

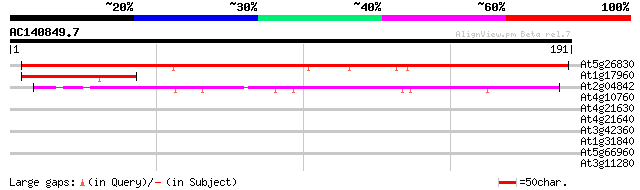
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140849.7 + phase: 0 /pseudo/partial
(191 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g26830 threonyl-tRNA synthetase 195 1e-50
At1g17960 unknown protein 50 6e-07
At2g04842 putative protein 49 2e-06
At4g10760 putative methyltransferase 36 0.014
At4g21630 serine protease - like protein 30 0.78
At4g21640 subtilisin-like protease 28 3.0
At3g42360 putative protein 28 3.0
At1g31840 hypothetical protein 27 5.0
At5g66960 protease-like 27 6.6
At3g11280 unknown protein 27 6.6
>At5g26830 threonyl-tRNA synthetase
Length = 676
Score = 195 bits (496), Expect = 1e-50
Identities = 112/207 (54%), Positives = 129/207 (62%), Gaps = 21/207 (10%)
Query: 5 KHGCKLCIGPCTTTENGFYYDAIYCNLGLKDYQFKIIESGAFKVVVEKQP---IEVTRDQ 61
++GC+LCIGPCTT GFYYD Y LGL D F IE+GA K E QP IEVT+DQ
Sbjct: 127 EYGCQLCIGPCTTRGEGFYYDGFYGELGLSDNHFPSIEAGAAKAAKEAQPFERIEVTKDQ 186
Query: 62 ALEIFSDDKFKVEIINNLAPDETITVYRCGPLVDLYRAC----LEFPGFFSILEGGQ--- 114
ALE+FS++ FKVE+IN L D TITVYRCGPLVDL R F F L
Sbjct: 187 ALEMFSENNFKVELINGLPADMTITVYRCGPLVDLCRGPHIPNTSFVKAFKCLRASSAYW 246
Query: 115 K*KKFTECLWHIVS*S----KEFK-------EAKKYDHRILGLKQELVSLHEWSPGSWFF 163
K K E L + S K+ K EAKKYDHR+LG KQEL H+ SPGS+FF
Sbjct: 247 KGDKDRESLQRVYGISYPDQKQLKKYLQFLEEAKKYDHRLLGQKQELFFSHQLSPGSYFF 306
Query: 164 LPHGALIYNKLMDFIRNQYRHRGYQEV 190
LP G +YN+LMDFI+NQY HRGY EV
Sbjct: 307 LPLGTRVYNRLMDFIKNQYWHRGYTEV 333
>At1g17960 unknown protein
Length = 458
Score = 50.4 bits (119), Expect = 6e-07
Identities = 22/40 (55%), Positives = 28/40 (70%), Gaps = 1/40 (2%)
Query: 5 KHGCKLCIGPCTTTENGFYYDAIYC-NLGLKDYQFKIIES 43
++GCKLCIGPC + GFYYD++Y LGL D F IE+
Sbjct: 129 EYGCKLCIGPCEPRDEGFYYDSLYYGELGLNDNHFPNIEA 168
>At2g04842 putative protein
Length = 650
Score = 48.5 bits (114), Expect = 2e-06
Identities = 53/206 (25%), Positives = 98/206 (46%), Gaps = 32/206 (15%)
Query: 9 KLCIGPCTTTENGFYYDAIYCNLGLKDYQFKIIESGAFKVVVEKQPI---EVTRDQALE- 64
K+ IGP +NGFYYD + L D K I+ +++ P+ EV+R++A +
Sbjct: 102 KVTIGPWI--DNGFYYD--FDMEPLTDKDLKRIKKEMDRIISRNLPLLREEVSREEAKKR 157
Query: 65 -IFSDDKFKVEIINNLAPDETITVYR---------CGPLVD----LYRACLEFPGFFSIL 110
+ ++ +K+EI++ + +E ITVY GP V+ + + +E
Sbjct: 158 IMAINEPYKMEILDGIK-EEPITVYHIGNEWWDLCAGPHVETTGKINKKAVELESVAGAY 216
Query: 111 EGGQK*KKFTECLWHIVS*SKE-------FKE-AKKYDHRILGLKQELVSLHEWSPGSW- 161
G + ++ + ++ S+E FKE AK+ DHR +G +L S+ + + G
Sbjct: 217 WRGDEKRQMLQRIYGTAWESEEQLKAYLHFKEEAKRRDHRRIGQDLDLFSIQDEAGGGLV 276
Query: 162 FFLPHGALIYNKLMDFIRNQYRHRGY 187
F+ P GA++ N + + + + GY
Sbjct: 277 FWHPKGAIVRNIIEESWKKMHVEHGY 302
>At4g10760 putative methyltransferase
Length = 685
Score = 35.8 bits (81), Expect = 0.014
Identities = 24/97 (24%), Positives = 47/97 (47%), Gaps = 5/97 (5%)
Query: 15 CTTTENGFYYDAIYCNLGLKDYQFKIIESGAFKVVVEKQPIEVTRDQALEIFSDDKFKVE 74
C+ E G +A + N ++ ++ I+ G F VV+ P ++ + +DD+ +
Sbjct: 450 CSEAELG---EAQWINCDIRSFRMDIL--GTFGVVMADPPWDIHMELPYGTMADDEMRTL 504
Query: 75 IINNLAPDETITVYRCGPLVDLYRACLEFPGFFSILE 111
+ +L D I ++ G ++L R CLE G+ + E
Sbjct: 505 NVPSLQTDGLIFLWVTGRAMELGRECLELWGYKRVEE 541
>At4g21630 serine protease - like protein
Length = 772
Score = 30.0 bits (66), Expect = 0.78
Identities = 26/100 (26%), Positives = 43/100 (43%), Gaps = 16/100 (16%)
Query: 12 IGPCTTTENGFYYDAIYCNLGLKDY-----QFKIIESGAFKVVVEKQPIEVTRDQALEIF 66
+ P + G YD +G+KDY I+S +V+ +K + + L+I
Sbjct: 618 VNPDKAAQPGLVYD-----MGIKDYINYMCSAGYIDSSISRVLGKKTKCTIPKPSILDI- 671
Query: 67 SDDKFKVEIINNLAPDETIT--VYRCGPLVDLYRACLEFP 104
I NL + T+T V GP+ +Y+A +E P
Sbjct: 672 ---NLPSITIPNLEKEVTLTRTVTNVGPIKSVYKAVIESP 708
>At4g21640 subtilisin-like protease
Length = 769
Score = 28.1 bits (61), Expect = 3.0
Identities = 26/100 (26%), Positives = 43/100 (43%), Gaps = 16/100 (16%)
Query: 12 IGPCTTTENGFYYDAIYCNLGLKDYQFKIIESG-----AFKVVVEKQPIEVTRDQALEIF 66
+ P + G YD +G+KDY + +G +V+ +K + + L+I
Sbjct: 615 VNPEKAAKPGLVYD-----MGIKDYINYMCSAGYNDSSISRVLGKKTKCPIPKPSMLDI- 668
Query: 67 SDDKFKVEIINNLAPDETIT--VYRCGPLVDLYRACLEFP 104
I NL + T+T V GP+ +YRA +E P
Sbjct: 669 ---NLPSITIPNLEKEVTLTRTVTNVGPIKSVYRAVIESP 705
>At3g42360 putative protein
Length = 228
Score = 28.1 bits (61), Expect = 3.0
Identities = 17/62 (27%), Positives = 29/62 (46%), Gaps = 4/62 (6%)
Query: 103 FPGFFSILEGGQK*KKFTECLWHIVS*SKEFKEAKKYDHRILGLKQELVSLHEWSPGSWF 162
F G + +EG + +F W +S K+ KKY L ++QE++ + P +
Sbjct: 136 FLGSSNQIEGNEWRARFVRNFWSTLS----LKDYKKYSVHFLEVQQEVLPIRGMEPVESY 191
Query: 163 FL 164
FL
Sbjct: 192 FL 193
>At1g31840 hypothetical protein
Length = 845
Score = 27.3 bits (59), Expect = 5.0
Identities = 29/107 (27%), Positives = 48/107 (44%), Gaps = 19/107 (17%)
Query: 6 HGCKLCIG---PCTTTENGFYYDAIYCN---LGLKDYQFKIIESGAFKVVVEKQPIEVTR 59
H KLC G P + +GF DA++C D+ ++E G F+V + V+
Sbjct: 203 HFDKLCRGGIEPSGVSAHGFVLDALFCKGEVTKALDFHRLVMERG-FRVGI------VSC 255
Query: 60 DQALEIFSDDKFKVEIINNLAPDETITVYRCGPLVDLYRACLEFPGF 106
++ L+ S D ++E+ + L V CGP ++ C GF
Sbjct: 256 NKVLKGLSVD--QIEVASRLLS----LVLDCGPAPNVVTFCTLINGF 296
>At5g66960 protease-like
Length = 792
Score = 26.9 bits (58), Expect = 6.6
Identities = 19/60 (31%), Positives = 30/60 (49%), Gaps = 5/60 (8%)
Query: 131 KEFKEAKKYD---HRILGLKQELVSLHEWSPGSWFFLPHGALIYNKLMDFIRNQYRHRGY 187
+ +E K+Y R+ L +E +S H+ SP + F G I KL+D+ + R GY
Sbjct: 126 RRVEEGKQYPVLCRRLASLHEEFIS-HK-SPAAGFDYTSGKRIEQKLLDYNQEAERFGGY 183
>At3g11280 unknown protein
Length = 263
Score = 26.9 bits (58), Expect = 6.6
Identities = 13/22 (59%), Positives = 15/22 (68%), Gaps = 2/22 (9%)
Query: 140 DHRILGLKQELVSLHEWSPGSW 161
DHR + QE+VSLH S GSW
Sbjct: 14 DHRFV--VQEMVSLHSSSSGSW 33
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.329 0.146 0.474
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,555,517
Number of Sequences: 26719
Number of extensions: 181169
Number of successful extensions: 461
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 455
Number of HSP's gapped (non-prelim): 10
length of query: 191
length of database: 11,318,596
effective HSP length: 94
effective length of query: 97
effective length of database: 8,807,010
effective search space: 854279970
effective search space used: 854279970
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC140849.7


