

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.13 + phase: 0 /pseudo
(676 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
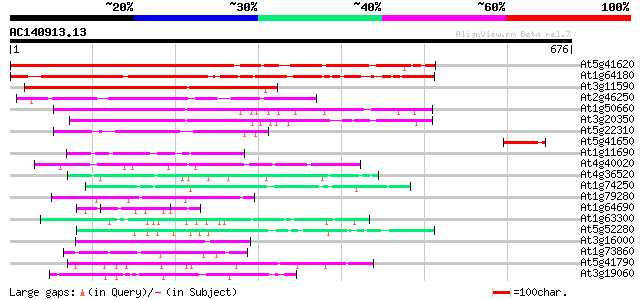
Score E
Sequences producing significant alignments: (bits) Value
At5g41620 unknown protein 443 e-124
At1g64180 unknown protein 348 6e-96
At3g11590 unknown protein 197 2e-50
At2g46250 unknown protein 183 2e-46
At1g50660 unknown protein 130 3e-30
At3g20350 unknown protein 127 2e-29
At5g22310 unknown protein 113 4e-25
At5g41650 unknown protein 79 8e-15
At1g11690 hypothetical protein 64 2e-10
At4g40020 putative protein 55 2e-07
At4g36520 trichohyalin like protein 54 4e-07
At1g74250 putative heat shock protein 51 2e-06
At1g79280 hypothetical protein 48 2e-05
At1g64690 hypothetical protein 48 2e-05
At1g63300 hypothetical protein 48 2e-05
At5g52280 hyaluronan mediated motility receptor-like protein 47 3e-05
At3g16000 myosin heavy chain like protein 47 5e-05
At1g73860 kinesin-related protein 47 5e-05
At5g41790 myosin heavy chain-like protein 46 8e-05
At3g19060 hypothetical protein 46 8e-05
>At5g41620 unknown protein
Length = 543
Score = 443 bits (1140), Expect = e-124
Identities = 258/521 (49%), Positives = 342/521 (65%), Gaps = 44/521 (8%)
Query: 1 MQHHRAIDRNNHALQPLSPASYGSSMEMTPYNPAATPNSSLDFKGRIG-EPHYSLKTSTE 59
++HH++IDRNNHALQP+SPASYGSS+E+T YN A TP+SSL+F+GR EPHY+LKTSTE
Sbjct: 59 IKHHQSIDRNNHALQPVSPASYGSSLEVTTYNKAVTPSSSLEFRGRPSREPHYNLKTSTE 118
Query: 60 LLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAED 119
LLKVLNRIWSLEEQH SNISLIKALKTE+ +R+++KELLR +QADRHE+D ++KQ+AE+
Sbjct: 119 LLKVLNRIWSLEEQHVSNISLIKALKTEVAHSRVRIKELLRYQQADRHELDSVVKQLAEE 178
Query: 120 KLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQ 179
KL+ K+KE +R+ +AVQSVR LEDERKLRKRSES+HRK+ARELSE+KSS ++ +K+LE+
Sbjct: 179 KLLSKNKEVERMSSAVQSVRKALEDERKLRKRSESLHRKMARELSEVKSSLSNCVKELER 238
Query: 180 ERTRRKLLEDLCDEFARGINEYEQEVHTLKQKS-EKDWVQRADHDRLVLHISESWLDERM 238
K++E LCDEFA+GI YE+E+H LK+K+ +KDW R D+LVLHI+ESWLDERM
Sbjct: 239 GSKSNKMMELLCDEFAKGIKSYEEEIHGLKKKNLDKDWAGRGGGDQLVLHIAESWLDERM 298
Query: 239 QMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVS 298
QM+LE S++DKL +EIETF++ K+ N++ R+RRNSLESVP N +
Sbjct: 299 QMRLEGGDTLNGKNRSVLDKLEVEIETFLQEKR------NEIPRNRRNSLESVPFNTLSA 352
Query: 299 APQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKKSVPGE 358
P+ + D ++DS GSDS+CFEL KP+ + DET K N K E
Sbjct: 353 PPRDV--DCEEDSGGSDSNCFELKKPA------------ESYGDETKKPNQHNKDGSIDE 398
Query: 359 GFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPEHFEISESGGFERKNNLSELH 418
K+ N + Q A L++N + I +E E ++ K +E
Sbjct: 399 KPKSPSSFQVNFEDQMAWA---LSSNGKKKTTRAIEDEEEEEDVKPENSNNNKKPENECA 455
Query: 419 STSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQASPVRQWMAS---- 474
+T+KN ++ +IR +E EASC+ R QASPVRQW++
Sbjct: 456 TTNKNDVMGEMIRTH--------RRLLSETREIDEASCNFPSSRRQASPVRQWISRTVAP 507
Query: 475 --LDISEASKVHSGSKDNTLKAKLLEARSKGQRSRLRTLKG 513
L SE + H G KDNTLK KL + +SRLR KG
Sbjct: 508 DLLGPSEIAIAH-GVKDNTLKTKL----ANSSKSRLRLFKG 543
>At1g64180 unknown protein
Length = 593
Score = 348 bits (893), Expect = 6e-96
Identities = 229/516 (44%), Positives = 318/516 (61%), Gaps = 72/516 (13%)
Query: 1 MQHHRAIDRNNHALQPLSPASYGSSMEMTPYNPAATPNSSLDFKGR--IGEPHYSLKTST 58
M+HH +RN+HALQP+SP SY +SSL+F+GR GEP+ ++KTST
Sbjct: 145 MKHHHLTERNDHALQPVSPTSY---------------DSSLEFRGRRRAGEPNNNIKTST 189
Query: 59 ELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAE 118
ELLKVLNRIW LEEQHS+NISLIK+LKTEL +R ++K+LLR +QAD+ ++DD +KQ+AE
Sbjct: 190 ELLKVLNRIWILEEQHSANISLIKSLKTELAHSRARIKDLLRCKQADKRDMDDFVKQLAE 249
Query: 119 DKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLE 178
+KL + +KE DRL +AVQS LEDERKLRKRSES++RKLA+ELSE+KS+ ++ +K++E
Sbjct: 250 EKLSKGTKEHDRLSSAVQS----LEDERKLRKRSESLYRKLAQELSEVKSTLSNCVKEME 305
Query: 179 QERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERM 238
+ +K+LE LCDEFA+GI YE+E+H LKQK +K+W + D ++L I+ESWLDER+
Sbjct: 306 RGTESKKILERLCDEFAKGIKSYEREIHGLKQKLDKNWKGWDEQDHMILCIAESWLDERI 365
Query: 239 QMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVS 298
Q + NG S ++KL EIETF+K QN+ S N++ R+RR SLESVP N A+S
Sbjct: 366 Q-----SGNG-----SALEKLEFEIETFLKTNQNADS--NEIARNRRTSLESVPFN-AMS 412
Query: 299 AP-QAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKKSVPG 357
AP + ++++DS GS S+CFEL K AK + DET K + K
Sbjct: 413 APIWEVDREEEEDSGGSGSNCFELKKHGSDVAKPPRG-------DETEKPELIKVGVSER 465
Query: 358 EGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISE-EPEHFEISESGGFERKNNLSE 416
++ PS+ K + A +++N + + TR +E EPE +E G E N + E
Sbjct: 466 PQRRSQSPSSLQVKFEDQMA-WAMSSNEK--KKTRANEMEPE----TEKCGKETNNVVGE 518
Query: 417 LHSTSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQASPVRQWMASLD 476
+ T + L+SE+ EASCS R + SP+RQW +
Sbjct: 519 MIRTHRR-----------LSSET---------REIDEASCSYPS-RRRESPIRQW-NTRT 556
Query: 477 ISEASKVHSGSKDNTLKAKLLEARSKGQRSRLRTLK 512
++ G KDNTLK KL EAR+ R R+R K
Sbjct: 557 VTPDLGAPRGVKDNTLKTKLSEARTTSSRPRVRLFK 592
>At3g11590 unknown protein
Length = 622
Score = 197 bits (501), Expect = 2e-50
Identities = 113/311 (36%), Positives = 193/311 (61%), Gaps = 8/311 (2%)
Query: 19 PASYGSSMEMTPYNPAATPN-SSLDFKGRIGEPHYSLKTSTELLKVLNRIWSLEEQHSSN 77
P + GS M++ + TP S++ K R+ + +L TS ELLK++NR+W +++ SS+
Sbjct: 194 PINSGSFMDIETRSRVETPTGSTVGVKTRLKDCSNALTTSKELLKIINRMWGQDDRPSSS 253
Query: 78 ISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQS 137
+SL+ AL +EL+R R++V +L+ + + + +++ LMK+ AE+K V KS EQ+ + AA++S
Sbjct: 254 MSLVSALHSELERARLQVNQLIHEHKPENNDISYLMKRFAEEKAVWKSNEQEVVEAAIES 313
Query: 138 VRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARG 197
V ELE ERKLR+R ES+++KL +EL+E KS+ A+K++E E+ R ++E +CDE AR
Sbjct: 314 VAGELEVERKLRRRFESLNKKLGKELAETKSALMKAVKEIENEKRARVMVEKVCDELARD 373
Query: 198 INEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVD 257
I+E + EV LK++S K + + +R +L ++++ +ER+QM+L A++ +KN+ VD
Sbjct: 374 ISEDKAEVEELKRESFK-VKEEVEKEREMLQLADALREERVQMKLSEAKHQLEEKNAAVD 432
Query: 258 KLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGDD------DDDDS 311
KL +++T++KAK+ N LN +S +D +++ S
Sbjct: 433 KLRNQLQTYLKAKRCKEKTREPPQTQLHNEEAGDYLNHHISFGSYNIEDGEVENGNEEGS 492
Query: 312 VGSDSHCFELN 322
SD H ELN
Sbjct: 493 GESDLHSIELN 503
>At2g46250 unknown protein
Length = 468
Score = 183 bits (465), Expect = 2e-46
Identities = 124/365 (33%), Positives = 193/365 (51%), Gaps = 47/365 (12%)
Query: 9 RNNHALQPLSPASYGSS---MEMTPYNPAATPNSSLDFKGRIGEPHYSLKTSTELLKVLN 65
RN LQP+SPAS SS +++ PA + S K Y L +ST+LLKVLN
Sbjct: 115 RNGDLLQPISPASCSSSSSSLQVVVRKPAFSQTGSSAVKSA----SYGLGSSTKLLKVLN 170
Query: 66 RIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKS 125
RIWSLEEQ+++N+SL++ALK ELD R ++KE+ + ++ D+ +RK
Sbjct: 171 RIWSLEEQNTANMSLVRALKMELDECRAEIKEVQQRKKLS-------------DRPLRKK 217
Query: 126 KEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRK 185
KE++ + +S++ EL+DERK+RK SE++HRKL REL E K + ALKDLE+E R
Sbjct: 218 KEEEEVKDVFRSIKRELDDERKVRKESETLHRKLTRELCEAKHCLSKALKDLEKETQERV 277
Query: 186 LLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAA 245
++E+LCDEFA+ + +YE +V + +KS D++++ I+E W D+R+QM+LE
Sbjct: 278 VVENLCDEFAKAVKDYEDKVRRIGKKSPVS-------DKVIVQIAEVWSDQRLQMKLEED 330
Query: 246 QNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGD 305
F+ ++AK+ SRS ++ R +SV ++ V +
Sbjct: 331 DKTFL----------------LRAKEKSRSQSSKGSGLRAKPDDSVSMHQRVCLKEL--- 371
Query: 306 DDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLDETSKSNVKKKKS-VPGEGFKNHI 364
++ + + +L K S + DK E+S NV +S F +
Sbjct: 372 EEGLEKRTRRDNKLQLKKSSGQVLNLSMSSEGDKIHPESSPRNVDDHESQEKSRTFSKNH 431
Query: 365 PSNSN 369
P N N
Sbjct: 432 PRNVN 436
>At1g50660 unknown protein
Length = 725
Score = 130 bits (326), Expect = 3e-30
Identities = 113/516 (21%), Positives = 242/516 (46%), Gaps = 70/516 (13%)
Query: 54 LKTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLM 113
L T E+ ++ + + +++Q ++ +SL+ +L+ EL+ ++++L ++++ + +++ +
Sbjct: 213 LDTMEEVHQIYSNMKRIDQQVNA-VSLVSSLEAELEEAHARIEDLESEKRSHKKKLEQFL 271
Query: 114 KQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSA 173
++++E++ +S+E +++ A + ++ ++ E+K R+R E ++ KL EL++ K +
Sbjct: 272 RKVSEERAAWRSREHEKVRAIIDDMKTDMNREKKTRQRLEIVNHKLVNELADSKLAVKRY 331
Query: 174 LKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESW 233
++D E+ER R+L+E++CDE A+ I E + E+ LK++S + D +R +L ++E W
Sbjct: 332 MQDYEKERKARELIEEVCDELAKEIGEDKAEIEALKRES-MSLREEVDDERRMLQMAEVW 390
Query: 234 LDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSA----ENQVIRDRRNSLE 289
+ER+QM+L A+ ++ S ++KL ++E+F++++ E +++R+ S+
Sbjct: 391 REERVQMKLIDAKVALEERYSQMNKLVGDLESFLRSRDIVTDVKEVREAELLRETAASVN 450
Query: 290 S--------VPLND----AVSAPQAMGDDDDDD----------SVGSDSHCFELN----- 322
VP N AV +G+ D + S S H L+
Sbjct: 451 IQEIKEFTYVPANPDDIYAVFEEMNLGEAHDREMEKSVAYSPISHDSKVHTVSLDANMMN 510
Query: 323 -KPSHMGAKVHKEEILDKNLD--------ETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQ 373
K H A H+ ++++ E S+ S+P KNH +SN S
Sbjct: 511 KKGRHSDAYTHQNGDIEEDDSGWETVSHLEEQGSSYSPDGSIPSVNNKNHNHRHSNASSG 570
Query: 374 SVDA-----EEGLTTNIRLVEGTRISEEPEHFEISESGGFERKNNLSELHSTSKNHIIDN 428
++ ++ +T + E I ++S R S S +I
Sbjct: 571 GTESLGKVWDDTMTPTTEISEVCSIPRRSSK-KVSSIAKLWRSTGASNGDRDSNYKVIS- 628
Query: 429 LIRGQLLASESGKNNKHAEHNNYGEASCSNAGWRNQASP----VRQWMASLDISEASKVH 484
+ +G + ++ G S + SP V QW +S + + V+
Sbjct: 629 ------MEGMNGGRVSNGRKSSAGMVSPDRVSSKGGFSPMMDLVGQWNSSPESANHPHVN 682
Query: 485 SG-----------SKDNTLKAKLLEARSKGQRSRLR 509
G ++ ++LK+KL+EAR + Q+ +L+
Sbjct: 683 RGGMKGCIEWPRGAQKSSLKSKLIEARIESQKVQLK 718
>At3g20350 unknown protein
Length = 673
Score = 127 bits (319), Expect = 2e-29
Identities = 113/481 (23%), Positives = 219/481 (45%), Gaps = 66/481 (13%)
Query: 73 QHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLH 132
Q +++SL +++ +L R +K+L ++++ + +++ +K+++E++ +S+E +++
Sbjct: 208 QQVNDVSLASSIELKLQEARACIKDLESEKRSQKKKLEQFLKKVSEERAAWRSREHEKVR 267
Query: 133 AAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCD 192
A + ++ ++ E+K R+R E ++ KL EL++ K + + D +QER R+L+E++CD
Sbjct: 268 AIIDDMKADMNQEKKTRQRLEIVNSKLVNELADSKLAVKRYMHDYQQERKARELIEEVCD 327
Query: 193 EFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDK 252
E A+ I E + E+ LK +S + + D +R +L ++E W +ER+QM+L A+ +K
Sbjct: 328 ELAKEIEEDKAEIEALKSES-MNLREEVDDERRMLQMAEVWREERVQMKLIDAKVTLEEK 386
Query: 253 NSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESV----PLNDAVSAP-------- 300
S ++KL ++E F+ ++ + E +V R + SV + + P
Sbjct: 387 YSQMNKLVGDMEAFLSSRNTTGVKEVRVAELLRETAASVDNIQEIKEFTYEPAKPDDILM 446
Query: 301 ----QAMGDDDDDDSV----------GSDSHCFE-----LNKPSHMGAKVHKE---EILD 338
MG++ D +S S +H +NK H A + E D
Sbjct: 447 LFEQMNMGENQDRESEQYVAYSPVSHASKAHTVSPDVNLINKGRHSNAFTDQNGEFEEDD 506
Query: 339 KNLDETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPE 398
+ S S P E N ++ + S++ E T +R
Sbjct: 507 SGWETVSHSEEHGSSYSPDESIPNISNTHHRNSNVSMNGTEYEKTLLR------------ 554
Query: 399 HFEISESGGFERKNNLSELHSTSKNHIIDNLIRGQLLASESGKNNKHAEHNNYGEASCSN 458
EI E R+ + +L S +K + G++ N + + + SN
Sbjct: 555 --EIKEVCSVPRRQS-KKLPSMAKLWSSLEGMNGRV------SNARKSTVEMVSPETGSN 605
Query: 459 AGWRNQASPVRQWMASLDISEASKVHSGSK----------DNTLKAKLLEARSKGQRSRL 508
G N V QW +S D + A+ G K N+LK KL+EA+ + Q+ +L
Sbjct: 606 KGGFNTLDLVGQWSSSPDSANANLNRGGRKGCIEWPRGAHKNSLKTKLIEAQIESQKVQL 665
Query: 509 R 509
+
Sbjct: 666 K 666
>At5g22310 unknown protein
Length = 481
Score = 113 bits (282), Expect = 4e-25
Identities = 83/284 (29%), Positives = 144/284 (50%), Gaps = 62/284 (21%)
Query: 54 LKTSTELLKVLNRIWSLEEQH-SSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDL 112
L TS EL+KVL RI L + H +++ LI AL ELDR R +K L+ E+D+
Sbjct: 186 LTTSKELVKVLKRIGELGDDHKTASNRLISALLCELDRARSSLKHLMS-------ELDE- 237
Query: 113 MKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTS 172
E++ ++S+++E ERKLR+R+E ++R+L REL+E K +
Sbjct: 238 --------------EEEEKRRLIESLQEEAMVERKLRRRTEKMNRRLGRELTEAKETERK 283
Query: 173 ALKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISES 232
+++++E+ + +LE++CDE +GI + ++E+ + +R ++HI++
Sbjct: 284 MKEEMKREKRAKDVLEEVCDELTKGIGDDKKEM---------------EKEREMMHIADV 328
Query: 233 WLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVI----------R 282
+ER+QM+L A+ F DK + V++L E+ + ++ S+E + I
Sbjct: 329 LREERVQMKLTEAKFEFEDKYAAVERLKKELRRVLDGEEGKGSSEIRRILEVIDGSGSDD 388
Query: 283 DRRNSLESVPLN--------------DAVSAPQAMGDDDDDDSV 312
D + L+S+ LN D + GDDDDDD V
Sbjct: 389 DEESDLKSIELNMESGSKWGYVDSLKDRRRFDGSGGDDDDDDPV 432
>At5g41650 unknown protein
Length = 117
Score = 79.0 bits (193), Expect = 8e-15
Identities = 40/50 (80%), Positives = 44/50 (88%), Gaps = 4/50 (8%)
Query: 596 QKGYSSLLSFTVTDINSTVTKLMALGAELDGPIKYEVHGKVLSSNHAMQC 645
+KGYSSLLSFTVTDIN+TVTKLMALGAELDG IKYE+HGKV AM+C
Sbjct: 60 EKGYSSLLSFTVTDINTTVTKLMALGAELDGTIKYEIHGKVA----AMKC 105
>At1g11690 hypothetical protein
Length = 247
Score = 64.3 bits (155), Expect = 2e-10
Identities = 42/214 (19%), Positives = 114/214 (52%), Gaps = 22/214 (10%)
Query: 69 SLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQ 128
+ +E + +L+ L+TEL + + ++KEL +A++ + ++ ++ + ++ +++++
Sbjct: 23 NFQEDEFLDFNLVPCLQTELWKAQTRIKEL----EAEKFKSEETIRCLIRNQ---RNEKE 75
Query: 129 DRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLE 188
+ + V ++++L ER+ +KR ++ + +L +++ +M+SS + + R R +E
Sbjct: 76 ETTNPFVDYLKEKLSKEREEKKRVKAENSRLKKKILDMESS-------VNRLRRERDTME 128
Query: 189 DLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNG 248
+C+E I+E LK + + W + +R +L ++E W +ER++++ A+
Sbjct: 129 KVCEELVTRIDE-------LKVNTRRVW-DETEEERQMLQMAEMWREERVRVKFMDAKLA 180
Query: 249 FMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIR 282
+K ++ +E+E ++ + E + +R
Sbjct: 181 LQEKYEEMNLFVVELEKCLETAREVGGIEEKRLR 214
>At4g40020 putative protein
Length = 615
Score = 54.7 bits (130), Expect = 2e-07
Identities = 98/442 (22%), Positives = 182/442 (41%), Gaps = 70/442 (15%)
Query: 30 PYNPAATPNSSLDFKGRIGEPHYSLKTSTE------------LLKVLNRIWSLEEQHSSN 77
P A+P S + IG Y +K S E LLK I +LE
Sbjct: 8 PSIDVASPRYSSNKVADIGTELYKMKASLENRENEVVSLKQELLKKDIFIKNLEAAEKKL 67
Query: 78 ISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQ- 136
+ K EL+ T+ V+E + E+ L ++I + S E+D ++VQ
Sbjct: 68 LDSFKDQSRELEETKALVEE-------SKVEIASLKEKIDTSYNSQDSSEEDEDDSSVQD 120
Query: 137 ----SVRDELEDER----KLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLE 188
S++ E+E + + + +++ K++ L EMKS + E T K ++
Sbjct: 121 FDIESLKTEMESTKESLAQAHEAAQASSLKVSELLEEMKSVKNELKSATDAEMTNEKAMD 180
Query: 189 DL----------CDEFARGINEYEQEVHTLKQKSEKDWVQRADH------------DRLV 226
DL C + + E E+ + +S++ W + + +RL
Sbjct: 181 DLALALKEVATDCSQTKEKLVIVETELEAARIESQQ-WKDKYEEVRKDAELLKNTSERLR 239
Query: 227 LHISESWL--DERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSA--ENQVIR 282
+ ES L + + + + + G +KNS++D+ + +E + A+ S+ A EN +R
Sbjct: 240 IEAEESLLAWNGKESVFVTCIKRGEDEKNSLLDENNRLLEALVAAENLSKKAKEENHKVR 299
Query: 283 DRRNSLESVPLNDAVSAPQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHKEEILDKNLD 342
D + +N+A A +A G ++S D+ L+K + + + E + N +
Sbjct: 300 D----ILKQAINEANVAKEAAGIARAENSNLKDA---LLDKEEELQFALKEIERVKVN-E 351
Query: 343 ETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLTTNIRLVEGTRISEEPEHFEI 402
+ N+KK K + E + +K +S++ +E + + V +I EE E E
Sbjct: 352 AVANDNIKKLKKMLSE----IEVAMEEEKQRSLNRQESMPKEVVEVVEKKI-EEKEKKEE 406
Query: 403 SESGGFERKNNLSEL--HSTSK 422
+ E+K + E HS K
Sbjct: 407 KKENKKEKKESKKEKKEHSEKK 428
>At4g36520 trichohyalin like protein
Length = 1432
Score = 53.5 bits (127), Expect = 4e-07
Identities = 91/437 (20%), Positives = 174/437 (38%), Gaps = 71/437 (16%)
Query: 70 LEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHE------------VDDLMKQIA 117
+E + S L + LK + TRIK L + DR E + ++Q
Sbjct: 615 MEMRSQSETKLNEPLKRMEEETRIKEARLR--EENDRRERVAVEKAENEKRLKAALEQEE 672
Query: 118 EDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSE-SIHRKLARELSEMKSSFTSALKD 176
+++ +++++E+ R++ E ERK++++ E + K A E E A
Sbjct: 673 KERKIKEAREKAENERRAVEAREKAEQERKMKEQQELELQLKEAFEKEEENRRMREAFA- 731
Query: 177 LEQERTRRKLLEDLCDEFARGINEYEQEVH-------TLKQKSE----KDWVQRADHDRL 225
LEQE+ RR +E R I E ++ TL+Q+ + K+ +R +++R
Sbjct: 732 LEQEKERRIKEAREKEENERRIKEAREKAELEQRLKATLEQEEKERQIKERQEREENERR 791
Query: 226 VLHISESWLDERM----------QMQLEAAQNGFMDKNSIVDKLSLE------IETFIKA 269
+ E +ER + +L+ + +K + + + LE IE F +A
Sbjct: 792 AKEVLEQAENERKLKEALEQKENERRLKETREKEENKKKLREAIELEEKEKRLIEAFERA 851
Query: 270 KQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGDDDD--------DDSVGSDSHCFEL 321
+ R E+ + R L+ + + +++ ++S + E+
Sbjct: 852 EIERRLKEDLEQEEMRMRLQEAKERERLHRENQEHQENERKQHEYSGEESDEKERDACEM 911
Query: 322 NKPSHMGAKVHKEEILDKNLDETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSV------ 375
K + H E+ +++L +T + N V K ++S S
Sbjct: 912 EKTCETTKEAHGEQSSNESLSDTLEENESIDNDVSVNKQKKEEEGTRQRESMSAETCPWK 971
Query: 376 -------DAEEGLTTNIRLVEGTRISEEPEHF-EISESGGFERKNNLSELHSTSKNHIID 427
DA + TN + TR+ E E + E+GG ++N S STS + +
Sbjct: 972 VFEKTLKDASQKEGTN-EMDADTRLFERNEETPRLGENGGCNQQNGESGEESTS---VTE 1027
Query: 428 NLIRGQLLASESGKNNK 444
N+I G+L + KN++
Sbjct: 1028 NIIGGKL--EQKSKNSE 1042
>At1g74250 putative heat shock protein
Length = 630
Score = 50.8 bits (120), Expect = 2e-06
Identities = 80/400 (20%), Positives = 159/400 (39%), Gaps = 30/400 (7%)
Query: 92 RIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKR 151
R+ +E + R+ + E +D ++ +AE R + D L + + E+ER+ +K+
Sbjct: 202 RMMEEENKKSRKKAKREYNDTVRGLAEFVKKRDKRVIDMLVKKNAEMEKKKEEERERKKK 261
Query: 152 SESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQK 211
E +L R ++ + + A + E E LE+ D+ R + V + K K
Sbjct: 262 MEK--ERLERAMNYEEPEWAKA-HEGEDEGAGLSELEEEDDDAKRKNEQLYCIVCSKKFK 318
Query: 212 SEKDW---VQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIK 268
SEK W Q H V + ES+ D + + E +G +D V++L +++ +
Sbjct: 319 SEKQWKNHEQSKKHKEKVAELRESFTDYEEENE-EEEIDGPLDSPESVEELHEKLQEELN 377
Query: 269 AKQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGDDDDDDSVGSDSHCFELNKPSHMG 328
R + +V+ + + + + + + +D+DD+ L K G
Sbjct: 378 IDNEERDVKKEVVGEADETDDEYFVAEEDMQGSSESEDEDDE--------MTLLKKMVSG 429
Query: 329 AKVHKEEILDKNLDETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLTTNIRLV 388
K ++ ++ K DE + + F N + NK+++ EE N
Sbjct: 430 QKNKQKNVVSKEEDEDETEVEIEGDTAEFSEFDNQKSTGRNKEAK----EERNKQN---- 481
Query: 389 EGTRISEEPEHFEISESGGFERKN-NLSE-----LHSTSKNHIIDNLIRGQLLASESGKN 442
G ++++ +I GG +N N +E L + K+ + ++
Sbjct: 482 AGNDMADDTSKVQIPGEGGNPDENMNATESASGALADSQKDEANSMEYDNRKSTGRRRRS 541
Query: 443 NKHAEHNNYGEASCSNAGWRNQASPVRQWMASLDISEASK 482
K + NN GE + + + V + M S D +A +
Sbjct: 542 KKGKDKNNQGELN-EKSSEADDTQYVNRDMESQDYKKAPR 580
>At1g79280 hypothetical protein
Length = 2111
Score = 48.1 bits (113), Expect = 2e-05
Identities = 61/265 (23%), Positives = 111/265 (41%), Gaps = 24/265 (9%)
Query: 51 HYSLKTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQ---ADRH 107
H S S++L V R L S + +A + +R RI ++ + R A R
Sbjct: 653 HSSDSRSSDLSPVPGRKNFLHLLEDSEEATKRAQEKAFERIRILEEDFAKARSEVIAIRS 712
Query: 108 EVDDLMKQ--IAEDKLVRKSKEQDRLHAAVQSVRDE--------LEDERKLRKRSESIHR 157
E D L + A +KL KE +R + SV ++ +RKLR+ SES+H
Sbjct: 713 ERDKLAMEANFAREKLEGIMKESERKREEMNSVLARNIEFSQLIIDHQRKLRESSESLH- 771
Query: 158 KLARELSEMKSSFTSALK-DLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQ----KS 212
A E+S S S LK + E K D ++ + + + T++ +
Sbjct: 772 -AAEEISRKLSMEVSVLKQEKELLSNAEKRASDEVSALSQRVYRLQATLDTVQSTEEVRE 830
Query: 213 EKDWVQRADHDRLVLHISESWLDERMQMQLEA--AQNGFMDKNSIVDKLSLEIETFIKAK 270
E +R + + + W + + ++Q E A++ D+N ++ +++E K
Sbjct: 831 ETRAAERRKQEEHIKQLQREWAEAKKELQEERSNARDFTSDRNQTLNNAVMQVEEMGKEL 890
Query: 271 QNSRSAENQVIRDRRNSLESVPLND 295
N+ A + + + R S+ L+D
Sbjct: 891 ANALKAVS--VAESRASVAEARLSD 913
Score = 32.7 bits (73), Expect = 0.68
Identities = 29/137 (21%), Positives = 66/137 (48%), Gaps = 11/137 (8%)
Query: 79 SLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQSV 138
+L+K +TELD ++++L + + VD+L + + + +RL V+ +
Sbjct: 1332 NLLKTKQTELDLCMKEMEKLRMETDLHKKRVDELRETYRNIDIA----DYNRLKDEVRQL 1387
Query: 139 RDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARGI 198
++L+ + + + + + ++S ++ T+ KDL + R K L+D A
Sbjct: 1388 EEKLKAKDAHAEDCKKVLLEKQNKISLLEKELTNCKKDLSE---REKRLDDAQQAQATMQ 1444
Query: 199 NEYEQEVHTLKQKSEKD 215
+E+ ++ KQ+ EK+
Sbjct: 1445 SEFNKQ----KQELEKN 1457
Score = 29.6 bits (65), Expect = 5.8
Identities = 23/111 (20%), Positives = 47/111 (41%), Gaps = 5/111 (4%)
Query: 118 EDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLAR---ELSEMKSSFTSAL 174
E +L + Q + + + ELE +K+ RK + ELS+ S L
Sbjct: 1430 EKRLDDAQQAQATMQSEFNKQKQELEKNKKIHYTLNMTKRKYEKEKDELSKQNQSLAKQL 1489
Query: 175 KDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQ--KSEKDWVQRADHD 223
++ ++E +R + + ++ + E E+ + L + KD V++ D
Sbjct: 1490 EEAKEEAGKRTTTDAVVEQSVKEREEKEKRIQILDKYVHQLKDEVRKKTED 1540
Score = 29.3 bits (64), Expect = 7.5
Identities = 31/149 (20%), Positives = 66/149 (43%), Gaps = 6/149 (4%)
Query: 85 KTELDRTRIKVKELLRDRQADRHEVDDLMKQIAE-DKLVRKSKEQDRLHAAVQSVRDELE 143
K EL++ + L ++ E D+L KQ K + ++KE+ +V ++
Sbjct: 1451 KQELEKNKKIHYTLNMTKRKYEKEKDELSKQNQSLAKQLEEAKEEAGKRTTTDAVVEQSV 1510
Query: 144 DERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTR-RKLLEDLCDEFARGINEYE 202
ER+ +++ I K +L + T LK ++E T+ R + + E + + +
Sbjct: 1511 KEREEKEKRIQILDKYVHQLKDEVRKKTEDLKKKDEELTKERSERKSVEKEVGDSLTKIK 1570
Query: 203 QEVHTLKQKSEKDWVQRADHDRLVLHISE 231
+E K K +++ + + + H+SE
Sbjct: 1571 KE----KTKVDEELAKLERYQTALTHLSE 1595
>At1g64690 hypothetical protein
Length = 273
Score = 47.8 bits (112), Expect = 2e-05
Identities = 47/177 (26%), Positives = 80/177 (44%), Gaps = 34/177 (19%)
Query: 81 IKALKTELD-------RTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHA 133
IK LK ELD R + +K+L +D + +R + ++ E + R KE +
Sbjct: 84 IKELKAELDYERKARRRAELMIKKLAKDVEEER-----MAREAEEMQNKRLFKELSSEKS 138
Query: 134 AVQSVRDELEDERKLRK-----RSESIHRKLARE---LSEMKSSFTSALKDLEQERTRR- 184
+ ++ +LE+ER++ + R E + KL L E S A + E+ER R
Sbjct: 139 EMVRMKRDLEEERQMHRLAEVLREERVQMKLMDARLFLEEKLSELEEANRQGERERNRMM 198
Query: 185 --KLLEDLCD----------EFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHI 229
K+LE C + RGIN + + + ++ KSEK W + + ++ L I
Sbjct: 199 KPKILERACSSPARRRCENPQIKRGINPFPRVMRAIRSKSEK-WGSKLECQKVQLKI 254
Score = 43.9 bits (102), Expect = 3e-04
Identities = 26/84 (30%), Positives = 47/84 (55%), Gaps = 2/84 (2%)
Query: 110 DDLMKQIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSS 169
D++ + AE RK++ + L ++ + ++E+ER R+ E +++L +ELS KS
Sbjct: 82 DEIKELKAELDYERKARRRAEL--MIKKLAKDVEEERMAREAEEMQNKRLFKELSSEKSE 139
Query: 170 FTSALKDLEQERTRRKLLEDLCDE 193
+DLE+ER +L E L +E
Sbjct: 140 MVRMKRDLEEERQMHRLAEVLREE 163
>At1g63300 hypothetical protein
Length = 1029
Score = 47.8 bits (112), Expect = 2e-05
Identities = 106/474 (22%), Positives = 189/474 (39%), Gaps = 91/474 (19%)
Query: 38 NSSLDFKGRIGEPHYSLKTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKE 97
N + +K + LK E L + LE Q S + +K E + ++KE
Sbjct: 506 NHDISYKLEQSQLQEQLKIQYECSSSLVDVTELENQVESLEAELKKQSEEFSESLCRIKE 565
Query: 98 LLRDRQADRHEVDDLMKQIAE---DKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSES 154
L + E++ Q+ E D + R EQ++ A+Q+ E RK R ++ S
Sbjct: 566 LESQMETLEEEMEK-QAQVFEADIDAVTRGKVEQEQ--RAIQAE----ETLRKTRWKNAS 618
Query: 155 IHRKLARELS----EMKSSFTS----ALKDLE-------QERTRRKLLEDLCDEFARGIN 199
+ KL E +M S FTS A+K + Q+R ++++D DE
Sbjct: 619 VAGKLQDEFKRLSEQMDSMFTSNEKMAMKAMTEANELRMQKRQLEEMIKDANDELRANQA 678
Query: 200 EYEQEVHTLKQK-------------------SEKDWVQRADHD-----RLVLHISESWLD 235
EYE ++H L +K +E D +R + D + I + ++
Sbjct: 679 EYEAKLHELSEKLSFKTSQMERMLENLDEKSNEIDNQKRHEEDVTANLNQEIKILKEEIE 738
Query: 236 ------ERMQMQLEAAQNGFMD----KNSIVD-----------KLSLEIETFIKAKQN-S 273
+ + +Q E A+N +D K S+++ K+ LE + + K++ S
Sbjct: 739 NLKKNQDSLMLQAEQAENLRVDLEKTKKSVMEAEASLQRENMKKIELESKISLMRKESES 798
Query: 274 RSAENQVIRDRRNSLESVPLNDAVSAPQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVHK 333
+AE QVI+ ++ E+ ++ + + + DD + E+ K A V K
Sbjct: 799 LAAELQVIKLAKDEKETA-ISLLQTELETVRSQCDDLKHSLSENDLEMEKHKKQVAHV-K 856
Query: 334 EEILDKNLDETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEG------LTTNIRL 387
E+ K E + +N++KK N+ K V A G + I+L
Sbjct: 857 SELKKK---EETMANLEKKLKESRTAITKTAQRNNINKGSPVGAHGGSKEVAVMKDKIKL 913
Query: 388 VEGTRISEEPEHFEISESGGFERKNNL--------SELHSTSKNHIIDNLIRGQ 433
+EG +I + E S + E++ NL ++L S+ + L+ GQ
Sbjct: 914 LEG-QIKLKETALESSSNMFIEKEKNLKNRIEELETKLDQNSQEMSENELLNGQ 966
Score = 33.9 bits (76), Expect = 0.31
Identities = 38/166 (22%), Positives = 72/166 (42%), Gaps = 13/166 (7%)
Query: 20 ASYGSSMEMTPYNPAATPNSSLDFKGRIGEPHYSLKTSTELLKVLNRIWSLEEQHSSNIS 79
AS S M+ PN+ + + P L ++ L + +RI E SS+
Sbjct: 202 ASIESDSTMSSSGSVIEPNTPEEVAKPLRHPTKHLHSAKSLFEEPSRISESEWSGSSD-- 259
Query: 80 LIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQSVR 139
+ + D T + RD + + D++ K +++LV +++ D +QS+R
Sbjct: 260 --HGISSTDDSTNSSNDIVARDTAINSSDEDEVEK--LKNELVGLTRQADLSELELQSLR 315
Query: 140 DELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRK 185
++ E K RS+ L RE++ +K S +D E+++ K
Sbjct: 316 KQIVKETK---RSQD----LLREVNSLKQERDSLKEDCERQKVSDK 354
Score = 28.9 bits (63), Expect = 9.8
Identities = 56/239 (23%), Positives = 101/239 (41%), Gaps = 35/239 (14%)
Query: 55 KTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMK 114
K S L L I +++ + ISL L+TEL+ R + +L + E++ K
Sbjct: 794 KESESLAAELQVIKLAKDEKETAISL---LQTELETVRSQCDDLKHSLSENDLEMEKHKK 850
Query: 115 QIAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSA- 173
Q+A K K KE+ + E+KL K S + K A+ + K S A
Sbjct: 851 QVAHVKSELKKKEETMANL-----------EKKL-KESRTAITKTAQRNNINKGSPVGAH 898
Query: 174 --LKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISE 231
K++ + + KLLE + E + K+K+ K+ ++ E
Sbjct: 899 GGSKEVAVMKDKIKLLEGQI-KLKETALESSSNMFIEKEKNLKNRIEEL----------E 947
Query: 232 SWLDERMQMQLEAAQNGFMD--KNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSL 288
+ LD+ Q E ++N ++ +N + L EIE+ ++ S E + +R+R + +
Sbjct: 948 TKLDQNSQ---EMSENELLNGQENEDIGVLVAEIES-LRECNGSMEMELKEMRERYSEI 1002
>At5g52280 hyaluronan mediated motility receptor-like protein
Length = 853
Score = 47.0 bits (110), Expect = 3e-05
Identities = 105/490 (21%), Positives = 196/490 (39%), Gaps = 92/490 (18%)
Query: 81 IKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQSVRD 140
++AL+ + + + ++ + L + + + +L K+++ K R ++ +Q+ RD
Sbjct: 273 LEALRRQSELSELEKQSLRKQAIKESKRIQELSKEVSCLKGERDGAMEECEKLRLQNSRD 332
Query: 141 ELEDERKLR---KRSESIHRKLARELS-----------------EMKSSFTSALKD---- 176
E + E +LR + S ++ ++ ELS E S+ A++D
Sbjct: 333 EADAESRLRCISEDSSNMIEEIRDELSCEKDLTSNLKLQLQRTQESNSNLILAVRDLNEM 392
Query: 177 LEQERTRRKLLEDLCDEFA-----RGINEYEQEVHTLKQKSEK-DW----VQRADHDRLV 226
LEQ+ L L +E +G++ E+ TLKQ+ E DW ++ + ++ +
Sbjct: 393 LEQKNNEISSLNSLLEEAKKLEEHKGMDSGNNEIDTLKQQIEDLDWELDSYKKKNEEQEI 452
Query: 227 L-----HISESWLDERM--------QMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNS 273
L ES +E Q + A++ ++D I+D+L +IE ++ K
Sbjct: 453 LLDELTQEYESLKEENYKNVSSKLEQQECSNAEDEYLDSKDIIDELKSQIE-ILEGKLKQ 511
Query: 274 RSAENQVIRDRRNSLES-VPLNDAVSAPQAMGDDDDDDSVGSDSHCFELNKPSHMGAKVH 332
+S E N LES V QA D+D D++ + + + +
Sbjct: 512 QSLEYSECLITVNELESQVKELKKELEDQAQAYDEDIDTM--------MREKTEQEQRAI 563
Query: 333 KEEILDKNLDETSKSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEEGLT-------TNI 385
K E +NL +T +N ++ E + S + + E LT N+
Sbjct: 564 KAE---ENLRKTRWNN-----AITAERLQEKCKRLSLEMESKLSEHENLTKKTLAEANNL 615
Query: 386 RLVEGT-RISEEPEHFEISESGGFERKNNLSELHSTSKNHIIDNLIRGQLLASESGKNNK 444
RL T +E H EI++ +RK H KN + ++ Q+L SE K K
Sbjct: 616 RLQNKTLEEMQEKTHTEITQEKE-QRK------HVEEKNKALS--MKVQMLESEVLKLTK 666
Query: 445 HAEHNNYGEASCSNAGWRNQASPVRQWMASLDISE--ASKVHSGSKDNTLKAKLLEARSK 502
+ S+A +++W D E S +K + L ++ +
Sbjct: 667 LRDE--------SSAAATETEKIIQEWRKERDEFERKLSLAKEVAKTAQKELTLTKSSND 718
Query: 503 GQRSRLRTLK 512
+ +RLR LK
Sbjct: 719 DKETRLRNLK 728
Score = 36.6 bits (83), Expect = 0.047
Identities = 31/118 (26%), Positives = 59/118 (49%), Gaps = 11/118 (9%)
Query: 81 IKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQIAEDK--LVRKSKEQDRLHAAVQSV 138
++ LKTE++ ++ EL ++ E D+L KQ++ K + RK +E ++ A
Sbjct: 724 LRNLKTEVEGLSLQYSELQNSFVQEKMENDELRKQVSNLKVDIRRKEEEMTKILDARMEA 783
Query: 139 RDEL--EDERKLRKRSESI------HRKLARELSEMKSSFTS-ALKDLEQERTRRKLL 187
R + E L K S+ + + + REL EM+ ++ +L+ E E R++L+
Sbjct: 784 RSQENGHKEENLSKLSDELAYCKNKNSSMERELKEMEERYSEISLRFAEVEGERQQLV 841
>At3g16000 myosin heavy chain like protein
Length = 727
Score = 46.6 bits (109), Expect = 5e-05
Identities = 49/215 (22%), Positives = 94/215 (42%), Gaps = 8/215 (3%)
Query: 80 LIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQI--AEDKLVRKSKEQDRLHAAVQS 137
L + +K EL+ ++KE Q+ + E+ ++ K++ + +L + K L+ V+
Sbjct: 517 LRRRVKDELEGVTHELKESSVKNQSLQKELVEIYKKVETSNKELEEEKKTVLSLNKEVKG 576
Query: 138 VRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRRKLLEDLCDEFARG 197
+ ++ ER+ RK E+ + + L EM + + ++LE+ T LED + R
Sbjct: 577 MEKQILMEREARKSLETDLEEAVKSLDEMNKNTSILSRELEKVNTHASNLEDEKEVLQRS 636
Query: 198 INEYEQEVHTLKQKSEKD--WVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSI 255
+ E + K+ E V +R VL L+E L +A+ + S
Sbjct: 637 LGEAKNASKEAKENVEDAHILVMSLGKEREVLEKKVKKLEE----DLGSAKGEILRMRSQ 692
Query: 256 VDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLES 290
D + T K K ++ +V+R R++S S
Sbjct: 693 PDSVKAVNSTDNKEKSDNTVTVKKVVRRRKSSTSS 727
Score = 41.2 bits (95), Expect = 0.002
Identities = 77/347 (22%), Positives = 136/347 (39%), Gaps = 61/347 (17%)
Query: 42 DFKGRIGEPHYSLKTSTELLKVLNRIWSLEEQHS--SNISLIKALKTELDRTRIKVKELL 99
DF+ ++ K E K EEQ S + ++ K L TEL R K+L
Sbjct: 179 DFEAKLQHEQEERKKEVEKAK--------EEQLSLINQLNSAKDLVTELGRELSSEKKLC 230
Query: 100 RDRQADRHEVDDLMKQIAEDKLVRKSKEQDRLHAAVQSVRD-------ELEDERKLRKRS 152
+ +++ + + EDK ++K +++L V+ ++D EL+D + +R
Sbjct: 231 EKLKDQIESLENSLSKAGEDKEALETKLREKLDL-VEGLQDRINLLSLELKDSEEKAQRF 289
Query: 153 ESIHRKLARELSEMKSSFTSALKDL-------EQERTRRKLLEDLCDEFARGINEYEQEV 205
+ K EL E+ S +T +DL +Q++ + D I E +
Sbjct: 290 NASLAKKEAELKELNSIYTQTSRDLAEAKLEIKQQKEELIRTQSELDSKNSAIEELNTRI 349
Query: 206 HTLKQKSEKDWVQRAD---HDRLVLHISE-----------SWLDERMQMQLEAAQNGFMD 251
TL + E ++Q+ D D L ++ S ++ +Q E D
Sbjct: 350 TTLVAEKE-SYIQKLDSISKDYSALKLTSETQAAADAELISRKEQEIQQLNENLDRALDD 408
Query: 252 KNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLESV-----PLNDAVSAPQAMGDD 306
N DK++ E + +K+ E +++ R+ LE D VS + M D+
Sbjct: 409 VNKSKDKVADLTEKYEDSKR-MLDIELTTVKNLRHELEGTKKTLQASRDRVSDLETMLDE 467
Query: 307 DDDDSVGSDSHCFELNKPSHMGAKVHKE-----EILDKNLDETSKSN 348
S + C +L A VH+E E ++NLD + N
Sbjct: 468 -------SRALCSKLESEL---AIVHEEWKEAKERYERNLDAEKQKN 504
>At1g73860 kinesin-related protein
Length = 1050
Score = 46.6 bits (109), Expect = 5e-05
Identities = 51/243 (20%), Positives = 107/243 (43%), Gaps = 32/243 (13%)
Query: 66 RIWSLEEQHSSNISLIKALKTELDRTRIKVKELLR--DRQADRHEVDDLMKQIAEDKLVR 123
++ +QH+ IS ALK EL+ T+ K ++ + Q + + ++ K ED + +
Sbjct: 255 KLMKQNDQHNLEIS---ALKQELETTKRKYEQQYSQIESQTKKSKWEEQKKNEEED-MDK 310
Query: 124 KSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQE--- 180
KE D+ + + ++R ELE RK ++ S ++ + + S LK+LEQE
Sbjct: 311 LLKENDQFNLQISALRQELETTRKAYEQQCS---QMESQTMVATTGLESRLKELEQEGKV 367
Query: 181 --------RTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISES 232
R K LE + E N E+++ L+Q ++ + + + ++
Sbjct: 368 VNTAKNALEERVKELEQMGKEAHSAKNALEEKIKQLQQMEKETKTANTSLEGKIQELEQN 427
Query: 233 ---WLDERMQMQLEAAQN------GFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRD 283
W + +M+ ++ N + S +D S + ++ + SRS + ++++
Sbjct: 428 LVMWKTKVREMEKKSESNHQRWSQKELSYKSFIDNQS---QALLELRSYSRSIKQEILKV 484
Query: 284 RRN 286
+ N
Sbjct: 485 QEN 487
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 45.8 bits (107), Expect = 8e-05
Identities = 92/416 (22%), Positives = 167/416 (40%), Gaps = 51/416 (12%)
Query: 70 LEEQHSSN----ISLIKALKTELDRTRIKVKELLRDRQADRHEVDDL---MKQIAEDKLV 122
L+E HS SL +T + +V EL ++ + DL +K E+
Sbjct: 303 LKESHSVKDRDLFSLRDIHETHQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEENKA 362
Query: 123 RKSKE---QDRLHAAVQSVR---DELEDERKLRKRSESIHRKLARELSEMKSSFTSALKD 176
SK D+L A +++ DEL + + K ES L + + + +L +
Sbjct: 363 ISSKNLEIMDKLEQAQNTIKELMDELGELKDRHKEKESELSSLVKSADQQVADMKQSLDN 422
Query: 177 LEQER---TRRKL-LEDLCDEFARGINEYEQEVHTLKQK---SEKDWV---------QRA 220
E+E+ ++R L + + E + I E+ E LK+ E++ QR
Sbjct: 423 AEEEKKMLSQRILDISNEIQEAQKTIQEHMSESEQLKESHGVKERELTGLRDIHETHQRE 482
Query: 221 DHDRLVLHISESWLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQN------SR 274
RL ++ L E+ + L A+ N ++ + + LEI +K Q+ +
Sbjct: 483 SSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVTE 542
Query: 275 SAENQ-VIRDRRNSLES-VPLNDAVSAPQAMGDDDDDDSVGS-DSHCFELNKPSHMGAKV 331
AE++ + + N L S V +++A + + + V S + ELN+ ++ +
Sbjct: 543 LAESKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQVKELNQ--NLNSSE 600
Query: 332 HKEEILDKNLDETS-----KSNVKKKKSVPGEGFKNHIPSNSNKKSQSVDAEE----GLT 382
+++IL + + E S + ++ S E K N+ D E L+
Sbjct: 601 EEKKILSQQISEMSIKIKRAESTIQELSSESERLKGSHAEKDNELFSLRDIHETHQRELS 660
Query: 383 TNIRLVEGTRISEEPEHFEISESGGFERKNNLSELHSTSKNHIIDNLIRGQLLASE 438
T +R +E S E E+SES + S ST + D L R Q++ E
Sbjct: 661 TQLRGLEAQLESSEHRVLELSES--LKAAEEESRTMSTKISETSDELERTQIMVQE 714
Score = 40.8 bits (94), Expect = 0.002
Identities = 47/240 (19%), Positives = 99/240 (40%), Gaps = 20/240 (8%)
Query: 55 KTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMK 114
++ST L ++ ++ LE++ + + A + E + E+ + + + +V +L+
Sbjct: 482 ESSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSKVQELVT 541
Query: 115 QIAEDKLVRKSKEQD-----RLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSS 169
++AE K KE + +H A + RD ++L R ES ++ +EL++
Sbjct: 542 ELAESKDTLTQKENELSSFVEVHEAHK--RDSSSQVKELEARVESAEEQV-KELNQ---- 594
Query: 170 FTSALKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHI 229
+L +K+L E + I E + L +SE+ A+ D + +
Sbjct: 595 ------NLNSSEEEKKILSQQISEMSIKIKRAESTIQELSSESERLKGSHAEKDNELFSL 648
Query: 230 SESWLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAENQVIRDRRNSLE 289
+ + E Q +L G + + LE+ +KA + + I + + LE
Sbjct: 649 RD--IHETHQRELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRTMSTKISETSDELE 706
Score = 40.4 bits (93), Expect = 0.003
Identities = 40/171 (23%), Positives = 81/171 (46%), Gaps = 13/171 (7%)
Query: 55 KTSTELLKVLNRIWSLEEQHSSNISLIKALKTELD--RTRIKVKEL-LRDRQADRHEVDD 111
K S E+ + L++I +L+E+ + + + +++ E++ +IK +EL L R E+D+
Sbjct: 897 KKSEEISEYLSQITNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQRSELDE 956
Query: 112 LMKQIAE------DKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSE 165
++ E DK+ S E L + ++++EL+ + + +E+ + +E SE
Sbjct: 957 ELRTKKEENVQMHDKINVASSEIMALTELINNLKNELDSLQVQKSETEAELEREKQEKSE 1016
Query: 166 MKSSFTSALKDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDW 216
+ + T K L ++ LE E + INE +E K D+
Sbjct: 1017 LSNQITDVQKALVEQEAAYNTLE----EEHKQINELFKETEATLNKVTVDY 1063
Score = 37.7 bits (86), Expect = 0.021
Identities = 55/273 (20%), Positives = 106/273 (38%), Gaps = 32/273 (11%)
Query: 56 TSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQ 115
+ST ++ ++ S ++Q S + +KA + E K E + + ++ + +LM +
Sbjct: 152 SSTRASELEAQLESSKQQVSDLSASLKAAEEENKAISSKNVETMNKLEQTQNTIQELMAE 211
Query: 116 IAEDKLVRKSKEQDRLHAAVQSVRDELEDERKLRKRSESIHRKLARELSEMKSSFTSAL- 174
+ + K + KE S L + + +R SIH K E E + L
Sbjct: 212 LGKLKDSHREKE---------SELSSLVEVHETHQRDSSIHVKELEEQVESSKKLVAELN 262
Query: 175 KDLEQERTRRKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISE--- 231
+ L +K+L E + I E + + L +S + + DR + + +
Sbjct: 263 QTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLKESHSVKDRDLFSLRDIHE 322
Query: 232 ------SWLDERMQMQLEAAQNGFMD----------KNSIVDKLSLEIETFIKAKQNSRS 275
S ++ QLE+++ D +N + +LEI ++ QN+
Sbjct: 323 THQRESSTRVSELEAQLESSEQRISDLTVDLKDAEEENKAISSKNLEIMDKLEQAQNTIK 382
Query: 276 A---ENQVIRDRRNSLESVPLNDAVSAPQAMGD 305
E ++DR ES + SA Q + D
Sbjct: 383 ELMDELGELKDRHKEKESELSSLVKSADQQVAD 415
Score = 35.4 bits (80), Expect = 0.10
Identities = 47/253 (18%), Positives = 112/253 (43%), Gaps = 18/253 (7%)
Query: 60 LLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHEVDDLMKQ--IA 117
+L++ + + EE+ + + I EL+RT+I V+EL D + ++ + + +
Sbjct: 677 VLELSESLKAAEEESRTMSTKISETSDELERTQIMVQELTADSSKLKEQLAEKESKLFLL 736
Query: 118 EDKLVRKSKEQDRLHAAVQSVRDELED--------ERKLRKRSESIHRKLARELSEMKSS 169
+K + + L A V ++ ELE E ++ ++ ++ +L + EM +
Sbjct: 737 TEKDSKSQVQIKELEATVATLELELESVRARIIDLETEIASKT-TVVEQLEAQNREMVAR 795
Query: 170 FTSALKDLEQERTR----RKLLEDLCDEFARGINEYEQEVHTLKQKSEKDWVQRADHDRL 225
+ K +E+ T + LED + + I E+ L+ + + VQ+ + ++
Sbjct: 796 ISELEKTMEERGTELSALTQKLEDNDKQSSSSIETLTAEIDGLRAELDSMSVQKEEVEKQ 855
Query: 226 VLHISESWLDERMQMQLEAAQNGFMDKNSIVDKLSLEIETFIKAKQNSRSAE-NQVIRDR 284
++ SE + ++ E NG + + +D E+E ++ K S +Q+ +
Sbjct: 856 MVCKSEEASVKIKRLDDEV--NGLRQQVASLDSQRAELEIQLEKKSEEISEYLSQITNLK 913
Query: 285 RNSLESVPLNDAV 297
+ V +++++
Sbjct: 914 EEIINKVKVHESI 926
>At3g19060 hypothetical protein
Length = 1647
Score = 45.8 bits (107), Expect = 8e-05
Identities = 70/334 (20%), Positives = 145/334 (42%), Gaps = 62/334 (18%)
Query: 49 EPHYSLKTSTELLKVLNRIWSLEEQHSSNISLIKALKTELDRTRIKVKELLRDRQADRHE 108
E +LKT EL ++ LE + + + + L+ + ++ R ++L + + R E
Sbjct: 1267 EKTLALKTF-ELEDAVSHAQMLEVRLQESKEITRNLEVDTEKARKCQEKLSAENKDIRAE 1325
Query: 109 VDDLM-------KQIAEDKLVRKSKE--------------------QDRLHAAVQSVRDE 141
+DL+ +++ + K V +S E Q +L+ A+ RD
Sbjct: 1326 AEDLLAEKCSLEEEMIQTKKVSESMEMELFNLRNALGQLNDTVAFTQRKLNDAIDE-RDN 1384
Query: 142 LEDERKLRKRSESIHRKLARELSEMKSSFTSALKDLEQERTRR-------KLLEDLCDEF 194
L+DE L + E K+ E EM++ + A + E +T KLLE +E
Sbjct: 1385 LQDE-VLNLKEE--FGKMKSEAKEMEARYIEAQQIAESRKTYADEREEEVKLLEGSVEEL 1441
Query: 195 ARGINEYEQEVHTLKQKSEKDWVQRADHDRLVLHISESWLDERMQMQLEAAQNGFMDKNS 254
IN E +V+ +K ++E+ +QR + + + LH ++ Q+E+A+N +
Sbjct: 1442 EYTINVLENKVNVVKDEAERQRLQREELE-MELH--------TIRQQMESARNADEEMKR 1492
Query: 255 IVDKLSLEI---ETFIKAKQNSRSAENQVIRDRRNSLESVPLNDAVSAPQAMGDDDDDDS 311
I+D+ +++ + I+A + + + + I + + L+ A + M + ++
Sbjct: 1493 ILDEKHMDLAQAKKHIEALERNTADQKTEITQLSEHISELNLHAEAQASEYMHKFKELEA 1552
Query: 312 VGSDSHCFELNKPSHMGAKVHKEEILDKNLDETS 345
+ E KP ++H + +D +L + S
Sbjct: 1553 MA------EQVKP-----EIHVSQAIDSSLSKGS 1575
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.131 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,379,701
Number of Sequences: 26719
Number of extensions: 618330
Number of successful extensions: 3765
Number of sequences better than 10.0: 331
Number of HSP's better than 10.0 without gapping: 38
Number of HSP's successfully gapped in prelim test: 299
Number of HSP's that attempted gapping in prelim test: 3239
Number of HSP's gapped (non-prelim): 630
length of query: 676
length of database: 11,318,596
effective HSP length: 106
effective length of query: 570
effective length of database: 8,486,382
effective search space: 4837237740
effective search space used: 4837237740
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140913.13


