFLPPDTTTKPGRNQETR 746
>At4g08890 putative transposon protein
Length = 1028
Score = 55.8 bits (133), Expect = 6e-08
Identities = 59/267 (22%), Positives = 106/267 (39%), Gaps = 35/267 (13%)
Query: 316 ALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKG 375
A V F+ Y G++++ + + ++++I E+WT+ + G+ + M+SN ES N V
Sbjct: 607 AYRVRDFEKYFGLLREKSAKCAKYLEDIGFEHWTRVHCRGKHYNIMSSNNSESMNHVLTK 666
Query: 376 TRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVK-------- 427
+ P+ +++ L +FA R +K +C++++ ++ +
Sbjct: 667 AKTYPIVYMIEFIRDVLMRWFASRRKK-----------VAWCKSSVTPEVDERFLQELPA 715
Query: 428 SNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAA 487
S + V+ F Y V + G F V L C C ++ L +PC H +AA
Sbjct: 716 SGKYAVKMFGPWSYQ------VTSKSG---EHFHVVLDQCTCTCLRYTKLRIPCEHALAA 766
Query: 488 CSSFRHDYTTLIPGVLKNESVYSIYNTTFKVVQD-KSYWLPYDGPVLCHNPNMRRLKKGR 546
D + + ++ + + D K +P L P R GR
Sbjct: 767 AIEHGIDPKSCVGWWYGLQTFSDSFQEPILPIADPKDVVIPQHIVDLILIPPYSRRPPGR 826
Query: 547 PNSTRI------RTEMDEEVVERTPIP 567
P S RI R E V+R P
Sbjct: 827 PLSKRIPSRGENRNNRIEATVDRRLAP 853
>At2g12720 putative protein
Length = 819
Score = 55.5 bits (132), Expect = 8e-08
Identities = 56/240 (23%), Positives = 97/240 (40%), Gaps = 31/240 (12%)
Query: 346 ENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADR---AQK 402
E W +A+ G R+ MT+NI ES N +NLP+ ++++ + +FA+R A K
Sbjct: 593 EMWARAHFPGDRYNLMTTNIAESMNRALSQAKNLPIVRMLEAIRQMMTRWFAERRDDASK 652
Query: 403 GFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKV 462
++ G + ++ + S V+ + R + ++ V
Sbjct: 653 QHTQLTPG------VEKLLQTRVTSSRLLDVQTIDASRVQVAYEASLHV----------V 696
Query: 463 DLRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESVYSIYNTTFKVVQDK 522
++ C C F LPC H IAA +L K+ + ++Y + D
Sbjct: 697 NVDEKQCTCRLFNKEKLPCIHAIAAAEHMGVSRISLCSPYYKSSHLVNVYACAI-MPSDT 755
Query: 523 SYWLP---YDGPVLCHNPNMRRLKKGRPNSTRIRTEMDEEVVERTPIPRQ---CWLCRHT 576
+P D P L P + + GRP R+++ + EV T PR+ C C+ T
Sbjct: 756 EVPVPQIVIDQPCL---PPIVANQPGRPKKLRMKSAL--EVAVETKRPRKEHACSRCKET 810
>At1g37170 hypothetical protein
Length = 598
Score = 55.5 bits (132), Expect = 8e-08
Identities = 41/148 (27%), Positives = 62/148 (41%), Gaps = 8/148 (5%)
Query: 344 PKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADRAQKG 403
P ENW + Y EG R+ TSN ES N F+ R + P+ + +++ +F ++ +K
Sbjct: 455 PVENWARCYFEGDRYNIDTSNACESLNSTFERARKYFLLPLFDAIIEKISEWF-NKHRKE 513
Query: 404 FVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKVD 463
V+ +N I + T V + N +SV G ++ VD
Sbjct: 514 SVKYSETRGLVPVVENEIHSLCPIAKTMPVTELNSFERQYSV-------IGEGGMSYSVD 566
Query: 464 LRAGWCDCGKFQALHLPCSHVIAACSSF 491
L C C +F PC H IAA F
Sbjct: 567 LLRKTCSCRRFDVDKYPCVHAIAAARHF 594
>At4g08610 predicted transposon protein
Length = 907
Score = 55.1 bits (131), Expect = 1e-07
Identities = 48/194 (24%), Positives = 80/194 (40%), Gaps = 10/194 (5%)
Query: 344 PKENWTQAYDEGRRWGHMTSNIVESWNFVFKGTRNLPVTPIVQSTYYRLACFFADRAQKG 403
P ENW Y EG R+ TSN+ ES N F+ R + P++ + +++ +F ++ +K
Sbjct: 557 PVENWAMCYFEGDRYNIDTSNVCESLNSTFESARKYFLLPLLDAIIEKISEWF-NKHRKK 615
Query: 404 FVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQFNRERYTFSVREIVNYREGRPMRTFKVD 463
V+ +N I + T V + N ++V G ++ VD
Sbjct: 616 SVKYSETRGLVPVVENEIHSLCPIAKTMPVTELNSFERQYNV-------IGEGGMSYSVD 668
Query: 464 LRAGWCDCGKFQALHLPCSHVIAACSSFRHDYTTLIPGVLKNESVYSIYNTTFKVVQDKS 523
L C C +F P H IAA +F T I V+ + S++S + + + +
Sbjct: 669 LLRKTCSCRRFDVDKYPYVHAIAAAQNFVG--TEWISDVIFSHSLFSADSRRTVLTNNTT 726
Query: 524 YWLPYDGPVLCHNP 537
P L H P
Sbjct: 727 NLRRRRPPNLSHQP 740
>At1g34700 hypothetical protein
Length = 990
Score = 55.1 bits (131), Expect = 1e-07
Identities = 39/173 (22%), Positives = 79/173 (45%), Gaps = 14/173 (8%)
Query: 316 ALNVPLFDYYRGVIKKANQRALDWVDNIPKENWTQAYDEGRRWGHMTSNIVESWNFVFKG 375
A V F+ Y ++++ + + ++++I E+WT+A+ G R+ M+SN ES N V
Sbjct: 689 AYRVKDFEKYFELLREKSAKCGKYLEDIGFEHWTRAHCRGERYNIMSSNNFESMNHVLTK 748
Query: 376 TRNLPVTPIVQSTYYRLACFFADRAQKGFVRVGSGDLFTEYCQNAIKDDIVKSNTHHVEQ 435
+ P+ +++ L +FA R +K C++++ ++ + H +
Sbjct: 749 AKTYPIVYMIEFIRDVLMRWFASRRKK-----------VARCKSSVTPEVDERFLHELPA 797
Query: 436 FNRERYTFSVREIVNYR-EGRPMRTFKVDLRAGWCDCGKFQALHLPCSHVIAA 487
+Y + +Y+ + F V L C C ++ L +PC H +AA
Sbjct: 798 SG--KYVVKMSGPWSYQVTSKSGEHFHVVLDQCTCTCLRYTKLRIPCEHALAA 848
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.339 0.146 0.487
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,104,798
Number of Sequences: 26719
Number of extensions: 486446
Number of successful extensions: 1496
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 63
Number of HSP's successfully gapped in prelim test: 5
Number of HSP's that attempted gapping in prelim test: 1351
Number of HSP's gapped (non-prelim): 110
length of query: 584
length of database: 11,318,596
effective HSP length: 105
effective length of query: 479
effective length of database: 8,513,101
effective search space: 4077775379
effective search space used: 4077775379
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140913.5


