

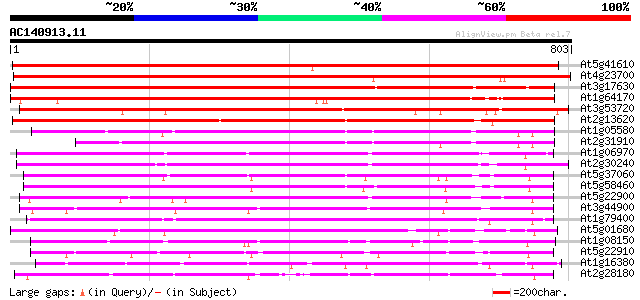
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140913.11 - phase: 0
(803 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g41610 Na+/H+ antiporter-like protein 1043 0.0
At4g23700 putative Na+/H+-exchanging protein 955 0.0
At3g17630 unknown protein 944 0.0
At1g64170 hypothetical protein 801 0.0
At3g53720 unknown protein 739 0.0
At2g13620 putative Na/H antiporter 732 0.0
At1g05580 unknown protein 431 e-121
At2g31910 putative Na+/H+ antiporter 377 e-104
At1g06970 hypothetical protein 372 e-103
At2g30240 putative Na/H antiporter 369 e-102
At5g37060 putative transporter protein 295 5e-80
At5g58460 putative protein 295 7e-80
At5g22900 Na+/H+ antiporter-like protein 285 5e-77
At3g44900 putative protein 277 1e-74
At1g79400 hypothetical protein 274 2e-73
At5g01680 putative transporter protein 261 1e-69
At1g08150 hypothetical protein 261 1e-69
At5g22910 Na+/H+ antiporter-like protein 252 5e-67
At1g16380 putative Na/H antiporter 245 8e-65
At2g28180 hypothetical protein 244 2e-64
>At5g41610 Na+/H+ antiporter-like protein
Length = 810
Score = 1043 bits (2697), Expect = 0.0
Identities = 529/789 (67%), Positives = 653/789 (82%), Gaps = 9/789 (1%)
Query: 5 TTSGNVCPPPMQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIA 64
T S CP PM+ SNGVFQGD+P+D+ALPLAILQI +V+V+TR LAYLL PLRQPRVIA
Sbjct: 3 TNSTKACPAPMKATSNGVFQGDNPIDFALPLAILQIVIVIVLTRVLAYLLRPLRQPRVIA 62
Query: 65 EIVGGILLGPSALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTG 124
E++GGI+LGPS LGR+K +L AVFP KSL VL+TLANLGL+FFLFLAG+E+D K+LR+TG
Sbjct: 63 EVIGGIMLGPSLLGRSKAFLDAVFPKKSLTVLETLANLGLLFFLFLAGLEIDTKALRRTG 122
Query: 125 GRVLAIAIAGISLPFALGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAE 184
+ L IA+AGI+LPFALGIGSSFVL+ TI+KGVN++AFLV+MGVALSITAFPVLARILAE
Sbjct: 123 KKALGIALAGITLPFALGIGSSFVLKATISKGVNSTAFLVFMGVALSITAFPVLARILAE 182
Query: 185 LKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSILI 244
LKLLTT +GR+AMSAAAVNDVAAWILLALA+ALSG++ SP VSLWVFLSGC FV+ + I
Sbjct: 183 LKLLTTEIGRLAMSAAAVNDVAAWILLALAIALSGSNTSPLVSLWVFLSGCAFVIGASFI 242
Query: 245 VLPIFKWMAQQCHEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGA 304
+ PIF+W++++CHEGEP++E YICATLA VL GF+TDAIGIH+MFGAFV G+L+PK+G
Sbjct: 243 IPPIFRWISRRCHEGEPIEETYICATLAVVLVCGFITDAIGIHSMFGAFVVGVLIPKEGP 302
Query: 305 FAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTIV 364
FAGALVEK+EDLVSGL LPLYFV+SGLKT++ATIQG QSWGLLV VT TACFGKI+GT+
Sbjct: 303 FAGALVEKVEDLVSGLFLPLYFVASGLKTNVATIQGAQSWGLLVLVTATACFGKILGTLG 362
Query: 365 VSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTP 424
VSL K+P E++ LGFLMN+KGLVELIVLNIGKDRKVLNDQTFAIMVLMAL TTF+TTP
Sbjct: 363 VSLAFKIPMREAITLGFLMNTKGLVELIVLNIGKDRKVLNDQTFAIMVLMALFTTFITTP 422
Query: 425 FVLAAYK---RKERKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKH 481
V+A YK R +++ YK+R +ER+N + QLRIL CFHG+ +IPS+INL+EASRGI+K
Sbjct: 423 VVMAVYKPARRAKKEGEYKHRAVERENTNTQLRILTCFHGAGSIPSMINLLEASRGIEKG 482
Query: 482 DALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWD-KGRHGDSLHVIVAFEAYQKLSQVF 540
+ LCVYA+HL+E ERSS+ILM KVR+NG+PFW+ +G + D+ V+VAF+A+Q+LS+V
Sbjct: 483 EGLCVYALHLRELSERSSAILMVHKVRKNGMPFWNRRGVNADADQVVVAFQAFQQLSRVN 542
Query: 541 VRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEH 600
VRPM AISSM++IHEDICTTA RK+AA++ILPFHK Q++DGSL+ R D+R VN+RVL
Sbjct: 543 VRPMTAISSMSDIHEDICTTAVRKKAAIVILPFHKHQQLDGSLETTRGDYRWVNRRVLLQ 602
Query: 601 APCSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIH 660
APCSVGIFVDRGLGGS VSA +VSY + VLFFGG DDREALAYG RMAEHPGI L V
Sbjct: 603 APCSVGIFVDRGLGGSSQVSAQDVSYSVVVLFFGGPDDREALAYGLRMAEHPGIVLTVFR 662
Query: 661 FLVEPNIAGKITKVDVGDSSS-NNSISD-SEDGEFLAEFKLKTANDDSIIYEERIVKDAE 718
F+V P G+I V+V ++++ N S+ + D E ++E + ++ D+S+ + E+ +++A
Sbjct: 663 FVVSPERVGEIVNVEVSNNNNENQSVKNLKSDEEIMSEIRKISSVDESVKFVEKQIENAA 722
Query: 719 ETV-ATIREINFCNLFLVGRRPAGELGFAL-ERSECPELGPVGGLLASQDFRTTASVLVM 776
V + I E+ NLFLVGR P GE+ A+ E SECPELGPVG LL S + T ASVLV+
Sbjct: 723 VDVRSAIEEVRRSNLFLVGRMPGGEIALAIRENSECPELGPVGSLLISPESSTKASVLVI 782
Query: 777 QQYHNGVPI 785
QQY NG I
Sbjct: 783 QQY-NGTGI 790
>At4g23700 putative Na+/H+-exchanging protein
Length = 820
Score = 955 bits (2469), Expect = 0.0
Identities = 502/817 (61%), Positives = 620/817 (75%), Gaps = 20/817 (2%)
Query: 6 TSGNVCPPPMQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAE 65
T+G CP PM+ SNGVFQG++PL++ALPL ILQIC+VL++TR LA+LL PLRQPRVIAE
Sbjct: 3 TNGTTCPGPMKATSNGVFQGENPLEHALPLLILQICIVLLLTRLLAFLLRPLRQPRVIAE 62
Query: 66 IVGGILLGPSALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGG 125
IVGGILLGPSALG++ +++ VFPPKSL VLDTLANLGLIFFLFL G+ELDPKSL++TG
Sbjct: 63 IVGGILLGPSALGKSTKFINTVFPPKSLTVLDTLANLGLIFFLFLVGLELDPKSLKRTGK 122
Query: 126 RVLAIAIAGISLPFALGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAEL 185
R L+IA+AGI+LPF LGIG+SF L+ +IA G + + FLV+MGVALSITAFPVLARILAE+
Sbjct: 123 RALSIALAGITLPFVLGIGTSFALRSSIADGASKAPFLVFMGVALSITAFPVLARILAEI 182
Query: 186 KLLTTSVGRMAMSAAAVNDVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSILIV 245
KLLTT +G++A+SAAAVNDVAAWILLALAVALSG SP SLWVFLSGCGFV+ I +V
Sbjct: 183 KLLTTDIGKIALSAAAVNDVAAWILLALAVALSGEGSSPLTSLWVFLSGCGFVLFCIFVV 242
Query: 246 LPIFKWMAQQCHEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAF 305
P K +A++C EGEPV+ELY+C TL VLAA FVTD IGIHA+FGAFV G++ PK+G F
Sbjct: 243 QPGIKLIAKRCPEGEPVNELYVCCTLGIVLAASFVTDFIGIHALFGAFVIGVIFPKEGNF 302
Query: 306 AGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTIVV 365
A ALVEK+EDLVSGL LPLYFVSSGLKT++ATIQG QSWGLLV V F ACFGKI+GT++V
Sbjct: 303 ANALVEKVEDLVSGLFLPLYFVSSGLKTNVATIQGAQSWGLLVLVIFNACFGKIIGTVLV 362
Query: 366 SLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPF 425
SL CKVP ++SL LGFLMN+KGLVELIVLNIGKDR VLNDQ FAIMVLMA+ TTFMTTP
Sbjct: 363 SLYCKVPLDQSLALGFLMNTKGLVELIVLNIGKDRGVLNDQIFAIMVLMAIFTTFMTTPL 422
Query: 426 VLAAYK--RKERKSNYKYRTIERKNADGQ-LRILACFHGSRNIPSLINLIEASRGIKKHD 482
VLA YK + K++YK RT+E N + L ++ CF NIP+++NLIEASRGI + +
Sbjct: 423 VLAVYKPGKSLTKADYKNRTVEETNRSNKPLCLMFCFQSIMNIPTIVNLIEASRGINRKE 482
Query: 483 ALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGR----HGDSLHVIVAFEAYQKLSQ 538
L VYAMHL E ERSS+ILMA KVR+NGLPFW+K + S V+VAFEA+++LS+
Sbjct: 483 NLSVYAMHLMELSERSSAILMAHKVRRNGLPFWNKDKSENNSSSSDMVVVAFEAFRRLSR 542
Query: 539 VFVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVL 598
V VRPM AIS MA IHEDIC +A+RK+ A++ILPFHK R+D + + RND+R +NK+V+
Sbjct: 543 VSVRPMTAISPMATIHEDICQSAERKKTAMVILPFHKHVRLDRTWETTRNDYRWINKKVM 602
Query: 599 EHAPCSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVV 658
E +PCSV I VDRGLGG+ V++S+ S I VLFFGG DDREALA+ RMAEHPGI L V
Sbjct: 603 EESPCSVAILVDRGLGGTTRVASSDFSLTITVLFFGGNDDREALAFAVRMAEHPGISLTV 662
Query: 659 IHFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLK-------TANDDS---II 708
+ F+ + ++++ + + + D E + E K K +N DS II
Sbjct: 663 VRFIPSDEFKPENVRIEITEDQLCSGATRLIDIEAITELKAKIKEKESSRSNSDSESHII 722
Query: 709 YEERIVKDAEETVATIREINFCNLFLVGRRPAGELGFALE-RSECPELGPVGGLLA-SQD 766
YEE+IVK EE + I+E + NLFLVG+ P G + + RS+ PELGP+G LL S+
Sbjct: 723 YEEKIVKCYEEVIEVIKEYSKSNLFLVGKSPEGSVASGINVRSDTPELGPIGNLLTESES 782
Query: 767 FRTTASVLVMQQYHNGVPINFVPEM-EEHSRDGDTES 802
T ASVLV+QQY P+ + E S D+ES
Sbjct: 783 VSTVASVLVVQQYIASRPVGISKNVTTEESLVEDSES 819
>At3g17630 unknown protein
Length = 857
Score = 944 bits (2439), Expect = 0.0
Identities = 480/779 (61%), Positives = 592/779 (75%), Gaps = 7/779 (0%)
Query: 1 MTSNTTSGNVCPPPMQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQP 60
M S +G CP PM+ SNG FQ + PLD+ALPL ILQI LV+V TR LAY L PL+QP
Sbjct: 58 MASTNVTGQ-CPGPMKATSNGAFQNESPLDFALPLIILQIVLVVVFTRLLAYFLKPLKQP 116
Query: 61 RVIAEIVGGILLGPSALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSL 120
RVIAEI+GGILLGPSALGR+K YL +FP KSL VLDTLAN+GL+FFLFL G+ELD ++
Sbjct: 117 RVIAEIIGGILLGPSALGRSKAYLDTIFPKKSLTVLDTLANIGLLFFLFLVGLELDFAAI 176
Query: 121 RKTGGRVLAIAIAGISLPFALGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLAR 180
+KTG + L IAIAGISLPF +G+G+SFVL TI+KGV+ F+V+MGVALSITAFPVLAR
Sbjct: 177 KKTGKKSLLIAIAGISLPFIVGVGTSFVLSATISKGVDQLPFIVFMGVALSITAFPVLAR 236
Query: 181 ILAELKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVC 240
ILAELKLLTT +GRMAMSAA VNDVAAWILLALA+ALSG+ SP VS+WV L G GFV+
Sbjct: 237 ILAELKLLTTDIGRMAMSAAGVNDVAAWILLALAIALSGDGTSPLVSVWVLLCGTGFVIF 296
Query: 241 SILIVLPIFKWMAQQCHEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVP 300
+++ + P+ +MA++C EGEPV ELY+C TL VLAA FVTD IGIHA+FGAFV GI+ P
Sbjct: 297 AVVAIKPLLAYMARRCPEGEPVKELYVCVTLTVVLAASFVTDTIGIHALFGAFVVGIVAP 356
Query: 301 KDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIV 360
K+G F L EKIEDLVSGLLLPLYF +SGLKTD+ TI+G QSWGLLV V T CFGKIV
Sbjct: 357 KEGPFCRILTEKIEDLVSGLLLPLYFAASGLKTDVTTIRGAQSWGLLVLVILTTCFGKIV 416
Query: 361 GTIVVSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTF 420
GT+ S++CKVPF E++ LGFLMN+KGLVELIVLNIGKDRKVLNDQ FAI+VLMAL TTF
Sbjct: 417 GTVGSSMLCKVPFREAVTLGFLMNTKGLVELIVLNIGKDRKVLNDQAFAILVLMALFTTF 476
Query: 421 MTTPFVLAAYKRKERKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKK 480
+TTP V+ YK + + YK+RTI+RK+ D +LRILACFH +RNIP+LINLIE+SRG K
Sbjct: 477 ITTPIVMLIYKPARKGAPYKHRTIQRKDHDSELRILACFHSTRNIPTLINLIESSRGTGK 536
Query: 481 HDALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIVAFEAYQKLSQVF 540
LCVYAMHL E ERSS+I M K R NGLP W+K +++AFEAYQ L V
Sbjct: 537 KGRLCVYAMHLMELSERSSAIAMVHKARNNGLPIWNKIERSTD-QMVIAFEAYQHLRAVA 595
Query: 541 VRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEH 600
VRPM AIS +++IHEDICT+A +KR A+I+LPFHK QR+DG+++ I + F VN+RVL+
Sbjct: 596 VRPMTAISGLSSIHEDICTSAHQKRVAMILLPFHKHQRMDGAMESIGHRFHEVNQRVLQR 655
Query: 601 APCSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIH 660
APCSVGI VDRGLGG+ V AS V+Y + + FFGG DDREALAYG +M EHPGI L V
Sbjct: 656 APCSVGILVDRGLGGTSQVVASEVAYKVVIPFFGGLDDREALAYGMKMVEHPGITLTVYK 715
Query: 661 FLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEET 720
F+ G + + + + D EF+ E ++S+ YEER+V+ ++
Sbjct: 716 FVA---ARGTLKRFEKSEHDEKEKKEKETDEEFVRELMNDPRGNESLAYEERVVESKDDI 772
Query: 721 VATIREINFCNLFLVGRRPAGELGFALERSECPELGPVGGLLASQDFRTTASVLVMQQY 779
+AT++ ++ CNLF+VGR A + ++ ++CPELGPVG LL+S +F TTASVLV+Q Y
Sbjct: 773 IATLKSMSKCNLFVVGRNAA--VASLVKSTDCPELGPVGRLLSSSEFSTTASVLVVQGY 829
>At1g64170 hypothetical protein
Length = 847
Score = 801 bits (2068), Expect = 0.0
Identities = 450/837 (53%), Positives = 571/837 (67%), Gaps = 70/837 (8%)
Query: 1 MTSNTTSGNVCPPP---MQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPL 57
+ + T CP M+ SNGVF G+ PLD+A PL ILQICLV+ VTR LA+LL P+
Sbjct: 4 LVNGTIPAMKCPKNVAMMKTTSNGVFDGESPLDFAFPLVILQICLVVAVTRSLAFLLRPM 63
Query: 58 RQPRVIAEIV------------------------------------GGILLGPSALGRNK 81
RQPRV+AEI+ GGILLGPSALGR
Sbjct: 64 RQPRVVAEIIVSPPSTGLGQSYSFRFNKYPTRLKYELYRDSVQLITGGILLGPSALGRIT 123
Query: 82 GYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFAL 141
Y +++FP +SL VLDTLANLGL+ FLFL G+E+D SLR+TG + ++IA AG+ LPF +
Sbjct: 124 SYKNSIFPARSLTVLDTLANLGLLLFLFLVGLEIDLTSLRRTGKKAISIAAAGMLLPFGM 183
Query: 142 GIGSSFVLQRTIAKGVNTSA--FLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSA 199
GI +SF + G N+ F+++MGVALSITAF VLARILAELKLLTT +GR++M+A
Sbjct: 184 GIVTSFAFPEASSSGDNSKVLPFIIFMGVALSITAFGVLARILAELKLLTTDLGRISMNA 243
Query: 200 AAVNDVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQCHEG 259
AA+NDVAAW+LLALAV+LSG+ SP V LWV LSG FV+ LIV IFK+++++C EG
Sbjct: 244 AAINDVAAWVLLALAVSLSGDRNSPLVPLWVLLSGIAFVIACFLIVPRIFKFISRRCPEG 303
Query: 260 EPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSG 319
EP+ E+Y+C L AVL AGF TDAIGIHA+FGAFV G+L PK G F+ A+VEKIEDLV G
Sbjct: 304 EPIGEMYVCVALCAVLLAGFATDAIGIHAIFGAFVMGVLFPK-GHFSDAIVEKIEDLVMG 362
Query: 320 LLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVL 379
LLLPLYFV SGLKTDI TIQG++SWG L V TACFGKIVGT+ V+L+CKV ES+VL
Sbjct: 363 LLLPLYFVMSGLKTDITTIQGVKSWGRLALVIVTACFGKIVGTVSVALLCKVRLRESVVL 422
Query: 380 GFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKS-- 437
G LMN+KGLVELIVLNIGKDRKVL+DQTFAIMVLMA+ TTF+TTP VLA YK E
Sbjct: 423 GVLMNTKGLVELIVLNIGKDRKVLSDQTFAIMVLMAIFTTFITTPIVLALYKPSETTQTH 482
Query: 438 ---NYKYRTIERK---NADG----QLRILACFHGSRNIPSLINLIEASRGIKK-HDALCV 486
+YK R RK + +G QL++L C S++I ++ ++EA+RG + + CV
Sbjct: 483 SSVSYKNRKHRRKIENDEEGEKMQQLKVLVCLQSSKDIDPMMKIMEATRGSNETKERFCV 542
Query: 487 YAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIVAFEAYQKLSQVFVRPMIA 546
Y MHL + ER SSI M QKVR NGLPFW+K R S V VAFEA KLS V VR + A
Sbjct: 543 YVMHLTQLSERPSSIRMVQKVRSNGLPFWNKKRENSSA-VTVAFEASSKLSSVSVRSVTA 601
Query: 547 ISSMANIHEDICTTADRKRAAVIILPFHKQQR-VDGSLDIIRNDFRLVNKRVLEHAPCSV 605
IS ++ IHEDIC++AD K A +ILPFHKQ R ++ + +R++++ +NKRVLE++PCSV
Sbjct: 602 ISPLSTIHEDICSSADSKCTAFVILPFHKQWRSLEKEFETVRSEYQGINKRVLENSPCSV 661
Query: 606 GIFVDRGLG-GSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFLVE 664
GI VDRGLG + V++SN S + VLFFGG DDREAL YG RMAEHPG+ L V+ ++
Sbjct: 662 GILVDRGLGDNNSPVASSNFSLSVNVLFFGGCDDREALVYGLRMAEHPGVNLTVV--VIS 719
Query: 665 PNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVATI 724
+ + +++ ++S S D +FLA K K AN + +EER V EE V I
Sbjct: 720 GPESARFDRLEAQETS-----LCSLDEQFLAAIK-KRAN--AARFEERTVNSTEEVVEII 771
Query: 725 REINFCNLFLVGRRPAGELGFALE--RSECPELGPVGGLLASQDFRTTASVLVMQQY 779
R+ C++ LVG+ G + L + ECPELGPVG L+ S + T+ SVLV+QQY
Sbjct: 772 RQFYECDILLVGKSSKGPMVSRLPVMKIECPELGPVGNLIVSNEISTSVSVLVVQQY 828
>At3g53720 unknown protein
Length = 842
Score = 739 bits (1908), Expect = 0.0
Identities = 414/836 (49%), Positives = 563/836 (66%), Gaps = 53/836 (6%)
Query: 15 MQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGGILLGP 74
++ +SNGV+QGD+PL++A PL I+Q L++ V+R LA L PLRQP+VIAEIVGGILLGP
Sbjct: 8 VKTSSNGVWQGDNPLNFAFPLLIVQTALIIAVSRFLAVLFKPLRQPKVIAEIVGGILLGP 67
Query: 75 SALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAG 134
SALGRN Y+ +FP S+P+L+++A++GL+FFLFL G+ELD S+R++G R IA+AG
Sbjct: 68 SALGRNMAYMDRIFPKWSMPILESVASIGLLFFLFLVGLELDLSSIRRSGKRAFGIAVAG 127
Query: 135 ISLPFALGIGSSFVLQRTIAKGVNT---SAFLVYMGVALSITAFPVLARILAELKLLTTS 191
I+LPF G+G +FV++ T+ + + FLV+MGVALSITAFPVLARILAELKLLTT
Sbjct: 128 ITLPFIAGVGVAFVIRNTLYTAADKPGYAEFLVFMGVALSITAFPVLARILAELKLLTTQ 187
Query: 192 VGRMAMSAAAVNDVAAWILLALAVALSGNS-------QSPFVSLWVFLSGCGFVVCSILI 244
+G AM+AAA NDVAAWILLALAVAL+GN +SP VSLWV LSG GFVV +++
Sbjct: 188 IGETAMAAAAFNDVAAWILLALAVALAGNGGEGGGEKKSPLVSLWVLLSGAGFVVFMLVV 247
Query: 245 VLPIFKWMAQQ-CHEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDG 303
+ P KW+A++ E + V E Y+C TLA V+ +GF TD IGIH++FGAFVFG+ +PKDG
Sbjct: 248 IRPGMKWVAKRGSPENDVVRESYVCLTLAGVMVSGFATDLIGIHSIFGAFVFGLTIPKDG 307
Query: 304 AFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTI 363
F L+E+IED VSGLLLPLYF +SGLKTD+A I+G +SWG+L V TAC GKIVGT
Sbjct: 308 EFGQRLIERIEDFVSGLLLPLYFATSGLKTDVAKIRGAESWGMLGLVVVTACAGKIVGTF 367
Query: 364 VVSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTT 423
VV+++ KVP E+L LGFLMN+KGLVELIVLNIGK++KVLND+TFAI+VLMAL TTF+TT
Sbjct: 368 VVAVMVKVPAREALTLGFLMNTKGLVELIVLNIGKEKKVLNDETFAILVLMALFTTFITT 427
Query: 424 PFVLAAYK-RKERKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHD 482
P V+A YK + K + + + +LRILAC HG N+ SLI+L+E+ R K
Sbjct: 428 PTVMAIYKPARGTHRKLKDLSASQDSTKEELRILACLHGPANVSSLISLVESIR-TTKIL 486
Query: 483 ALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGD-SLHVIVAFEAYQKLSQVFV 541
L ++ MHL E ERSSSI+M Q+ R+NGLPF + RHG+ +VI FEAY++L +V V
Sbjct: 487 RLKLFVMHLMELTERSSSIIMVQRARKNGLPFVHRYRHGERHSNVIGGFEAYRQLGRVAV 546
Query: 542 RPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRV-------------DGSL-DIIR 587
RP+ A+S + +HEDIC AD KR +IILPFHK+ DG++ + +
Sbjct: 547 RPITAVSPLPTMHEDICHMADTKRVTMIILPFHKRWNADHGHSHHHQDGGGDGNVPENVG 606
Query: 588 NDFRLVNKRVLEHAPCSVGIFVDRGLGG----SCHVSASNVSYCIAVLFFGGGDDREALA 643
+ +RLVN+RVL++APCSV + VDRGLG + + SNV + V+FFGG DDRE++
Sbjct: 607 HGWRLVNQRVLKNAPCSVAVLVDRGLGSIEAQTLSLDGSNVVERVCVIFFGGPDDRESIE 666
Query: 644 YGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSS--------NNSISDSEDGEF-- 693
G RMAEHP +++ VI FLV + + S ++ ++ E
Sbjct: 667 LGGRMAEHPAVKVTVIRFLVRETLRSTAVTLRPAPSKGKEKNYAFLTTNVDPEKEKELDE 726
Query: 694 --LAEFKLKTANDDSIIYEERIVKDAEETVATIREINFCNLFLVGRR--PAGELGFALER 749
L +FK K + + Y+E+ + E + +I + +L +VGR P+ E+ ER
Sbjct: 727 GALEDFKSKW--KEMVEYKEKEPNNIIEEILSIGQSKDFDLIVVGRGRIPSAEVAALAER 784
Query: 750 -SECPELGPVGGLLASQDFRTTASVLVMQQYHNG----VPINFVPEMEEHSRDGDT 800
+E PELGP+G +LAS S+LV+QQ++ + ++ + S +GDT
Sbjct: 785 QAEHPELGPIGDVLASSINHIIPSILVVQQHNKAHVEDITVSKIVSESSLSINGDT 840
>At2g13620 putative Na/H antiporter
Length = 821
Score = 732 bits (1890), Expect = 0.0
Identities = 381/795 (47%), Positives = 545/795 (67%), Gaps = 32/795 (4%)
Query: 4 NTTSGNVCPPPMQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVI 63
+T + +C P +NGV+QGD+PLD++LPL +LQ+ LV+VVTR ++L P RQPRVI
Sbjct: 8 STDASIICYAPSMITTNGVWQGDNPLDFSLPLFVLQLTLVVVVTRFFVFILKPFRQPRVI 67
Query: 64 AEIVGGILLGPSALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKT 123
+EI+GGI+LGPS LGR+ + H +FP +S+ VL+T+AN+GL++FLFL G+E+D +RKT
Sbjct: 68 SEILGGIVLGPSVLGRSTKFAHTIFPQRSVMVLETMANVGLLYFLFLVGVEMDIMVVRKT 127
Query: 124 GGRVLAIAIAGISLPFALGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILA 183
G R L IAI G+ LPF +G SF + R+ + +++++GVALS+TAFPVLARILA
Sbjct: 128 GKRALTIAIGGMVLPFLIGAAFSFSMHRS-EDHLGQGTYILFLGVALSVTAFPVLARILA 186
Query: 184 ELKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSIL 243
ELKL+ T +GR++MSAA VND+ AWILLALA+AL+ + ++ F SLWV +S F+ +
Sbjct: 187 ELKLINTEIGRISMSAALVNDMFAWILLALAIALAESDKTSFASLWVMISSAVFIAVCVF 246
Query: 244 IVLPIFKWMAQQCHEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDG 303
+V P W+ ++ EGE E +IC L V+ +GF+TDAIG H++FGAFVFG+++P +G
Sbjct: 247 VVRPGIAWIIRKTPEGENFSEFHICLILTGVMISGFITDAIGTHSVFGAFVFGLVIP-NG 305
Query: 304 AFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTI 363
L+EK+ED VSGLLLPL+F SGLKT+IA IQG +W L V F AC GK++GT+
Sbjct: 306 PLGLTLIEKLEDFVSGLLLPLFFAISGLKTNIAAIQGPATWLTLFLVIFLACAGKVIGTV 365
Query: 364 VVSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTT 423
+V+ +P E + LG L+N+KGLVE+IVLN+GKD+KVL+D+TFA MVL+AL+ T + T
Sbjct: 366 IVAFFHGMPVREGITLGLLLNTKGLVEMIVLNVGKDQKVLDDETFATMVLVALVMTGVIT 425
Query: 424 PFVLAAYKRKERKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDA 483
P V YK ++ +YK RTI++ D +LR+L C H RN+P++INL+EAS K+
Sbjct: 426 PIVTILYKPVKKSVSYKRRTIQQTKPDSELRVLVCVHTPRNVPTIINLLEASHPTKR-SP 484
Query: 484 LCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIVAFEAY-QKLSQVFVR 542
+C+Y +HL E R+S++L+ R++G P ++ S H+I AFE Y Q + V V+
Sbjct: 485 ICIYVLHLVELTGRASAMLIVHNTRKSGRPALNR-TQAQSDHIINAFENYEQHAAFVAVQ 543
Query: 543 PMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEHAP 602
P+ AIS + +HED+C+ A+ KR + II+PFHKQQ VDG ++ +RLVN+ +LE++P
Sbjct: 544 PLTAISPYSTMHEDVCSLAEDKRVSFIIIPFHKQQTVDGGMESTNPAYRLVNQNLLENSP 603
Query: 603 CSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFL 662
CSVGI VDRGL G+ ++++ VS +AVLFFGG DDREALAY RMA+HPGI L V+ F+
Sbjct: 604 CSVGILVDRGLNGATRLNSNTVSLQVAVLFFGGPDDREALAYAWRMAQHPGITLTVLRFI 663
Query: 663 VEPNIAGKITKVDVGDSSSNNSISDSE--------------DGEFLAEFKLKTANDDSII 708
+ D D++S + +DS+ D +++ F+ + A +SI+
Sbjct: 664 HDE---------DEADTASTRATNDSDLKIPKMDHRKQRQLDDDYINLFRAENAEYESIV 714
Query: 709 YEERIVKDAEETVATIREINFC-NLFLVGRRP--AGELGFAL-ERSECPELGPVGGLLAS 764
Y E++V + EETVA +R ++ +LF+VGR + L L + SECPELG +G LLAS
Sbjct: 715 YIEKLVSNGEETVAAVRSMDSSHDLFIVGRGEGMSSPLTAGLTDWSECPELGAIGDLLAS 774
Query: 765 QDFRTTASVLVMQQY 779
DF T SVLV+QQY
Sbjct: 775 SDFAATVSVLVVQQY 789
>At1g05580 unknown protein
Length = 867
Score = 431 bits (1109), Expect = e-121
Identities = 266/763 (34%), Positives = 417/763 (53%), Gaps = 28/763 (3%)
Query: 32 ALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGGILLGPSALGRNKGYLHAVFPPK 91
+LP I Q+ + + R L YL PL P +A+I+ G+L PS LG + + VFP +
Sbjct: 50 SLPFFITQLFVANLSYRVLYYLTRPLYLPPFVAQILCGLLFSPSVLGNTRFIIAHVFPYR 109
Query: 92 SLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFALGIGSSFVLQR 151
VL+T ANL L++ +FL G+ +D + +R T + + IA G+ + AL +G+
Sbjct: 110 FTMVLETFANLALVYNIFLLGLGMDLRMVRITELKPVIIAFTGLLV--ALPVGAFLYYLP 167
Query: 152 TIAKGVNTSAFLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSAAAVNDVAAWILL 211
+ V+ VAL+ T FP LARILA+LKLL + +GR AM AA V D+ W+LL
Sbjct: 168 GNGHPDKIISGCVFWSVALACTNFPDLARILADLKLLRSDMGRTAMCAAIVTDLCTWVLL 227
Query: 212 ALAVAL-----SGNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQCHEGEPVDELY 266
A + N PFV + + FV+ I ++ P W+ + + V + +
Sbjct: 228 VFGFASFSKSGTWNKMMPFVIITTAI----FVLLCIFVIRPGIAWIFAKTVKAGHVGDTH 283
Query: 267 ICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLPLYF 326
+ L V+ G +TDA G+H++ GAF+FG+ +P D + EK+ D +SG+L+PL++
Sbjct: 284 VWFILGGVVLCGLITDACGVHSITGAFLFGLSIPHDHIIRNMIEEKLHDFLSGILMPLFY 343
Query: 327 VSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFLMNSK 386
+ GL+ DI + ++V V ++ KIV T++ SL +P ++ +G LMN+K
Sbjct: 344 IICGLRADIGFMLQFTDKFMMVVVICSSFLVKIVTTVITSLFMHIPMRDAFAIGALMNTK 403
Query: 387 GLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYRTIER 446
G + L+VLN G+D K L+ + M + L+ + + P + AYK K++ ++YK+RT+++
Sbjct: 404 GTLSLVVLNAGRDTKALDSPMYTHMTIALLVMSLVVEPLLAFAYKPKKKLAHYKHRTVQK 463
Query: 447 KNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDALCVYAMHLKEFCER-SSSILMAQ 505
+ +LR+LAC H N+ + NL++ S K+ L V+A+HL E R ++S+L+
Sbjct: 464 IKGETELRVLACVHVLPNVSGITNLLQVSNATKQ-SPLSVFAIHLVELTGRTTASLLIMN 522
Query: 506 KVRQNGLPFWDKGRHGDSLHVIVAFEAYQKLSQVF-VRPMIAISSMANIHEDICTTADRK 564
+ F D+ R +S + FEA + + V+ + A+S A +HEDIC A+ K
Sbjct: 523 DECKPKANFSDRVR-AESDQIAETFEAMEVNNDAMTVQTITAVSPYATMHEDICVLAEDK 581
Query: 565 RAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEHAPCSVGIFVDRGLG--GSCHVSAS 622
R IILP+HK DG + + +N+ VL HAPCSVGI VDRG+ S
Sbjct: 582 RVCFIILPYHKHLTPDGRMGEGNSSHAEINQNVLSHAPCSVGILVDRGMAMVRSESFRGE 641
Query: 623 NVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSN 682
++ +A+LF GG DDREAL+Y RM I+L V+ F + G+ + G ++
Sbjct: 642 SMKREVAMLFVGGPDDREALSYAWRMVGQHVIKLTVVRF-----VPGREALISSGKVAAE 696
Query: 683 NSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVATIREI---NFCNLFLVGRRP 739
D E + EF KT ND S+ Y E++V D ++T+ATIRE+ N +L++VGR
Sbjct: 697 YEREKQVDDECIYEFNFKTMNDSSVKYIEKVVNDGQDTIATIREMEDNNSYDLYVVGRGY 756
Query: 740 AGELGFAL---ERSECPELGPVGGLLASQDFRTTASVLVMQQY 779
+ + S PELG +G LAS +F ASVLV+QQY
Sbjct: 757 NSDSPVTAGLNDWSSSPELGTIGDTLASSNFTMHASVLVIQQY 799
>At2g31910 putative Na+/H+ antiporter
Length = 735
Score = 377 bits (969), Expect = e-104
Identities = 237/698 (33%), Positives = 369/698 (51%), Gaps = 22/698 (3%)
Query: 95 VLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFALGIGSSFVLQRTIA 154
+L+T ANL L++ +FL G+ LD + ++ + + IAI G+ G G ++ A
Sbjct: 2 LLETFANLALVYNVFLLGLGLDLRMIKIKDIKPVIIAIVGLLAALLAGAGLYYLPSNGEA 61
Query: 155 KGVNTSAFLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSAAAVNDVAAWILLALA 214
+ A +Y +A T FP LARILA+LKLL T +G AM AA V D+ WIL
Sbjct: 62 DKI--LAGCMYWSIAFGCTNFPDLARILADLKLLRTDMGHTAMCAAVVTDLCTWILFIFG 119
Query: 215 VALSGNSQSPFVSL-WVFLSGCGFVVCSILIVLPIFKWMAQQCHEGEPVDELYICATLAA 273
+A+ S L + S FV+ ++ P W+ EG V + ++ TLA
Sbjct: 120 MAIFSKSGVRNEMLPYSLASTIAFVLLCYFVIQPGVAWIFNNTVEGGQVGDTHVWYTLAG 179
Query: 274 VLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKT 333
V+ +T+ G+H++ GAF+FG+ +P D + EK+ D +SG+L+PL+++ GL+
Sbjct: 180 VIICSLITEVCGVHSITGAFLFGLSIPHDHIIRKMIEEKLHDFLSGMLMPLFYIICGLRA 239
Query: 334 DIATIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFLMNSKGLVELIV 393
DI + S G++ VT + KI+ T+ S+ ++P + L +G LMN+KG + L++
Sbjct: 240 DIGYMNRTVSVGMMAVVTSASVMVKILSTMFCSIFLRIPLRDGLAIGALMNTKGTMALVI 299
Query: 394 LNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYRTIERKNADGQL 453
LN G+D K L+ + + L L+ + + P + AYK K++ YK RTI++ + +L
Sbjct: 300 LNAGRDTKALDVIMYTHLTLAFLVMSMVVQPLLAIAYKPKKKLIFYKNRTIQKHKGESEL 359
Query: 454 RILACFHGSRNIPSLINLIEASRGIKKHDALCVYAMHLKEFCER-SSSILMAQKVRQNGL 512
+L C H N+ + NL++ S KK L V+A+HL E R ++S+L+ +
Sbjct: 360 CVLTCVHVLPNVSGITNLLQLSNPTKK-SPLNVFAIHLVELTGRTTASLLIMNDEAKPKA 418
Query: 513 PFWDKGRHGDSLHVIVAFEAYQKLSQ-VFVRPMIAISSMANIHEDICTTADRKRAAVIIL 571
F D+ R +S + F A + + V V+ + A+S A + EDIC A+ K+A I+L
Sbjct: 419 NFADRVR-AESDQIAEMFTALEVNNDGVMVQTITAVSPYATMDEDICLLAEDKQACFILL 477
Query: 572 PFHKQQRVDGSLDIIRNDFRLVNKRVLEHAPCSVGIFVDRGLG----GSCHVSASNVSYC 627
P+HK DG L+ +N+ V+ HAPCSVGI VDRG+ S
Sbjct: 478 PYHKNMTSDGRLNEGNAVHAEINQNVMSHAPCSVGILVDRGMTTVRFESFMFQGETTKKE 537
Query: 628 IAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISD 687
IA+LF GG DDREALAY RM +QL V+ F + + V G+++
Sbjct: 538 IAMLFLGGRDDREALAYAWRMVGQEMVQLTVVRF-----VPSQEALVSAGEAADEYEKDK 592
Query: 688 SEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVATIREI---NFCNLFLVGRRPAGELG 744
D E + EF KT ND S+ Y E++VK+ +ET+ I E+ N +L++VGR E
Sbjct: 593 HVDEESIYEFNFKTMNDPSVTYVEKVVKNGQETITAILELEDNNSYDLYIVGRGYQVETP 652
Query: 745 FA---LERSECPELGPVGGLLASQDFRTTASVLVMQQY 779
+ + P+LG +G L S +F ASVLV+QQY
Sbjct: 653 VTSGLTDWNSTPDLGIIGDTLISSNFTMQASVLVVQQY 690
>At1g06970 hypothetical protein
Length = 829
Score = 372 bits (955), Expect = e-103
Identities = 241/780 (30%), Positives = 400/780 (50%), Gaps = 33/780 (4%)
Query: 10 VCPPPMQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGG 69
VC S GVF G PL YA+PL +LQ+ ++++ +R L LL PL+Q + A+++ G
Sbjct: 27 VCQKNHMLTSKGVFLGSDPLKYAMPLMLLQMSVIIITSRLLYRLLKPLKQGMISAQVLAG 86
Query: 70 ILLGPSALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLA 129
I+LGPS G++ Y+ P L TL+NLG LFL G+ +D +RK G + +
Sbjct: 87 IILGPSLFGQSSAYMQMFLPISGKITLQTLSNLGFFIHLFLLGLRIDASIIRKAGSKAIL 146
Query: 130 IAIAGISLPFALGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAELKLLT 189
I A +LPF+LG + L+ T + + + ++T+FPV +LAEL +L
Sbjct: 147 IGTASYALPFSLGNLTVLFLKNTYNLPPDVVHCISTVISLNAMTSFPVTTTVLAELNILN 206
Query: 190 TSVGRMAMSAAAVNDVAAWIL-LALAVALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPI 248
+ +GR+A + + V + +WI+ L + L + S+W F+ ++ + P
Sbjct: 207 SDLGRLATNCSIVCEAFSWIVALVFRMFLRDGT---LASVWSFVWVTALILVIFFVCRPA 263
Query: 249 FKWMAQQCHEG-EPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAG 307
W+ ++ + E+ + +L ++ +G+HA FGAF G+ +P
Sbjct: 264 IIWLTERRSISIDKAGEIPFFPIIMVLLTISLTSEVLGVHAAFGAFWLGVSLPDGPPLGT 323
Query: 308 ALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFG-KIVGTIVVS 366
L K+E + L+LP + SGL+T+ I +S ++ +G K +GT S
Sbjct: 324 GLTTKLEMFATSLMLPCFISISGLQTNFFIIG--ESHVKIIEAVILITYGCKFLGTAAAS 381
Query: 367 LICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFV 426
C + ++ L LM +G++E+ + KD KVLN + F ++++ L+ T ++ V
Sbjct: 382 AYCNIQIGDAFSLALLMCCQGVIEIYTCVMWKDEKVLNTECFNLLIITLLLVTGISRFLV 441
Query: 427 LAAYKRKERKSNYKYRTI-ERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDALC 485
+ Y +R + RTI + + + Q R+L C + N+PS++NL+EAS + +
Sbjct: 442 VCLYDPSKRYRSKSKRTILDTRQRNLQFRLLLCVYNVENVPSMVNLLEASYP-SRFSPIS 500
Query: 486 VYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIVAFEAYQKLSQ--VFVRP 543
V+ +HL E R+ ++L+ P S H++ F+ +++ +Q + +
Sbjct: 501 VFTLHLVELKGRAHAVLVPHHQMNKLDP-----NTVQSTHIVNGFQRFEQQNQGTLMAQH 555
Query: 544 MIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEHAPC 603
A + ++I++DICT A K+A +I++PFHKQ +DG++D + R +N VLE APC
Sbjct: 556 FTAAAPFSSINDDICTLALDKKATLIVIPFHKQYAIDGTVDHVNPSIRNINLNVLEKAPC 615
Query: 604 SVGIFVDRG-LGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFL 662
SVGIF+DRG G V S +AV+F G DD EALA+ R+AEHP + + +IHF
Sbjct: 616 SVGIFIDRGETEGRRSVLMSYTWRNVAVIFIEGRDDAEALAFSMRIAEHPEVSVTMIHFR 675
Query: 663 VEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVA 722
+ ++ VDV S+ + + +FK + I Y E IV+D ET
Sbjct: 676 HKSSLQQNHV-VDVE--------SELAESYLINDFKNFAMSKPKISYREEIVRDGVETTQ 726
Query: 723 TIREI-NFCNLFLVGR---RPAGELGFALERSECPELGPVGGLLASQDFRTTASVLVMQQ 778
I + + +L +VGR + L + SECPELG +G + AS DF SVLV+ Q
Sbjct: 727 VISSLGDSFDLVVVGRDHDLESSVLYGLTDWSECPELGVIGDMFASSDFH--FSVLVIHQ 784
>At2g30240 putative Na/H antiporter
Length = 831
Score = 369 bits (948), Expect = e-102
Identities = 247/804 (30%), Positives = 413/804 (50%), Gaps = 34/804 (4%)
Query: 10 VCPPPMQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGG 69
VC S G+F +PL YALPL +LQ+ +++V +R + +L PL+Q + A+++ G
Sbjct: 29 VCQAQNMLTSRGIFMKSNPLKYALPLLLLQMSVIIVTSRLIFRVLQPLKQGMISAQVLTG 88
Query: 70 ILLGPSALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLA 129
++LGPS LG N Y++ P ++ TL+N+G + LFL G+++D +RK G + +
Sbjct: 89 VVLGPSFLGHNVIYMNMFLPAGGKIIIQTLSNVGFVIHLFLLGLKIDGSIIRKAGSKAIL 148
Query: 130 IAIAGISLPFALGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAELKLLT 189
I A + PF+LG + + +T+ + + S+T+FPV +LAEL +L
Sbjct: 149 IGTASYAFPFSLGNLTIMFISKTMGLPSDVISCTSSAISLSSMTSFPVTTTVLAELNILN 208
Query: 190 TSVGRMAMSAAAVNDVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPIF 249
+ +GR+A + V +V +W +ALA L ++ SL+ G ++ + PI
Sbjct: 209 SELGRLATHCSMVCEVCSW-FVALAFNLYTRDRT-MTSLYALSMIIGLLLVIYFVFRPII 266
Query: 250 KWMAQQCHEG-EPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGA 308
W+ Q+ + + D + L + A +A+G+HA FGAF G+ +P
Sbjct: 267 VWLTQRKTKSMDKKDVVPFFPVLLLLSIASLSGEAMGVHAAFGAFWLGVSLPDGPPLGTE 326
Query: 309 LVEKIEDLVSGLLLPLYFVSSGLKTDIATI-QGLQSWGLLVFVTFTACFG-KIVGTIVVS 366
L K+E S L LP + SGL+T+ I + + +++ + +G K +GT S
Sbjct: 327 LAAKLEMFASNLFLPCFIAISGLQTNFFEITESHEHHVVMIEIILLITYGCKFLGTAAAS 386
Query: 367 LICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFV 426
C+ ++L L FLM +G++E+ + KD +V++ + F ++++ L T ++ V
Sbjct: 387 AYCQTQIGDALCLAFLMCCQGIIEVYTTIVWKDAQVVDTECFNLVIITILFVTGISRFLV 446
Query: 427 LAAYKRKER-KSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDALC 485
+ Y +R KS K + + + QLR+L + N+PS++NL+EA+ + + +
Sbjct: 447 VYLYDPSKRYKSKSKRTILNTRQHNLQLRLLLGLYNVENVPSMVNLLEATYP-TRFNPIS 505
Query: 486 VYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIVAFEAYQKLSQ--VFVRP 543
+ +HL E R+ ++L P S H++ AF+ +++ Q + +
Sbjct: 506 FFTLHLVELKGRAHALLTPHHQMNKLDP-----NTAQSTHIVNAFQRFEQKYQGALMAQH 560
Query: 544 MIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEHAPC 603
A + ++I+ DICT A K+A +I++PFHKQ +DG++ + R +N VL+ APC
Sbjct: 561 FTAAAPYSSINNDICTLALDKKATLIVIPFHKQYAIDGTVGQVNGPIRTINLNVLDAAPC 620
Query: 604 SVGIFVDRG-LGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFL 662
SV IF+DRG G V +N +A+LF GG DD EALA RMAE P + + +IHF
Sbjct: 621 SVAIFIDRGETEGRRSVLMTNTWQNVAMLFIGGKDDAEALALCMRMAEKPDLNVTMIHFR 680
Query: 663 VEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVA 722
+ + + D D S N ISD FK AN I Y E IV+D ET
Sbjct: 681 HKSALQDE----DYSDMSEYNLISD---------FKSYAANKGKIHYVEEIVRDGVETTQ 727
Query: 723 TIREI-NFCNLFLVGR---RPAGELGFALERSECPELGPVGGLLASQDFRTTASVLVMQQ 778
I + + ++ LVGR + L + SECPELG +G +L S DF + V+ QQ
Sbjct: 728 VISSLGDAYDMVLVGRDHDLESSVLYGLTDWSECPELGVIGDMLTSPDFHFSVLVVHQQQ 787
Query: 779 YHNGVPINFVPEME--EHSRDGDT 800
+ + ++ ++ EH + GDT
Sbjct: 788 GDDLLAMDDSYKLPNVEHQKIGDT 811
>At5g37060 putative transporter protein
Length = 859
Score = 295 bits (756), Expect = 5e-80
Identities = 214/795 (26%), Positives = 387/795 (47%), Gaps = 55/795 (6%)
Query: 21 GVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGGILLGPSALGRN 80
G+F+G++ ++YA +++ +++ + ++ L P RQPR+++EI+GG+++GPS G
Sbjct: 53 GMFKGENAMNYAFSTFLIEAIIIIFFIKVVSIALRPFRQPRIVSEIIGGMMIGPSMFGGI 112
Query: 81 KGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFA 140
+ + + +FPP + + + +G +FLFL + D ++ K + IA G+ +P
Sbjct: 113 RNFNYYLFPPIANYICANIGLMGFFYFLFLTAAKTDVGAIGKAPRKHKYIAAIGVIVPII 172
Query: 141 LGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSAA 200
++ + + + + + + ALS T+FPV+ +L ++ LL + VG+ AMS A
Sbjct: 173 CVGSVGMAMRDQMDENLQKPSSIGGVVFALSFTSFPVIYTVLRDMNLLNSEVGKFAMSVA 232
Query: 201 AVNDVAAWILLALAVALS----GNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQC 256
+ D+A ++ + A++ G + S F W +S F +L+V F W+ Q
Sbjct: 233 LLGDMAGVYVIVIFEAMTHADVGGAYSVF---WFLVSVVIFAAFMLLVVRRAFDWIVSQT 289
Query: 257 HEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDL 316
EG V++ YI L VLA+ F+TD G+ G G+LVP L + E
Sbjct: 290 PEGTLVNQNYIVMILMGVLASCFLTDMFGLSIAVGPIWLGLLVPHGPPLGSTLAVRSETF 349
Query: 317 VSGLLLPLYFVSSGLKTDIATIQGLQSW----GLLVFVTFTACFGKIVGTIVVSLICKVP 372
+ L+P + G T+I ++ ++W L ++T K + T +L KVP
Sbjct: 350 IYEFLMPFTYALVGQGTNIHFLRD-ETWRNQLSPLFYMTVVGFITKFLSTAFAALFFKVP 408
Query: 373 FNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAYKR 432
ES+ LG +MN +G ++L+V D++++ + +MVL ++ T +TTP + Y
Sbjct: 409 ARESITLGLMMNLRGQMDLLVYLHWIDKRIVGFPGYTVMVLHTVVVTAVTTPLINFFYDP 468
Query: 433 KERKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDALCVYAMHLK 492
+ K+RTI+ + ++ ++ + LI ++ + K L ++A+ L
Sbjct: 469 TRPYRSSKHRTIQHTPQNTEMGLVLAVSDHETLSGLITFLDFAYPTKS-SPLSIFAVQLV 527
Query: 493 EFCERSSSILMAQKVR--------QNGLPFWDKGRHGDSLHVIVAFEAYQKLSQ--VFVR 542
E R++ + + + R + ++ + G V AF+ Y++ V +R
Sbjct: 528 ELAGRATPLFIDHEQRKEEEEEEYEEEEEEPERKQSGRIDQVQSAFKLYEEKRNECVTLR 587
Query: 543 PMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRL-VNKRVLEHA 601
A + +++DIC A K+ A I+LP+ K++ D + +R+ L VN VLEH
Sbjct: 588 SYTAHAPKRLMYQDICELALGKKTAFILLPYQKERLEDAAPTELRDSGMLSVNADVLEHT 647
Query: 602 PCSVGIFVDRG--------LGGSCHVSASNV-----SYCIAVLFFGGGDDREALAYGARM 648
PCSV I+ D+G L S +++ +Y VLF GG D+REAL RM
Sbjct: 648 PCSVCIYFDKGRLKNAVVRLSMDLQHSTNSIRMRQETYRFVVLFLGGADNREALHLADRM 707
Query: 649 AEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSII 708
+ +P + L VI FL + G+ + D + F +K +++ +
Sbjct: 708 STNPDVTLTVIRFL---------SYNHEGEDEREKKLDDG----VVTWFWVKNESNERVS 754
Query: 709 YEERIVKDAEETVATIR--EINFCNLFLVGRRPAGE---LGFALERSECPELGPVGGLLA 763
Y+E +VK+ ET+A I+ +N +L++ GRR L SE +LG +G +A
Sbjct: 755 YKEVVVKNGAETLAAIQAMNVNDYDLWITGRREGINPKILEGLSTWSEDHQLGVIGDTVA 814
Query: 764 SQDFRTTASVLVMQQ 778
+ F + SVLV+QQ
Sbjct: 815 ASVFASEGSVLVVQQ 829
>At5g58460 putative protein
Length = 857
Score = 295 bits (755), Expect = 7e-80
Identities = 217/792 (27%), Positives = 379/792 (47%), Gaps = 51/792 (6%)
Query: 21 GVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGGILLGPSALGRN 80
G+F+G++ ++Y +++ L++ + + LL PLRQPR++ EI+GG+++GPS LGRN
Sbjct: 53 GMFKGENGMNYTFSTFLIEAILIIFFIKIVYVLLRPLRQPRIVCEIIGGMMIGPSMLGRN 112
Query: 81 KGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFA 140
+ + + +FPP + + + +G +F FL + D + K + IA + +P A
Sbjct: 113 RNFNYYLFPPIANYICANIGLMGFFYFFFLTAAKTDVAEIFKAPRKHKYIAAVSVLVPIA 172
Query: 141 LGIGSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSAA 200
+ L+ + + + + + AL T+FPV+ +L ++ LL + +G+ AMS
Sbjct: 173 CVGSTGAALKHKMDIRLQKPSSIGGVTFALGFTSFPVIYTVLRDMNLLNSEIGKFAMSVT 232
Query: 201 AVNDVAAWILLAL--AVALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQCHE 258
+ D+ +L L A+A + + +W +S C +L+V F+W+ + E
Sbjct: 233 LLGDMVGVYVLVLFEAMAQADGGGGAYSVIWFLISAAIMAACLLLVVKRSFEWIVAKTPE 292
Query: 259 GEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVS 318
G V++ YI L VL + F+TD G+ G G++VP L + E V+
Sbjct: 293 GGLVNQNYIVNILMGVLVSCFLTDMFGMAIAVGPIWLGLVVPHGPPLGSTLAIRSETFVN 352
Query: 319 GLLLPLYFVSSGLKTDIATIQGLQSW----GLLVFVTFTACFGKIVGTIVVSLICKVPFN 374
L+P F G KT++ I ++W L++++ K V + +L KVP
Sbjct: 353 EFLMPFSFALVGQKTNVNLI-SKETWPKQISPLIYMSIVGFVTKFVSSTGAALFFKVPTR 411
Query: 375 ESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKE 434
+SL LG +MN +G +++++ D++++ +++MVL A++ T +T P + Y
Sbjct: 412 DSLTLGLMMNLRGQIDILLYLHWIDKQMVGLPGYSVMVLYAIVVTGVTAPLISFLYDPTR 471
Query: 435 RKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDALCVYAMHLKEF 494
+ K RTI+ + + ++ LI ++ + K V+A+ L E
Sbjct: 472 PYRSSKRRTIQHTPQNTETGLVLAVTDHDTFSGLITFLDFAYPTKT-SPFSVFAIQLVEL 530
Query: 495 CERSSSILMA------QKVRQNGLPFWDKG-RHGDSLHVIVAFEAYQKLSQ--VFVRPMI 545
R+ + +A ++ + P G R D V AF+ YQ+ V +
Sbjct: 531 EGRAQPLFIAHDKKREEEYEEEEEPAERMGSRRVD--QVQSAFKLYQEKRSECVTMHAYT 588
Query: 546 AISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRL-VNKRVLEHAPCS 604
A +S N++++IC A K+ A I+LP+ K++ D +L +R+ L VN VL H PCS
Sbjct: 589 AHASKHNMYQNICELALTKKTAFILLPYQKERLQDAALTELRDSGMLSVNADVLAHTPCS 648
Query: 605 VGIFVDRGLGGSCHVSAS-------------NVSYCIAVLFFGGGDDREALAYGARMAEH 651
V I+ ++G + V +S Y VLF GG D+REAL RM E+
Sbjct: 649 VCIYYEKGRLKNAMVRSSMDPQHTTNSSHMRQEMYRFVVLFLGGADNREALHLADRMTEN 708
Query: 652 PGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSIIYEE 711
P I L VI FL + G+ + D + F +K ++ + Y+E
Sbjct: 709 PFINLTVIRFLAHNH---------EGEDEREKKLDDG----VVTWFWVKNESNARVSYKE 755
Query: 712 RIVKDAEETVATIR--EINFCNLFLVGRRPAGE---LGFALERSECPELGPVGGLLASQD 766
+VK+ ET+A I+ +N +L++ GRR L SE +LG +G +A
Sbjct: 756 VVVKNGAETLAAIQAMNVNDYDLWITGRREGINPKILEGLSTWSEDHQLGVIGDTVAGSV 815
Query: 767 FRTTASVLVMQQ 778
F + SVLV+QQ
Sbjct: 816 FASEGSVLVVQQ 827
>At5g22900 Na+/H+ antiporter-like protein
Length = 822
Score = 285 bits (730), Expect = 5e-77
Identities = 217/812 (26%), Positives = 381/812 (46%), Gaps = 68/812 (8%)
Query: 14 PMQPASNGVFQGD---------HPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIA 64
P+ P+SNGV+ H +YA P + ++ + + L + L L R +
Sbjct: 27 PINPSSNGVWPQQKFSDPNINVHFWNYAFPHLQMIFLIISFLWQFLHFFLRRLGMIRFTS 86
Query: 65 EIVGGILLGPSALGRNKGYLHAVFPPKSLPVLDTL-ANLGLIFFLFLAGIELDPKSLRKT 123
++ G+LL S L N ++ +L A + F FL G+++D +R T
Sbjct: 87 HMLTGVLLSKSFLKENSAARRFFSTEDYKEIVFSLTAACSYMMFWFLMGVKMDTGLIRTT 146
Query: 124 GGRVLAIAIAGISLPFALGIGSSFVLQRTIAKG-----VNTSAFLVYMGVALSITAFPVL 178
G + + I ++ + L + F R + +N+ ++V + +++FPV+
Sbjct: 147 GRKAITIGLSSVLLSTLVCSVIFFGNLRDVGTKNSDHTLNSLEYVVIYSIQC-LSSFPVV 205
Query: 179 ARILAELKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSG--NSQSPFVSLWVF----- 231
+L EL+L + +GR+A+S+A ++D + IL ++ + + + Q+ S+++
Sbjct: 206 GNLLFELRLQNSELGRLAISSAVISDFSTSILASVLIFMKELKDEQTRLGSVFIGDVIAG 265
Query: 232 ---LSGCGFVVCSILIVLPIFK----WMAQQCHEGEPVDELYICATLAAVLAAGFVTDAI 284
L G VV + I + +F+ ++ +Q G PV +Y+ + V + + +
Sbjct: 266 NRPLMRAGIVVLFVCIAIYVFRPLMFYIIKQTPSGRPVKAIYLSTIIVMVSGSAILANWC 325
Query: 285 GIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSW 344
G F+ G+ VP A+++K E + G LP + SS + DI+ + G +
Sbjct: 326 KQSIFMGPFILGLAVPHGPPLGSAIIQKYESAIFGTFLPFFIASSSTEIDISALFGWEGL 385
Query: 345 GLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLN 404
++ + T+ K + T V +L +P + L +M+ KG+ EL + R +
Sbjct: 386 NGIILIMVTSFVVKFIFTTVPALFYGMPMEDCFALSLIMSFKGIFELGAYALAYQRGSVR 445
Query: 405 DQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYRTIERKNADGQLRILACFHGSRN 464
+TF + L + + + P + Y + Y+ R ++ + +LRIL+C + + +
Sbjct: 446 PETFTVACLYITLNSAIIPPILRYLYDPSRMYAGYEKRNMQHLKPNSELRILSCIYRTDD 505
Query: 465 IPSLINLIEASRGIKKHDALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSL 524
I +INL+EA + + Y +HL E +++ I ++ K++ S
Sbjct: 506 ISPMINLLEAICP-SRESPVATYVLHLMELVGQANPIFISHKLQTRRTE-----ETSYSN 559
Query: 525 HVIVAFEAYQK--LSQVFVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGS 582
+V+V+FE ++K VFV A+S +H DIC A ++I+LPFH+ DGS
Sbjct: 560 NVLVSFEKFRKDFYGSVFVSTYTALSMPDTMHGDICMLALNNTTSLILLPFHQTWSADGS 619
Query: 583 LDIIRND-FRLVNKRVLEHAPCSVGIFVDRGLGGSCHVSASN----------VSYCIAVL 631
I N+ R +NK VL+ APCSVG+FV R G ++S+ SY I ++
Sbjct: 620 ALISNNNMIRNLNKSVLDVAPCSVGVFVYRSSSGRKNISSGRKTINGTVPNLSSYNICMI 679
Query: 632 FFGGGDDREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISDSE-D 690
F GG DDREA+ RMA P I + ++ + + + N++ D D
Sbjct: 680 FLGGKDDREAVTLATRMARDPRINITIVRL------------ITTDEKARENTVWDKMLD 727
Query: 691 GEFLAEFKLKTANDDSIIYEERIVKDAEETVATIRE-INFCNLFLVGRRPAGELGFA--- 746
E L + K T D I Y E+ ++DA ET + +R ++ ++F+VGR F
Sbjct: 728 DELLRDVKSNTLVD--IFYSEKAIEDAAETSSLLRSMVSDFDMFIVGRGNGRTSVFTEGL 785
Query: 747 LERSECPELGPVGGLLASQDFRTTASVLVMQQ 778
E SE ELG +G LL SQDF ASVLV+QQ
Sbjct: 786 EEWSEFKELGIIGDLLTSQDFNCQASVLVIQQ 817
>At3g44900 putative protein
Length = 817
Score = 277 bits (709), Expect = 1e-74
Identities = 222/808 (27%), Positives = 383/808 (46%), Gaps = 64/808 (7%)
Query: 14 PMQPASNGVFQGDHPLD---------YALPLAILQICLVLVVTRGLAYLLNPLRQPRVIA 64
P+ P+S+G++ D Y P + +V ++ + + L L R +
Sbjct: 26 PINPSSSGLWPSPKLPDPQANIEFWNYMFPHVQIIFLIVTILWQFFHFFLRRLGMIRFTS 85
Query: 65 EIVGGILLGPSALGRN---KGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLR 121
++ GILL S L N + +L ++L + + F FL G+++D +R
Sbjct: 86 HMLTGILLSKSFLKENTPARKFLSTEDYKETL--FGLVGACSYMMFWFLMGVKMDLSLIR 143
Query: 122 KTGGRVLAIAIAGISLPFALGIGSSFVLQRTIA--KGVNTSAF--LVYMGVALSITAFPV 177
TG + +AI ++ + L + F++ R + KG +F ++++ + +++FPV
Sbjct: 144 STGRKAVAIGLSSVLLSITVCALIFFLILRDVGTKKGEPVMSFFEIIFIYLIQCLSSFPV 203
Query: 178 LARILAELKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSG--NSQSPFVSLWVFLSGC 235
+ +L EL+L + +GR+AMS+A ++D + IL A+ V L + +S S+++
Sbjct: 204 IGNLLFELRLQNSELGRLAMSSAVISDFSTSILSAVLVFLKELKDDKSRLGSVFIGDVIV 263
Query: 236 G------------FVVCSILIVLPIFKWMAQQCHEGEPVDELYICATLAAVLAAGFVTDA 283
G FV +I I P+ ++ ++ G PV + YI A + V + + D
Sbjct: 264 GNRPMKRAGTVVLFVCFAIYIFRPLMFFIIKRTPSGRPVKKFYIYAIIILVFGSAILADW 323
Query: 284 IGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQG--- 340
G F+ G+ VP A+++K E +V G LP + +S + D + +Q
Sbjct: 324 CKQSIFIGPFILGLAVPHGPPLGSAILQKFESVVFGTFLPFFVATSAEEIDTSILQSWID 383
Query: 341 LQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDR 400
L+S +LV V+F K T + + + +P + + L +M+ KG+ E R
Sbjct: 384 LKSIVILVSVSFIV---KFALTTLPAFLYGMPAKDCIALSLIMSFKGIFEFGAYGYAYQR 440
Query: 401 KVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYRTIERKNADGQLRILACFH 460
+ TF ++ L L+ + + P + Y + Y+ R + + +LRIL+C +
Sbjct: 441 GTIRPVTFTVLSLYILLNSAVIPPLLKRIYDPSRMYAGYEKRNMLHMKPNSELRILSCIY 500
Query: 461 GSRNIPSLINLIEASRGIKKHDALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRH 520
+ +I +INL+EA+ + + + Y +HL E +++ +L++ +++ +
Sbjct: 501 KTDDIRPMINLLEATCP-SRENPVATYVLHLMELVGQANPVLISHRLQTRK----SENMS 555
Query: 521 GDSLHVIVAFEAYQK--LSQVFVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQR 578
+S +V+V+FE + VFV A+S +H DIC A ++IILPFH+
Sbjct: 556 YNSENVVVSFEQFHNDFFGSVFVSTYTALSVPKMMHGDICMLALNNTTSLIILPFHQTWS 615
Query: 579 VDGSLDIIRN-DFRLVNKRVLEHAPCSVGIFVDRGLGGSCHV---SASNVSYCIAVLFFG 634
DGS + + R +NK VL+ +PCSVGIFV R G + +A+ SY + +LF G
Sbjct: 616 ADGSAIVSDSLMIRQLNKSVLDLSPCSVGIFVYRSSNGRRTIKETAANFSSYQVCMLFLG 675
Query: 635 GGDDREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFL 694
G DDREAL+ RMA I + V+ + A + T D D E L
Sbjct: 676 GKDDREALSLAKRMARDSRITITVVSLISSEQRANQATDWD-----------RMLDLELL 724
Query: 695 AEFKLKTANDDSIIYEERIVKDAEETVATIREI-NFCNLFLVGRRPAGELGFA---LERS 750
+ K I++ E +V DA +T ++ I N +LF+VGR + F E S
Sbjct: 725 RDVKSNVLAGADIVFSEEVVNDANQTSQLLKSIANEYDLFIVGREKGRKSVFTEGLEEWS 784
Query: 751 ECPELGPVGGLLASQDFRTTASVLVMQQ 778
E ELG +G LL SQD ASVLV+QQ
Sbjct: 785 EFEELGIIGDLLTSQDLNCQASVLVIQQ 812
>At1g79400 hypothetical protein
Length = 783
Score = 274 bits (700), Expect = 2e-73
Identities = 215/774 (27%), Positives = 365/774 (46%), Gaps = 31/774 (4%)
Query: 24 QGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGGILLGPSALGRNKGY 83
QGD L L +Q+ +LV ++ LL P Q +A+I+ GI+L P L R
Sbjct: 11 QGDE-LFNPLNTMFIQMACILVFSQLFYLLLKPCGQAGPVAQILAGIVLSPVLLSRIPKV 69
Query: 84 LHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFALGI 143
+ + F+FL G+E+D +R+ + I ++ + L
Sbjct: 70 KEFFLQKNAADYYSFFSFALRTSFMFLIGLEVDLHFMRRNFKKAAVITLSSFVVSGLLSF 129
Query: 144 GSSFVLQRTIAKGVNTSAFLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSAAAVN 203
S + + F + + V LS TA PV+ R +A+ KL T +GR+ +S A
Sbjct: 130 ASLMLFIPLFGIKEDYFTFFLVLLVTLSNTASPVVVRSIADWKLNTCEIGRLTISCALFI 189
Query: 204 DVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQCHEGEPVD 263
++ +L + +A S + + L++FL V ILI + + W+ ++ + + +
Sbjct: 190 ELTNVVLYTIIMAFI--SGTIILELFLFLLA---TVALILINMVLAPWLPKRNPKEKYLS 244
Query: 264 ELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLP 323
+ +L G ++ +++ F GI+ P+ G L++++ + +LP
Sbjct: 245 KAETLVFFIFLLIIGITIESYDVNSSVSVFAIGIMFPRQGKTHRTLIQRLSYPIHEFVLP 304
Query: 324 LYFVSSGLKTDIATIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFLM 383
+YF G + I + G+++ V T GK +G I + K+P L L ++
Sbjct: 305 VYFGYIGFRFSIIALTKRFYLGIVIIVIVTIA-GKFIGVISACMYLKIPKKYWLFLPTIL 363
Query: 384 NSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYR- 442
+ KG V L++L+ K T M++ AL+ T + + VLA++ K R+ ++ Y
Sbjct: 364 SVKGHVGLLLLDSNYSEKKWWTTTIHDMMVAALVITTLVSG-VLASFLLKTREKDFAYEK 422
Query: 443 -TIERKNADGQLRILACFHGSRNIPSLINLIEASRGIK-KHDALCVYAMHLKEFCERSSS 500
++E N + +LRIL+C +G R+ I+L+ A G + D MHL ++ S
Sbjct: 423 TSLESHNTNEELRILSCAYGVRHARGAISLVSALSGSRGASDPFTPLLMHLVPLPKKRKS 482
Query: 501 ILMAQKVRQNGLPFWDKGRHG--DSLHVIVAFEAYQKLSQVFVRPMIAISSMANIHEDIC 558
LM + ++G G + L + + +++ K S++ ++ + ++ M N+HE+IC
Sbjct: 483 ELMYHEHDEDGGNANGDDEFGTNEGLEINDSIDSFAKDSKILIQQVKLVTQMLNMHEEIC 542
Query: 559 TTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEHAPCSVGIFVDRGLGGSCH 618
+ R +++ LPFHK QR+DG FR +N+ VL H PCS+GIFVDR + G
Sbjct: 543 NATEDLRVSIVFLPFHKHQRIDGKTTNDGELFRQMNRNVLRHGPCSIGIFVDRNITGFQQ 602
Query: 619 VSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGD 678
+ +A LFFGG DDREALA +A + I L VI F+ E + A T V
Sbjct: 603 PHGFDSVQHVATLFFGGPDDREALALCRWLANNTLIHLTVIQFVSEESKAE--TPVGNAM 660
Query: 679 SSSNNSI----------SDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVATIREI- 727
+ NN + D FL EF + + + E++V + T+ +REI
Sbjct: 661 TRDNNEVFMEVLGRNQTEQETDRSFLEEFYNRFVTTGQVGFIEKLVSNGPHTLTILREIG 720
Query: 728 NFCNLFLVGRRPAGELGFAL---ERSECPELGPVGGLLASQDFRTTASVLVMQQ 778
+LF+VG + G+ + + ECPELG VG LAS ASVLV+Q+
Sbjct: 721 EMYSLFVVG-KSTGDCPMTVRMKDWEECPELGTVGDFLAS-SLDVNASVLVVQR 772
>At5g01680 putative transporter protein
Length = 780
Score = 261 bits (667), Expect = 1e-69
Identities = 203/805 (25%), Positives = 372/805 (45%), Gaps = 55/805 (6%)
Query: 1 MTSNTTSGNVCPPPMQPASNGVFQGDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQP 60
+TSN T VC + +S G+ +GD PL Y+ PL +L I LV ++ LL PL
Sbjct: 5 VTSNGTHEFVCEAWLGSSSGGLLRGDDPLKYSTPLLLLLISLVSSLSSVFQALLRPLANV 64
Query: 61 RVIAEIVGGILLGPSALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIELDPKSL 120
+ +I+ GI LGPSALG+N + +F +S ++++ + +F +++ ++D +
Sbjct: 65 DFVTQILAGIFLGPSALGQNIDLVKKLFNTRSYFIIESFEAISFMFISYISTAQVDMGVI 124
Query: 121 RKTGGRVLAIAIAGISLPFALGIGSSFV----LQRTIAKGVNTSAFLVYMGVALSITAFP 176
++ G + ++ P+ +G + V ++ T+AK N L + S+ F
Sbjct: 125 KRGGKLAIINGLSLFLFPYVVGAIACTVITSNIRGTVAK--NNPEQLHNLLTNQSVVYFQ 182
Query: 177 VLARILAELKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSG---NSQSPFVSLWVFLS 233
V +L+ LK+L + GR+A+S+ V + W L + + S L F
Sbjct: 183 VAYSVLSNLKMLNSEPGRLALSSIMVANCFGWGFFLLLITFDSFLHQNYSKTTYLPTFTK 242
Query: 234 GCGFVVCSILIVLPIFKWMAQQCHEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAF 293
+V +++ PIF W+ ++ EG+ + ++C + A F+++ +G + G+
Sbjct: 243 VL-LLVGIVVVCRPIFNWIVKRTPEGKKLKASHLCTICVMLCTATFLSETVGFPYVVGSV 301
Query: 294 VFGILVPKDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQGLQSWGLLVFVTFT 353
G++ PK F L +KI +L+P Y + G K D + L+ L F+ FT
Sbjct: 302 ALGLVTPKTPPFGTGLTDKIGSFCYAVLMPCYVIGIGNKVDFFSF-NLRDIISLEFLIFT 360
Query: 354 ACFGKIVGTIVVSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVL 413
K ++ SL +VP + ++++GF++ +G+ ++ + + K ++ + F IMV+
Sbjct: 361 ISAAKFASIVLPSLYFQVPISHAVIVGFIVCIQGIYDVQIFKQLLNYKNISHEAFGIMVI 420
Query: 414 MALITTFMTTPFVLAAYKRKERKS-NYKYRTIERKNADGQLRILACFHGSRNIPSLINLI 472
A++ + + T V Y +RK Y+ +T++ + L+IL CF+ +P ++ ++
Sbjct: 421 SAMVHSTIFTAIVKNLYGWVQRKHITYRRQTVQHYEPNKPLKILTCFYHRETVPPILTVL 480
Query: 473 EASRGIKKHDALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIVAFEA 532
E S + + +++L+E + + +L+ N R F +
Sbjct: 481 ELSTCPSSASSHSIVSVNLEELEQNNVPLLIQHHPGHNDESSTSSSRRDQISKAFEKFRS 540
Query: 533 YQKLSQ-VFVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFR 591
L + V V A++ +HED+C A K +II G D + R
Sbjct: 541 GHDLQENVSVECFTAVAPSKTMHEDVCALAFEKETDLIIF---------GMADGTAAERR 591
Query: 592 LVNKRVLEHAPCSVGIFVDRG-------LGGSCHVSASNVSYCIAVLFFGGGDDREALAY 644
L + V +P SV + +D+G +G + + ++ C +F GG DDRE LA+
Sbjct: 592 LC-RNVRNASPSSVAVLMDQGRLPDFKNMGTAMKNGSMRINIC--SIFLGGADDRETLAF 648
Query: 645 GARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTAND 704
RM P + L V+ + N+ S N+ + D + +F+ T N
Sbjct: 649 AVRMTNQPYVNLTVLKLVDGENV-----------SHLNDVVEKRLDFRTIEKFRQDTMNK 697
Query: 705 DSIIYEERIVKDAEETVATIREI-NFCNLFLVGRRPAGELGFALER-----SECPELGPV 758
++ E +A + V +RE N +L +VG R E F + + SE ELG +
Sbjct: 698 HNVALRE----EASDLVNLLREEGNNYDLIMVGIR--HEKSFEVLQGLSVWSEIEELGEI 751
Query: 759 GGLLASQDFRTTASVLVMQQYHNGV 783
G LL S+D + +ASVL +QQ + V
Sbjct: 752 GDLLVSRDLKLSASVLAVQQQLSSV 776
>At1g08150 hypothetical protein
Length = 815
Score = 261 bits (666), Expect = 1e-69
Identities = 217/780 (27%), Positives = 366/780 (46%), Gaps = 58/780 (7%)
Query: 30 DYALPLAILQICLVLVVTRGLAYLLNP--LRQPRVIAEIVGGILLGPSALGRNKGYL-HA 86
DY+LP I LVL + + L L P++ + ++ G L + L N + H
Sbjct: 62 DYSLPHLESVIVLVLCLWQFFYLSLKKIGLPVPKITSMMIAGAALSQTNLLPNDWTIQHI 121
Query: 87 VFPPKSLP-VLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFALGIGS 145
+FP + P V +TL +F+ F+ G+++D +RKTG +V+ IA + LP I +
Sbjct: 122 LFPDDTRPKVPETLGGFAFVFYWFIEGVKMDVGMVRKTGTKVIVTGIATVILPI---IAA 178
Query: 146 SFVLQRTIAKGVNTSAFLVYMGVAL--SITAFPVLARILAELKLLTTSVGRMAMSAAAVN 203
+ V + G + Y + SI+AF ++R+L +L++ + GR+ +S A V
Sbjct: 179 NMVFGKLRETGGKYLTGMEYRTILFMQSISAFTGISRLLRDLRINHSEFGRIVISTAMVA 238
Query: 204 DVAAW--ILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQCHEGEP 261
D + L AL + VS + G+V+ + +V P W+ ++ + P
Sbjct: 239 DGTGFGVNLFALVAWMDWR-----VSALQGVGIIGYVIFMVWVVRPAMFWVIKRTPQERP 293
Query: 262 VDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLL 321
V E +I L + I + G F+ G+ VP LVEK E +G+L
Sbjct: 294 VKECFIYIILILAFGGYYFLKEIHMFPAVGPFLLGLCVPHGPPLGSQLVEKFESFNTGIL 353
Query: 322 LPLYFVSSGLKTD---IATIQG--------LQSWGLLVFVTFTACFGKIVGTIVVSLICK 370
LPL+ S L+ D +A G L ++ V F A KI+ +++ +L+ K
Sbjct: 354 LPLFLFFSMLQIDGPWLANQIGQLRHFDGQLYEALTIIIVVFVA---KIIFSMIPALLAK 410
Query: 371 VPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAAY 430
+P +S V+ ++++KG+VEL G + VL+ ++F IM M L+++ ++ + Y
Sbjct: 411 MPLTDSFVMALILSNKGIVELCYFLYGVESNVLHVKSFTIMATMILVSSTISPVLIHYLY 470
Query: 431 KRKERKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIEASRGIKKHDALCVYAMH 490
+R +++ R + +L+ L C H + +I +INL+ S + + C Y +H
Sbjct: 471 DSSKRFISFQKRNLMSLKLGSELKFLVCIHKADHISGMINLLAQSFPLHESTISC-YVIH 529
Query: 491 LKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHVIVAFEAYQKL-SQVFVRPMIAISS 549
L E + + ++ ++++ + G S +V++AF+ ++ + + IS+
Sbjct: 530 LVELVGLDNPVFISHQMQKA-----EPGNRSYSNNVLIAFDNFKHYWKSISLELFTCISN 584
Query: 550 MANIHEDICTTADRKRAAVIILPFH----KQQRVDGSLDIIRNDFRLVNKRVLEHAPCSV 605
+H++I + A K+A+ ++LPFH Q S D++R R N VL APCSV
Sbjct: 585 PRYMHQEIYSLALDKQASFLMLPFHIIWSLDQTTVVSDDVMR---RNANLNVLRQAPCSV 641
Query: 606 GIFVDRGLGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFLVEP 665
GIFV R S S+ + C +F GG DDREALA G +M +P + L V+ +
Sbjct: 642 GIFVHRQKLLSAQKSSPSFEVC--AIFVGGKDDREALALGRQMMRNPNVNLTVLKLIP-- 697
Query: 666 NIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVATIR 725
K+D + + + +E E L + Y E V D +T +
Sbjct: 698 ------AKMDGMTTGWDQMLDSAEVKEVLRNNNNTVGQHSFVEYVEETVNDGSDTSTLLL 751
Query: 726 EI-NFCNLFLVGRRP---AGELGFALERSECPELGPVGGLLASQDFRTTASVLVMQQYHN 781
I N +LF+VGR + E +E ELG +G LL SQDF SVLV+QQ N
Sbjct: 752 SIANSFDLFVVGRSAGVGTDVVSALSEWTEFDELGVIGDLLVSQDFPRRGSVLVVQQQQN 811
>At5g22910 Na+/H+ antiporter-like protein
Length = 800
Score = 252 bits (644), Expect = 5e-67
Identities = 218/783 (27%), Positives = 359/783 (45%), Gaps = 69/783 (8%)
Query: 31 YALPLAILQICLVLVVTRGLAYLLNPLRQPRVIAEIVGGILLGPSALGR---NKGYLHAV 87
YALPL LQI L+ V L + PR ++ I+ G++LGP L + L
Sbjct: 47 YALPLLELQIILIFVCIVLSHMFLRRIGIPRFVSNILAGLILGPQLLDLLEYSSDRLSLD 106
Query: 88 FPPKSLPVLDTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFALGIG-SS 146
P L+ +A LGL+ F FL G++ + +++ + G R + IA++ + G+ +
Sbjct: 107 IPGNV--ALEGVARLGLVMFTFLMGVKTNKRAVYQIGKRPIVIAVSSFFVTMISGLAFRN 164
Query: 147 FVLQRTIAKGVNTSAFLVYMGVALSI---TAFPVLARILAELKLLTTSVGRMAMSAAAVN 203
F L + + V +SI T PV+ ++ ELK+ + +GR+A+S AAV+
Sbjct: 165 FRLDKVDPLYMPLRLAPTERSVIVSIQAVTLLPVITHLVYELKMSNSELGRIAISTAAVS 224
Query: 204 DVAAWILLALAVALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQC----HEG 259
D ++ L V +S +VS + ++ +L++L IFK MAQ+ EG
Sbjct: 225 DFLGFLTL---VCISYVGTYRYVSPGIANRDIVALIILVLVILFIFKPMAQRIVDMTPEG 281
Query: 260 EPVDELYICATLAAVLAAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSG 319
+PV ++Y+ T+ +AA + GA + G+ +P AL + E LV+
Sbjct: 282 KPVPKVYLYVTILTAIAASIYLSVFNQMYILGALLVGLAIPDGPPLGSALEARFESLVTN 341
Query: 320 LLLPLYFVSSGLKTDIATIQGLQSW----------GLLVFVTFTACFGKIVGTIVVSLIC 369
+ P+ +K D+ ++ L S+ GL V V +TA F + + C
Sbjct: 342 IFFPISIAVMAMKADV--VRALYSFDDISFNILLLGLTVVVKWTASF------VPCLIFC 393
Query: 370 KVPFNESLVLGFLMNSKGLVELIVLNIGKDRKVLNDQTFAIMVLMALITTFMTTPFVLAA 429
++P ES+++ +MN KG V+L ++ R+ L+ T+ +M++ L+ + + A
Sbjct: 394 ELPTRESVIIATIMNYKGFVDLCFFDVALRRRNLSRATYTVMIIYVLLNAGILPTIIKAL 453
Query: 430 YKRKERKSNYKYRTIERKNADGQLRILACFHGSRNIPSLINLIE-----ASRGIKKHDAL 484
Y K + Y R I + L+IL C H NI I+L+E + K +
Sbjct: 454 YDPKRKYIGYVKRDIMHLKTNSDLKILTCLHKPDNISGAISLLELLSSPLNNDNKDRGVI 513
Query: 485 CVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRHGDSLHV---IVAFEAYQK--LSQV 539
V A+HL + R+ IL+ R K R + ++ ++AF +Q+
Sbjct: 514 AVTALHLVKLAGRTFPILIPHDKR-------SKARLLQNSYIQTMMLAFTEFQQENWEST 566
Query: 540 FVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLE 599
V A S + +DIC A ++II+P ++ DG + R VN+ +L+
Sbjct: 567 TVSSFTAYSHENLMDQDICNLALDHLTSMIIVPSGRKWSPDGEYESDDIMIRRVNESLLD 626
Query: 600 HAPCSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVI 659
APCSVGI RG + S ++ + V+F GG DDREAL+ M ++ + L VI
Sbjct: 627 LAPCSVGILNYRGYNKGKKKTNSIIN--VGVIFIGGKDDREALSLAKWMGQNSRVCLTVI 684
Query: 660 HFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEE 719
FL + D S N D E L + K + ++ Y E++V
Sbjct: 685 RFLSGQEL----------DKSKNWDY--LVDDEVLNDLKATYSLANNFNYMEKVVNGGPA 732
Query: 720 TVATIREI-NFCNLFLVGRRPAG---ELGFALERSECPELGPVGGLLASQDFRTTASVLV 775
T+R + +L +VGR +L + E PELG +G LLAS+D R SVLV
Sbjct: 733 VATTVRLVAEDHDLMIVGRDHEDYSLDLTGLAQWMELPELGVIGDLLASKDLRARVSVLV 792
Query: 776 MQQ 778
+QQ
Sbjct: 793 VQQ 795
>At1g16380 putative Na/H antiporter
Length = 785
Score = 245 bits (625), Expect = 8e-65
Identities = 197/783 (25%), Positives = 375/783 (47%), Gaps = 51/783 (6%)
Query: 38 LQICLVLVVTRGLAYLLNPLRQPRVIAEIVGGILLGPSALGRNKGYLHAVFPPK-SLPVL 96
+Q+ +LV ++ L P Q +A+I+ GI+L + R +H F K S
Sbjct: 24 IQMACILVFSQFFYLFLKPCGQAGPVAQILAGIVLSLLTIIRK---VHEFFLQKDSASYY 80
Query: 97 DTLANLGLIFFLFLAGIELDPKSLRKTGGRVLAIAIAGISLPFALGIGSSFVLQRTIA-K 155
+ L F+FL G+E+D +++ + I + + + + + + L R + K
Sbjct: 81 IFFSFLLRTAFVFLIGLEIDLDFMKRNLKNSIVITLGSLVISGIIWLPFLWFLIRFMQIK 140
Query: 156 GVNTSAFLVYMGVALSITAFPVLARILAELKLLTTSVGRMAMSAAAVNDVAAWILLALAV 215
G + +L ++ + LS TA PV+ R + + KL T+ +GR+A+S ++ + + +
Sbjct: 141 GDFLTFYLAFL-ITLSNTAAPVVIRSIIDWKLHTSEIGRLAISCGLFIEITNIFIYTIVL 199
Query: 216 ALSGNSQSPFVSLWVFLSGCGFVVCSILIVLPIFKWMAQQCHEGEPVDELYICATLAAVL 275
+ + + + ++ F +G + L W+ ++ + + + + A + +L
Sbjct: 200 SFISGTMTADIFIYSFATGVIILTNRFLA-----SWLPKRNPKEKYLSKAETLAFIILIL 254
Query: 276 AAGFVTDAIGIHAMFGAFVFGILVPKDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKTDI 335
++ +++ F+ G++ P++G L++++ + +LP+YF G + +
Sbjct: 255 IIALTIESSNLNSTLFVFIIGLMFPREGKTYRTLIQRLSYPIHEFVLPVYFGYIGFRFSV 314
Query: 336 ATIQGLQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFLMNSKGLVELIVLN 395
++ + + +L + GK++G + K+P L L +++ KG + L++L+
Sbjct: 315 NSLTK-RHYLVLGMTVALSLLGKLLGVLFACSFLKIPKQYWLFLSTMLSVKGHIGLVLLD 373
Query: 396 IGKDRK-----VLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYRT-IERKNA 449
K V++D A +V+M L++ +T+ + R + KS +T +E +
Sbjct: 374 SNLMYKKWFTPVVHDMFVAALVIMTLLSGVITSLLL-----RSQEKSFAHIKTSLELFDT 428
Query: 450 DGQLRILACFHGSRNIPSLINLIEASRGIK---KHDALCVYAMHLKEFCERSSSILMAQK 506
+LR+L C +G R+ I+L+ A G Y MHL ++ + L+ +
Sbjct: 429 TEELRVLTCVYGVRHARGSISLVSALSGFSPGTSSSPFTPYLMHLIPLPKKRKTELLYHE 488
Query: 507 VRQ---NGLPFWDKGRHGDSLHVIVAFEAYQKLSQVFVRPMIAISSMANIHEDICTTADR 563
+ + N D+ + L + + +++ + ++ VR + ++ M N+HE+IC +
Sbjct: 489 LDEDAGNSNGGDDEFGTNEGLEINDSIDSFTRDRKIMVRQVKLVAPMENMHEEICNATED 548
Query: 564 KRAAVIILPFHKQQRVDGSLDIIRNDFRLVNKRVLEHAPCSVGIFVDRGLGGSCHVSASN 623
R +++ LPFHK QR+DG FR +N++VL+ A CS+GIFVDR + G + S+
Sbjct: 549 LRVSIVFLPFHKHQRIDGKTTNDGEVFRHMNRKVLKQAQCSIGIFVDRNITGFHQLHGSD 608
Query: 624 VSYCIAVLFFGGGDDREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGD--SSS 681
+A LFFGG DDREAL+ + + I L VI F+ + + KI VGD +
Sbjct: 609 SVQHVAALFFGGPDDREALSLCKWLTNNSQIHLTVIQFVADDSKTEKI----VGDAVTKE 664
Query: 682 NNSI----------SDSEDGEFLAEFKLKTANDDSIIYEERIVKDAEETVATIREI-NFC 730
NN + + D FL EF + + + E+ V + +T+ +REI
Sbjct: 665 NNEVFLEIVSEDQTENETDRIFLEEFYHRFVTTGQVGFIEKRVSNGMQTLTILREIGEMY 724
Query: 731 NLFLVGRRPAGELGFAL---ERSECPELGPVGGLLASQDFRTTASVLVMQQYHNGVPINF 787
+LF+VG+ G+ + ECPELG VG LAS + ASVLV+Q++ N +F
Sbjct: 725 SLFVVGKN-RGDCPMTSGMNDWEECPELGTVGDFLASSNMDVNASVLVVQRHRNSFD-SF 782
Query: 788 VPE 790
V E
Sbjct: 783 VDE 785
>At2g28180 hypothetical protein
Length = 822
Score = 244 bits (622), Expect = 2e-64
Identities = 220/806 (27%), Positives = 379/806 (46%), Gaps = 87/806 (10%)
Query: 8 GNVCPP-PMQPASNGVFQ-------GDHPLDYALPLAILQICLVLVVTRGLAYLLNPLRQ 59
G +C P + +S+G+++ G + Y LP + I LV + +G L L
Sbjct: 61 GLICEEHPPKLSSDGIWEKLIIKSAGLYFWQYRLPKLEIVILLVFFLWQGFNILFKKLGL 120
Query: 60 --PRVIAEIVGGILLGP--SALGRNKGYLHAVFPPKSLPVLDTLANLGLIFFLFLAGIEL 115
P++ + ++ G+LL + G N + + V L + G + F FL G+ +
Sbjct: 121 SIPKLSSMMLAGLLLNVLVTLSGENSIIADILVTKNRIDVAGCLGSFGFLIFWFLKGVRM 180
Query: 116 DPKSLRKTGGRVLAIAIAGISLPFALGIGSSFVLQRTIAKGVNTSAFLVYMGVAL--SIT 173
D K + K + +A ++ P +G F+L + F Y + L SIT
Sbjct: 181 DVKRIFKAEAKARVTGVAAVTFPIVVG----FLLFNLKSAKNRPLTFQEYDVMLLMESIT 236
Query: 174 AFPVLARILAELKLLTTSVGRMAMSAAAVNDVAAWILLALAVALSGNSQSPFVSLWVFLS 233
+F +AR+L +L + +S+GR+A+S+A V+D+ +LL V+ S + + +++ ++
Sbjct: 237 SFSGIARLLRDLGMNHSSIGRVALSSALVSDIVGLLLLIANVSRSSATLADGLAILTEIT 296
Query: 234 GCGFVVCSILIVLPIFKWMAQQCHEGEPVDELYICATLAAVLAAGFVTDAIGIHAMFGAF 293
F+V + +V PI + ++ EG P+++ YI L V + + + GAF
Sbjct: 297 L--FLVIAFAVVRPIMFKIIKRKGEGRPIEDKYIHGVLVLVCLSCMYWEDLSQFPPLGAF 354
Query: 294 VFGILVPKDGAFAGALVEKIEDLVSGLLLPLYFVSSGLKTDIATIQG------------- 340
G+ +P ALVE++E G++LPL+ + L+TD +G
Sbjct: 355 FLGLAIPNGPPIGSALVERLESFNFGIILPLFLTAVMLRTDTTAWKGALTFFSGDDKKFA 414
Query: 341 LQSWGLLVFVTFTACFGKIVGTIVVSLICKVPFNESLVLGFLMNSKGLVELIVLNIGKDR 400
+ S LL+F+ K+ +++V + K+P +S++L +M+ K V IVLN
Sbjct: 415 VASLVLLIFLL------KLSVSVIVPYLYKMPLRDSIILALIMSHK--VLSIVLN----- 461
Query: 401 KVLNDQTFAIMVLMALITTFMTTPFVLAAYKRKERKSNYKYRTIERKNADGQLRILACFH 460
++++ MA+ F+ Y ++ Y+ R + G+L+ L C H
Sbjct: 462 --------SLLIPMAI--GFL--------YDPSKQFICYQKRNLASMKNMGELKTLVCIH 503
Query: 461 GSRNIPSLINLIEASRGIKKHDALCVYAMHLKEFCERSSSILMAQKVRQNGLPFWDKGRH 520
+I S+INL+EAS + C Y +HL E + L++ KV++ G+ +K
Sbjct: 504 RPDHISSMINLLEASYQSEDSPLTC-YVLHLVELRGQDVPTLISHKVQKLGVGAGNK--- 559
Query: 521 GDSLHVIVAFEAYQK--LSQVFVRPMIAISSMANIHEDICTTADRKRAAVIILPFHKQQR 578
S +VI++FE + + S + + I++ ++ +DIC A K +IILPFH+
Sbjct: 560 -YSENVILSFEHFHRSVCSSISIDTFTCIANANHMQDDICWLALDKAVTLIILPFHRTWS 618
Query: 579 VDGSLDIIRND-FRLVNKRVLEHAPCSVGIFVDRGLGGSCHVSASNVSYCIAVLFFGGGD 637
+D + + + R +N VL+ APCSVGI ++R L ++ C V+F GG D
Sbjct: 619 LDRTSIVSDVEAIRFLNVNVLKQAPCSVGILIERHLVNKKQEPHESLKVC--VIFVGGKD 676
Query: 638 DREALAYGARMAEHPGIQLVVIHFLVEPNIAGKITKVDVGDSSSNNSISDSEDGEFLAEF 697
DREALA+ RMA + L V+ L + D+ + S + + E
Sbjct: 677 DREALAFAKRMARQENVTLTVLRLLASGKSKDATGWDQMLDTVELRELIKSNNAGMVKE- 735
Query: 698 KLKTANDDSIIYEERIVKDAEETVATIREINF-CNLFLVGRRPAGELGFALER----SEC 752
+ S IY E+ + D +T +R + F +LF+VG R GE A + E
Sbjct: 736 ------ETSTIYLEQEILDGADTSMLLRSMAFDYDLFVVG-RTCGENHEATKGIENWCEF 788
Query: 753 PELGPVGGLLASQDFRTTASVLVMQQ 778
ELG +G LAS DF + SVLV+QQ
Sbjct: 789 EELGVIGDFLASPDFPSKTSVLVVQQ 814
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.140 0.412
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,013,530
Number of Sequences: 26719
Number of extensions: 704741
Number of successful extensions: 2227
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1990
Number of HSP's gapped (non-prelim): 53
length of query: 803
length of database: 11,318,596
effective HSP length: 107
effective length of query: 696
effective length of database: 8,459,663
effective search space: 5887925448
effective search space used: 5887925448
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140913.11


