

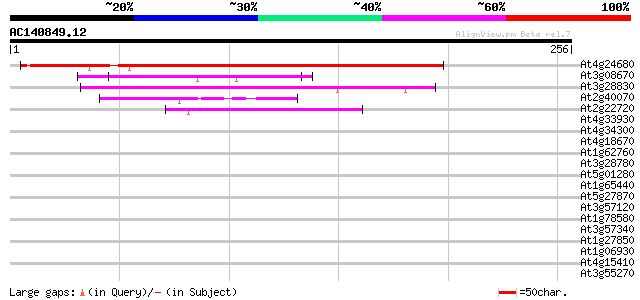
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140849.12 + phase: 0
(256 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g24680 unknown protein 214 5e-56
At3g08670 hypothetical protein 46 2e-05
At3g28830 hypothetical protein 45 4e-05
At2g40070 En/Spm-like transposon protein 44 6e-05
At2g22720 unknown protein 44 8e-05
At4g33930 putative protein 40 0.001
At4g34300 putative protein 39 0.004
At4g18670 extensin-like protein 39 0.004
At1g62760 hypothetical protein 39 0.004
At3g28780 histone-H4-like protein 38 0.006
At5g01280 putative protein 37 0.008
At1g65440 37 0.010
At5g27870 pectin methyl-esterase - like protein 36 0.018
At3g57120 unknown protein 36 0.018
At1g78580 trehalose-6-phosphate synthase 36 0.018
At3g57340 dnaJ-like protein 35 0.030
At1g27850 transposon protein, putative 35 0.039
At1g06930 hypothetical protein 35 0.051
At4g15410 phosphatase like protein 34 0.067
At3g55270 MAP kinase phosphatase (MKP1) 34 0.067
>At4g24680 unknown protein
Length = 1480
Score = 214 bits (544), Expect = 5e-56
Identities = 121/205 (59%), Positives = 137/205 (66%), Gaps = 16/205 (7%)
Query: 6 RRWTSSTRRGGMTVLGKVAVPKPINLPSQR-----------GTLGWGSKSSSSASNAWG- 53
RRW +TRR GMT+LGKVAVPKPINLPSQR GTL WGSKSS NAWG
Sbjct: 9 RRW-GTTRRSGMTILGKVAVPKPINLPSQRWFNFLLDIYFRGTLSWGSKSSL---NAWGT 64
Query: 54 SSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPP 113
SSLSP +G SPSHLS RPSSGGS TRPST+ S+ A + +++ AW S+SRPSSASG
Sbjct: 65 SSLSPRTESGPGSPSHLSNRPSSGGSVTRPSTADSNKAHDSSSSVAWDSNSRPSSASGVF 124
Query: 114 TSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGITQRKNDDFSLS 173
SNQ S RP SA+TRPGSS LSRFAE V E S W E LG+ KND FSL+
Sbjct: 125 PSNQPSVALQRPHSADTRPGSSQLSRFAEPVSETSATWGQHVAPEKLGVAPSKNDGFSLT 184
Query: 174 TGDFPTLGSEKDKSVHDVELQGSLL 198
+GDFP+LG+EKD S QG L
Sbjct: 185 SGDFPSLGAEKDTSEKSTRPQGDTL 209
Score = 30.0 bits (66), Expect = 1.3
Identities = 29/108 (26%), Positives = 44/108 (39%), Gaps = 13/108 (12%)
Query: 23 VAVPKPINLPSQRGT--LGWGSKSSSSASNAWGS-------SLSPNANAGASSPS----H 69
VA+ +P + ++ G+ L ++ S S WG ++P+ N G S S
Sbjct: 131 VALQRPHSADTRPGSSQLSRFAEPVSETSATWGQHVAPEKLGVAPSKNDGFSLTSGDFPS 190
Query: 70 LSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQ 117
L A + TRP DMA + G +RP S+SG Q
Sbjct: 191 LGAEKDTSEKSTRPQGDTLDMALRNYKFADAGPHARPPSSSGRSVEGQ 238
>At3g08670 hypothetical protein
Length = 567
Score = 45.8 bits (107), Expect = 2e-05
Identities = 37/110 (33%), Positives = 52/110 (46%), Gaps = 17/110 (15%)
Query: 46 SSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPS---------TSGSDMASELTT 96
+S++ A +S + + S + S+RP+ S TRPS TSG +S L T
Sbjct: 123 ASSARASSASKASRLSVSQSESGYHSSRPARSSSVTRPSISTSQYSSFTSGRSPSSILNT 182
Query: 97 TSAWGS--------SSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
+SA S SSR SS++ P T +TS S + RPGSS S
Sbjct: 183 SSASVSSYIRPSSPSSRSSSSARPSTPTRTSSASRSSTPSRIRPGSSSSS 232
Score = 40.8 bits (94), Expect = 7e-04
Identities = 35/102 (34%), Positives = 46/102 (44%), Gaps = 5/102 (4%)
Query: 32 PSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
PS R + + + S+A SS G+SS S ARPS +RPST S
Sbjct: 196 PSSRSSSSARPSTPTRTSSASRSSTPSRIRPGSSSSSMDKARPS---LSSRPSTPTS--R 250
Query: 92 SELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPG 133
+L+ +S +SRP+S PT S TSL S T G
Sbjct: 251 PQLSASSPNIIASRPNSRPSTPTRRSPSSTSLSATSGPTISG 292
Score = 33.1 bits (74), Expect = 0.15
Identities = 32/112 (28%), Positives = 47/112 (41%), Gaps = 1/112 (0%)
Query: 24 AVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRP 83
+V +P SQ + G SS + + S S + SS S SARPS+ +
Sbjct: 156 SVTRPSISTSQYSSFTSGRSPSSILNTSSASVSSYIRPSSPSSRSSSSARPSTPTRTSSA 215
Query: 84 STSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSL-RPRSAETRPGS 134
S S + ++S+ +RPS +S P T Q S P +RP S
Sbjct: 216 SRSSTPSRIRPGSSSSSMDKARPSLSSRPSTPTSRPQLSASSPNIIASRPNS 267
Score = 28.5 bits (62), Expect = 3.7
Identities = 30/98 (30%), Positives = 45/98 (45%), Gaps = 22/98 (22%)
Query: 44 SSSSASNAWGSSLSPNANA-------GASSPSHLSARPSSGGSGTRPSTSGSDMASELTT 96
SSSS+ + SLS + ASSP+ +++RP+S RPS T
Sbjct: 228 SSSSSMDKARPSLSSRPSTPTSRPQLSASSPNIIASRPNS-----RPS----------TP 272
Query: 97 TSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS 134
T SS+ S+ SGP S + ++ R + +RP S
Sbjct: 273 TRRSPSSTSLSATSGPTISGGRAASNGRTGPSLSRPSS 310
>At3g28830 hypothetical protein
Length = 536
Score = 45.1 bits (105), Expect = 4e-05
Identities = 43/174 (24%), Positives = 73/174 (41%), Gaps = 12/174 (6%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMAS 92
S G++ SK SSS+ ++ S++ + + + ++ SSGGS S S ++
Sbjct: 210 SSSGSVSTKSKESSSSGSSASGSVATKSKESSGGSAATKSKESSGGSAATKSKESSGGSA 269
Query: 93 ELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENS---- 148
TS S S +S SG + +S+ S + + + GS+ A G S
Sbjct: 270 TTGKTSGSPSGSPKASPSGSVSGKSSSKGSASAQGSASAQGSASAQGSASAQGSASAQRR 329
Query: 149 ----VAWNGARTTETLGITQRKNDDFSLSTGDFPT----LGSEKDKSVHDVELQ 194
+A + +R T+T Q K+ S S+ T + SE K V +Q
Sbjct: 330 ESGAMAMSKSRETKTSSQRQSKSSSESSSSSTTTTTVKQVESETSKEVMSFIMQ 383
Score = 35.4 bits (80), Expect = 0.030
Identities = 39/181 (21%), Positives = 68/181 (37%), Gaps = 12/181 (6%)
Query: 41 GSKSSSSASNAWGS-----SLSPNANAGASSPSHLS------ARPSSGGSGTRPSTSGSD 89
G+ SS S+S GS S+S + +SS S S ++ SSGGS S S
Sbjct: 195 GAASSESSSTKSGSVSSSGSVSTKSKESSSSGSSASGSVATKSKESSGGSAATKSKESSG 254
Query: 90 MASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSV 149
++ + + G S+ SG P+ + + S + GS+ A G S
Sbjct: 255 GSAATKSKESSGGSATTGKTSGSPSGSPKASPSGSVSGKSSSKGSASAQGSASAQGSAS- 313
Query: 150 AWNGARTTETLGITQRKNDDFSLSTGDFPTLGSEKDKSVHDVELQGSLLCITIISIDSSA 209
A A + +R++ ++S S++ S T+ ++S
Sbjct: 314 AQGSASAQGSASAQRRESGAMAMSKSRETKTSSQRQSKSSSESSSSSTTTTTVKQVESET 373
Query: 210 A 210
+
Sbjct: 374 S 374
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 44.3 bits (103), Expect = 6e-05
Identities = 35/91 (38%), Positives = 49/91 (53%), Gaps = 9/91 (9%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSS-GGSGTRPSTSGSDMASELTTTSAW 100
+ S+ SA+ +S ++ G SS S S RPSS GG G+RP+T + +S LT S
Sbjct: 117 NSSTESAARNHLTSRQQTSSPGLSSSSGASRRPSSSGGPGSRPATP-TGRSSTLTANS-- 173
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSAETR 131
SSRPS+ PTS T ++ RP +R
Sbjct: 174 -KSSRPST----PTSRATVSSATRPSLTNSR 199
Score = 36.6 bits (83), Expect = 0.014
Identities = 34/108 (31%), Positives = 51/108 (46%), Gaps = 4/108 (3%)
Query: 30 NLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGS- 88
+L S++ T G SSS AS SS P + ++P+ S+ ++ +RPST S
Sbjct: 127 HLTSRQQTSSPGLSSSSGASRRPSSSGGPGSRP--ATPTGRSSTLTANSKSSRPSTPTSR 184
Query: 89 DMASELTTTSAWGSSSRPSSASGP-PTSNQTSQTSLRPRSAETRPGSS 135
S T S S S S+ + P P S TS +S R ++P +S
Sbjct: 185 ATVSSATRPSLTNSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPTTS 232
Score = 32.7 bits (73), Expect = 0.20
Identities = 46/160 (28%), Positives = 70/160 (43%), Gaps = 22/160 (13%)
Query: 9 TSSTRRGGMTVLGKVAVPKPINLPSQRGTLGWGSKSS--SSASNAWGSSLSPNANAGASS 66
+ S+R T V+ +L + R T+ +K + S +++ S L+P A S
Sbjct: 173 SKSSRPSTPTSRATVSSATRPSLTNSRSTVSATTKPTPMSRSTSLSSSRLTPTA----SK 228
Query: 67 PSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPR 126
P+ +AR S GS TR + S TTT + G S S P S T+++S P
Sbjct: 229 PTTSTAR--SAGSVTRSTPS--------TTTKSAG-----PSRSTTPLSRSTARSS-TPT 272
Query: 127 SAETRPGSSHLSRFAEHVGENSVAWNGARTTETLGITQRK 166
S T P S +SR + + + A TT I+Q K
Sbjct: 273 SRPTLPPSKTISRSSTPTRRPIASASAATTTANPTISQIK 312
Score = 31.2 bits (69), Expect = 0.57
Identities = 24/88 (27%), Positives = 39/88 (44%), Gaps = 21/88 (23%)
Query: 68 SHLSARPSSGGSGTRPSTSGSDMASELT------------------TTSAWGSSSRPSSA 109
SH + +G S +RP+T S +A+ T +S+ G+S RPSS+
Sbjct: 93 SHRTMMSQTGDSKSRPATLTSRLANSSTESAARNHLTSRQQTSSPGLSSSSGASRRPSSS 152
Query: 110 SGP---PTSNQTSQTSLRPRSAETRPGS 134
GP P + ++L S +RP +
Sbjct: 153 GGPGSRPATPTGRSSTLTANSKSSRPST 180
>At2g22720 unknown protein
Length = 340
Score = 43.9 bits (102), Expect = 8e-05
Identities = 28/93 (30%), Positives = 45/93 (48%), Gaps = 3/93 (3%)
Query: 72 ARPSSGGSG---TRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSA 128
+RPSS GS +RP+TSGS M + +S SR S SG P S+ + + RP+++
Sbjct: 4 SRPSSSGSKMNHSRPATSGSQMPNSRPASSGSQMQSRAVSGSGRPASSGSQMQNSRPQNS 63
Query: 129 ETRPGSSHLSRFAEHVGENSVAWNGARTTETLG 161
S + + G A +G++ + G
Sbjct: 64 RPASAGSQMQQRPASSGSQRPASSGSQRPASSG 96
Score = 39.3 bits (90), Expect = 0.002
Identities = 36/100 (36%), Positives = 47/100 (47%), Gaps = 11/100 (11%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
GSK + S GS + PN+ ASS S + +R SG RP++SGS M +
Sbjct: 10 GSKMNHSRPATSGSQM-PNSRP-ASSGSQMQSRAVSGSG--RPASSGSQMQNSRP----- 60
Query: 101 GSSSRPSSASGPPTSNQTSQTSLRPRSA-ETRPGSSHLSR 139
+SRP+SA S S RP S+ RP SS R
Sbjct: 61 -QNSRPASAGSQMQQRPASSGSQRPASSGSQRPASSGSQR 99
Score = 36.2 bits (82), Expect = 0.018
Identities = 34/102 (33%), Positives = 48/102 (46%), Gaps = 7/102 (6%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSP-SHLSARPSSGGSGTRPSTSGSDM----ASELT 95
GS+ +S + GS + A +G+ P S S +S +RP+++GS M AS +
Sbjct: 22 GSQMPNSRPASSGSQMQSRAVSGSGRPASSGSQMQNSRPQNSRPASAGSQMQQRPASSGS 81
Query: 96 TTSAWGSSSRP-SSASGPPTSNQTSQTSLR-PRSAETRPGSS 135
A S RP SS S P S+ Q +R P S T G S
Sbjct: 82 QRPASSGSQRPASSGSQRPGSSTNRQAPMRPPGSGSTMNGQS 123
Score = 35.0 bits (79), Expect = 0.039
Identities = 34/89 (38%), Positives = 46/89 (51%), Gaps = 15/89 (16%)
Query: 59 NANAGASSPSHLS-ARPSSGGS---GTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPT 114
N + +SS S ++ +RP++ GS +RP++SGS M S A S RP+S SG
Sbjct: 2 NHSRPSSSGSKMNHSRPATSGSQMPNSRPASSGSQMQSR-----AVSGSGRPAS-SGSQM 55
Query: 115 SNQTSQTSLRPRSA----ETRPGSSHLSR 139
N Q S RP SA + RP SS R
Sbjct: 56 QNSRPQNS-RPASAGSQMQQRPASSGSQR 83
Score = 32.3 bits (72), Expect = 0.25
Identities = 28/130 (21%), Positives = 48/130 (36%), Gaps = 7/130 (5%)
Query: 27 KPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGS------- 79
+P + SQ + ++S + + P + AS+ S + RP+S GS
Sbjct: 29 RPASSGSQMQSRAVSGSGRPASSGSQMQNSRPQNSRPASAGSQMQQRPASSGSQRPASSG 88
Query: 80 GTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSR 139
RP++SGS T A S ++N+ Q + R S + P +
Sbjct: 89 SQRPASSGSQRPGSSTNRQAPMRPPGSGSTMNGQSANRNGQLNSRSDSRRSAPAKVPVDH 148
Query: 140 FAEHVGENSV 149
+ N V
Sbjct: 149 RKQMSSSNGV 158
>At4g33930 putative protein
Length = 343
Score = 40.4 bits (93), Expect = 0.001
Identities = 36/118 (30%), Positives = 51/118 (42%), Gaps = 7/118 (5%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDM-----ASELTT 96
S S+ + S +WGS S +N G S S S GSG S SG +S+ T
Sbjct: 22 SGSAEAWSWSWGSGQS-GSNGGWGWRSGNSGGSSGSGSGGSDSNSGGSSWGWGWSSDGTD 80
Query: 97 TS-AWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNG 153
T+ WGSSS + +SG +++ + SA + H S + H N NG
Sbjct: 81 TNWGWGSSSGSNHSSGTGSTHNGHSSGSNHSSATGSTHNGHTSTGSNHSSGNGSRHNG 138
Score = 37.0 bits (84), Expect = 0.010
Identities = 31/100 (31%), Positives = 46/100 (46%), Gaps = 11/100 (11%)
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
GWGS S S+ S+ GS+ N +S +H SA S+ T ST + + + +
Sbjct: 84 GWGSSSGSNHSSGTGST----HNGHSSGSNHSSATGSTHNGHT--STGSNHSSGNGSRHN 137
Query: 99 AWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLS 138
+ S S SS++G S+ T T S GS+H S
Sbjct: 138 GYSSGSNHSSSTGSNHSSSTGSTHNNHSS-----GSNHSS 172
Score = 32.0 bits (71), Expect = 0.33
Identities = 27/92 (29%), Positives = 43/92 (46%), Gaps = 2/92 (2%)
Query: 30 NLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPS--SGGSGTRPSTSG 87
N S G+ G S S+ S+A GS+ + + + G++ S +R + S GS ST
Sbjct: 92 NHSSGTGSTHNGHSSGSNHSSATGSTHNGHTSTGSNHSSGNGSRHNGYSSGSNHSSSTGS 151
Query: 88 SDMASELTTTSAWGSSSRPSSASGPPTSNQTS 119
+ +S +T + S S SS G N +S
Sbjct: 152 NHSSSTGSTHNNHSSGSNHSSILGSTHKNHSS 183
Score = 27.7 bits (60), Expect = 6.3
Identities = 26/90 (28%), Positives = 43/90 (46%)
Query: 30 NLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSD 89
N S G+ G S S+ S++ GS+ S + + ++ S S S GS + +SGS+
Sbjct: 127 NHSSGNGSRHNGYSSGSNHSSSTGSNHSSSTGSTHNNHSSGSNHSSILGSTHKNHSSGSN 186
Query: 90 MASELTTTSAWGSSSRPSSASGPPTSNQTS 119
+S + +T SS S+ T N T+
Sbjct: 187 HSSIVGSTHNNHSSGSNHSSITGSTHNHTA 216
>At4g34300 putative protein
Length = 313
Score = 38.5 bits (88), Expect = 0.004
Identities = 34/113 (30%), Positives = 51/113 (45%), Gaps = 6/113 (5%)
Query: 39 GWGSKSSSSASN-AWGSSLSPNANAGASSP--SHLSARPSSGGSGTRPS--TSGSDMASE 93
GWG S + +N WGSS N ++G S H S S G+G+ + +SGS+ +S
Sbjct: 67 GWGWSSDGTDTNWGWGSSSGSNHSSGTGSTHNGHSSGSNHSSGTGSTHNGHSSGSNHSSS 126
Query: 94 L-TTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVG 145
+T + S S SS G N S ++ + T G S S + +G
Sbjct: 127 TGSTHNNHSSGSNHSSIVGSTHKNHGSGSNHSSIAGPTHNGHSSGSNHSSIIG 179
Score = 29.6 bits (65), Expect = 1.7
Identities = 32/93 (34%), Positives = 44/93 (46%), Gaps = 7/93 (7%)
Query: 33 SQRGTLGWGSKSSSS----ASNAWG-SSLSPNANAGASSPSHLSARPSSGGSGTRPSTSG 87
S G+ G GS S S +S WG SS + N G S S S S GS +SG
Sbjct: 45 SSGGSFGSGSGGSDSNSGGSSWGWGWSSDGTDTNWGWGSSSG-SNHSSGTGSTHNGHSSG 103
Query: 88 SDMASEL-TTTSAWGSSSRPSSASGPPTSNQTS 119
S+ +S +T + S S SS++G +N +S
Sbjct: 104 SNHSSGTGSTHNGHSSGSNHSSSTGSTHNNHSS 136
>At4g18670 extensin-like protein
Length = 839
Score = 38.5 bits (88), Expect = 0.004
Identities = 40/119 (33%), Positives = 48/119 (39%), Gaps = 14/119 (11%)
Query: 23 VAVPKPINLPSQRGTLGWGS--KSSSSASNAWGSS-LSPNANAGASSPSHLSARPSSGGS 79
+ VP P PS G+ S S S + + GS SP SP P+ GGS
Sbjct: 434 ITVPSPPTTPSPGGSPPSPSIVPSPPSTTPSPGSPPTSPTTPTPGGSPPSSPTTPTPGGS 493
Query: 80 G----TRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS 134
T P+ GS +S TT S GS PS + PP T P S T PGS
Sbjct: 494 PPSSPTTPTPGGSPPSSP-TTPSPGGSPPSPSISPSPPI------TVPSPPSTPTSPGS 545
Score = 37.4 bits (85), Expect = 0.008
Identities = 37/127 (29%), Positives = 47/127 (36%), Gaps = 10/127 (7%)
Query: 16 GMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHL----S 71
G + V VP P PS G+ S S S +P+ SPS + S
Sbjct: 401 GRSTRPPVVVPSPPTTPSPGGSPPSPSISPSPPITVPSPPTTPSPGGSPPSPSIVPSPPS 460
Query: 72 ARPSSGGSGTRPST---SGSDMASELTTT---SAWGSSSRPSSASGPPTSNQTSQTSLRP 125
PS G T P+T GS +S T T S S + P+ PP+S T P
Sbjct: 461 TTPSPGSPPTSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSP 520
Query: 126 RSAETRP 132
S P
Sbjct: 521 PSPSISP 527
Score = 32.0 bits (71), Expect = 0.33
Identities = 33/94 (35%), Positives = 38/94 (40%), Gaps = 7/94 (7%)
Query: 41 GSKSSSSASNAWGSS--LSPNANAGASSPSHLSARPSSGGSGTRPSTSGS---DMASELT 95
GS SS + G S SP SP PS GGS PS S S + S +
Sbjct: 479 GSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPPSPSISPSPPITVPSPPS 538
Query: 96 TTSAWGSSSRPSS--ASGPPTSNQTSQTSLRPRS 127
T ++ GS PSS S P S T T P S
Sbjct: 539 TPTSPGSPPSPSSPTPSSPIPSPPTPSTPPTPIS 572
>At1g62760 hypothetical protein
Length = 312
Score = 38.5 bits (88), Expect = 0.004
Identities = 41/97 (42%), Positives = 46/97 (47%), Gaps = 14/97 (14%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S SSS S+A SSLSP SSP LS PS S P S S ++S + S
Sbjct: 39 SPPSSSPSSAPPSSLSP------SSPPPLSLSPS---SPPPPPPSSSPLSSLSPSLSPSP 89
Query: 102 SSSRPSSASGPPTSNQTSQ---TSLRPRSAETRPGSS 135
SS PSSA PP+S S SL P S P SS
Sbjct: 90 PSSSPSSA--PPSSLSPSSPPPLSLSPSSPPPPPPSS 124
Score = 30.4 bits (67), Expect = 0.97
Identities = 24/66 (36%), Positives = 31/66 (46%)
Query: 54 SSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPP 113
SSLSP+ + S S SA PSS + P S S + S+ SS S+S
Sbjct: 79 SSLSPSLSPSPPSSSPSSAPPSSLSPSSPPPLSLSPSSPPPPPPSSSPLSSLSPSSSSST 138
Query: 114 TSNQTS 119
SNQT+
Sbjct: 139 YSNQTN 144
>At3g28780 histone-H4-like protein
Length = 614
Score = 37.7 bits (86), Expect = 0.006
Identities = 28/103 (27%), Positives = 45/103 (43%)
Query: 33 SQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMAS 92
S+ + G S S+ A+ + S A +G S+ + S+ G S+SG A+
Sbjct: 454 SESASAGAASGGSTEANGGAAAGGSTEAGSGTSTETSSMGGGSAAAGGVSESSSGGSTAA 513
Query: 93 ELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
T+ SA G S+ ASG ++ T + SA GS+
Sbjct: 514 GGTSESASGGSATAGGASGGTYTDSTGGSPTGSPSAGGPSGSA 556
Score = 36.2 bits (82), Expect = 0.018
Identities = 29/100 (29%), Positives = 47/100 (47%), Gaps = 7/100 (7%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
G+ G +S+SS + GS+ S +A+AGA+S A + G+ + SG+ T
Sbjct: 433 GSAETGGESTSSGVASGGSTGSESASAGAASGGSTEANGGAAAGGSTEAGSGTS-----T 487
Query: 96 TTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
TS+ G S ++A G S+ T+ S GS+
Sbjct: 488 ETSSMGGGS--AAAGGVSESSSGGSTAAGGTSESASGGSA 525
Score = 32.3 bits (72), Expect = 0.25
Identities = 34/142 (23%), Positives = 56/142 (38%), Gaps = 10/142 (7%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGG--SGTRPSTSGSDMASE 93
G++ G +S+ + S + +A+ GA+S +SGG SG T G +S
Sbjct: 390 GSVESGGESTGATSGGSAETSDESASGGAAS----GGESASGGAASGGSAETGGESTSSG 445
Query: 94 LTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNG 153
+ ++ GS+ S+++G + T T GS S +G S A G
Sbjct: 446 V---ASGGSTGSESASAGAASGGSTEANGGAAAGGSTEAGSG-TSTETSSMGGGSAAAGG 501
Query: 154 ARTTETLGITQRKNDDFSLSTG 175
+ + G T S S G
Sbjct: 502 VSESSSGGSTAAGGTSESASGG 523
Score = 32.3 bits (72), Expect = 0.25
Identities = 40/140 (28%), Positives = 55/140 (38%), Gaps = 25/140 (17%)
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
G G+ S +SA S + + + + SA +SGGS SGS
Sbjct: 289 GAGAASGASAKTGGESGEAASGGSAETGGESASAGAASGGSAETGGESGSG--------- 339
Query: 99 AWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGEN---------SV 149
G++S SASG TS + +T SAET G S A GE+ SV
Sbjct: 340 --GAASGGESASGGATSGGSPETG---GSAET--GGESASGGAASGGESASGGAASSGSV 392
Query: 150 AWNGARTTETLGITQRKNDD 169
G T T G + +D+
Sbjct: 393 ESGGESTGATSGGSAETSDE 412
Score = 32.3 bits (72), Expect = 0.25
Identities = 27/92 (29%), Positives = 44/92 (47%), Gaps = 11/92 (11%)
Query: 36 GTLGWGSKSSSSASNAWGSSLS----PNANAGASSPSHLSARPSSGGSGTRPSTSGSDMA 91
G+ GS +S+ S+ G S + +++G S+ + ++ +SGGS T SG
Sbjct: 477 GSTEAGSGTSTETSSMGGGSAAAGGVSESSSGGSTAAGGTSESASGGSATAGGASGGTY- 535
Query: 92 SELTTTSAWGSSSRPSSASGPPTSNQTSQTSL 123
T S GS + SA GP S S++S+
Sbjct: 536 ----TDSTGGSPTGSPSAGGP--SGSASESSM 561
Score = 31.2 bits (69), Expect = 0.57
Identities = 33/123 (26%), Positives = 52/123 (41%), Gaps = 10/123 (8%)
Query: 15 GGMTVLGKVAVPKPINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSA-- 72
GG T G + ++ G S+SSS S A G + S +A+ G+++ S
Sbjct: 476 GGSTEAGSGTSTETSSMGGGSAAAGGVSESSSGGSTAAGGT-SESASGGSATAGGASGGT 534
Query: 73 -RPSSGGSGT------RPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRP 125
S+GGS T PS S S+ + E T + SAS T+ Q + +
Sbjct: 535 YTDSTGGSPTGSPSAGGPSGSASESSMEGGTFGGQSMGGQAGSASYQSTNYQKTHSKSAG 594
Query: 126 RSA 128
+S+
Sbjct: 595 KSS 597
Score = 30.8 bits (68), Expect = 0.74
Identities = 31/114 (27%), Positives = 50/114 (43%), Gaps = 8/114 (7%)
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTS 98
G +S S+A+ G + ++ A + S +A SGG+ + SG A++ + S
Sbjct: 215 GAAGESGSAATADSGDAAGADSGGAAGADSGGAASADSGGAAAGETASGGAAAADTSGGS 274
Query: 99 A--WGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVA 150
A G S+ +ASG ++ S S E G S AE GE++ A
Sbjct: 275 AETGGESASGGAASGAGAASGAS-AKTGGESGEAASGGS-----AETGGESASA 322
Score = 30.4 bits (67), Expect = 0.97
Identities = 30/86 (34%), Positives = 37/86 (42%), Gaps = 8/86 (9%)
Query: 38 LGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGS-GTRPSTSGSDMASELTT 96
LG GS S+ G S SP +++G SPS + P+ GGS G SGS S +
Sbjct: 127 LGGGSSGLGSS----GDSGSPGSDSG--SPSADTGSPTDGGSYGDTTGDSGSSAGSP-SY 179
Query: 97 TSAWGSSSRPSSASGPPTSNQTSQTS 122
S GS S SG T S
Sbjct: 180 PSDDGSGSTAGGPSGSTTDGSAGGES 205
Score = 29.6 bits (65), Expect = 1.7
Identities = 28/100 (28%), Positives = 44/100 (44%), Gaps = 8/100 (8%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
G G S ++SA + G++ A+ GA +A +SGGS S S A+
Sbjct: 238 GAAGADSGGAASADSG-GAAAGETASGGA------AAADTSGGSAETGGESASGGAAS-G 289
Query: 96 TTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
+A G+S++ SG S +++T SA G S
Sbjct: 290 AGAASGASAKTGGESGEAASGGSAETGGESASAGAASGGS 329
Score = 29.3 bits (64), Expect = 2.2
Identities = 35/123 (28%), Positives = 48/123 (38%), Gaps = 14/123 (11%)
Query: 41 GSKSSSSASNAWGSSLSP---NANAGASSPSHLSARPSSGGSG-TRPSTSGSDMASELTT 96
GS S+ + S G S ++ + A SPS+ PS GSG T SGS
Sbjct: 148 GSPSADTGSPTDGGSYGDTTGDSGSSAGSPSY----PSDDGSGSTAGGPSGSTTDGSAGG 203
Query: 97 TSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGART 156
S+ G S + A+G S T+ S + S + A+ G S GA
Sbjct: 204 ESSMGGDSSSAGAAGESGSAATAD------SGDAAGADSGGAAGADSGGAASADSGGAAA 257
Query: 157 TET 159
ET
Sbjct: 258 GET 260
Score = 28.9 bits (63), Expect = 2.8
Identities = 33/148 (22%), Positives = 61/148 (40%), Gaps = 3/148 (2%)
Query: 36 GTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELT 95
G+ G +S+S+ + + GS+ + + + S + SG P T GS A
Sbjct: 311 GSAETGGESASAGAASGGSAETGGESGSGGAASGGESASGGATSGGSPETGGS--AETGG 368
Query: 96 TTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAEHVGENSVAWNGAR 155
+++ G++S SASG S+ + ++ A T GS+ S + G S + +
Sbjct: 369 ESASGGAASGGESASGGAASSGSVESGGESTGA-TSGGSAETSDESASGGAASGGESASG 427
Query: 156 TTETLGITQRKNDDFSLSTGDFPTLGSE 183
+ G + + S + GSE
Sbjct: 428 GAASGGSAETGGESTSSGVASGGSTGSE 455
Score = 28.1 bits (61), Expect = 4.8
Identities = 20/79 (25%), Positives = 40/79 (50%), Gaps = 5/79 (6%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
G SS+ A+ GS+ + ++ A + S +A SGG+ + + SG A E T++
Sbjct: 209 GDSSSAGAAGESGSAATADSGDAAGADSGGAAGADSGGAAS--ADSGGAAAGE---TASG 263
Query: 101 GSSSRPSSASGPPTSNQTS 119
G+++ +S T +++
Sbjct: 264 GAAAADTSGGSAETGGESA 282
>At5g01280 putative protein
Length = 460
Score = 37.4 bits (85), Expect = 0.008
Identities = 27/78 (34%), Positives = 43/78 (54%), Gaps = 4/78 (5%)
Query: 41 GSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAW 100
G + SS+S++ +S P + +P+ RPS+ S +T+ + + S +TTS+
Sbjct: 81 GIRRPSSSSSSRSTSRPPTPTRKSKTPAK---RPSTPTSRATSTTTRATLTSS-STTSST 136
Query: 101 GSSSRPSSASGPPTSNQT 118
S SRPSS+SG TS T
Sbjct: 137 RSWSRPSSSSGTGTSRVT 154
Score = 34.7 bits (78), Expect = 0.051
Identities = 31/98 (31%), Positives = 49/98 (49%), Gaps = 12/98 (12%)
Query: 42 SKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWG 101
S S++S++ +W S S ++ G S + +AR TRP+TS + T++
Sbjct: 129 SSSTTSSTRSW-SRPSSSSGTGTSRVTLTAAR------ATRPTTSTDQQTTGSATSTR-- 179
Query: 102 SSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSR 139
S++RP SA P S S++S R T GSS + R
Sbjct: 180 SNNRPMSA---PNSKPGSRSSTPTRRPSTPNGSSTVLR 214
Score = 27.3 bits (59), Expect = 8.2
Identities = 31/116 (26%), Positives = 48/116 (40%), Gaps = 11/116 (9%)
Query: 35 RGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASEL 94
R T G S + + ++P + + +HL+A + + + S SE
Sbjct: 9 RRTAGENLLYSDGEKSDYEWLVTPPGSPSRNVTNHLNAPDDNLMTLISRLENYSKEESEH 68
Query: 95 TTTSAWGSSS-----RPSSASG------PPTSNQTSQTSLRPRSAETRPGSSHLSR 139
TTS SSS RPSS+S PPT + S+T + S T +S +R
Sbjct: 69 QTTSLHSSSSVSGIRRPSSSSSSRSTSRPPTPTRKSKTPAKRPSTPTSRATSTTTR 124
>At1g65440
Length = 1703
Score = 37.0 bits (84), Expect = 0.010
Identities = 24/83 (28%), Positives = 33/83 (38%), Gaps = 8/83 (9%)
Query: 37 TLGWGSKS---SSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSDMASE 93
T GWGS+S S + +WGS + G + S GG G+ GSD +E
Sbjct: 1601 TGGWGSESGGNKSDGAGSWGSGSGGGGSGGWGNDSGGKKSSEDGGFGSGSGGGGSDWGNE 1660
Query: 94 -----LTTTSAWGSSSRPSSASG 111
+ WGS S + G
Sbjct: 1661 SGGKKSSADGGWGSESGGKKSDG 1683
Score = 35.4 bits (80), Expect = 0.030
Identities = 24/89 (26%), Positives = 38/89 (41%), Gaps = 2/89 (2%)
Query: 29 INLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGS 88
+N Q G WG+ + +A WG+ S G+ S + S+GG G+ + S
Sbjct: 1556 MNSDRQDGNGDWGNNDTGTADGGWGN--SGGGGWGSESAGKKTGGGSTGGWGSESGGNKS 1613
Query: 89 DMASELTTTSAWGSSSRPSSASGPPTSNQ 117
D A + S G S + SG S++
Sbjct: 1614 DGAGSWGSGSGGGGSGGWGNDSGGKKSSE 1642
Score = 33.5 bits (75), Expect = 0.11
Identities = 27/85 (31%), Positives = 36/85 (41%), Gaps = 12/85 (14%)
Query: 39 GWGSKSSSS-----ASNAWGSSLSPNANAGASSPSHLSARPSSGG-----SGTRPSTSGS 88
GWGS+S+ ++ WGS N + GA S S SGG G + S G
Sbjct: 1586 GWGSESAGKKTGGGSTGGWGSESGGNKSDGAGSWGSGSGGGGSGGWGNDSGGKKSSEDGG 1645
Query: 89 DMASELTTTSAWGSSS--RPSSASG 111
+ S WG+ S + SSA G
Sbjct: 1646 FGSGSGGGGSDWGNESGGKKSSADG 1670
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 36.2 bits (82), Expect = 0.018
Identities = 36/112 (32%), Positives = 49/112 (43%), Gaps = 13/112 (11%)
Query: 38 LGWGSKSSSSASNAWGSSLSPNANAGASSPSHL---SARPSSGGSGTRPSTSGSDMASEL 94
LG S + S+ S GSSLS N + SPS + S P +G G+ PS + S + S
Sbjct: 560 LGLFSGNGSTNSTVTGSSLSSNTTESSDSPSTVVTPSTSPPAGHLGS-PSDTPSSVVSPS 618
Query: 95 TTTSAWGSSSRPSSAS---GPPTS------NQTSQTSLRPRSAETRPGSSHL 137
T+ A + P++ S P TS S T S T P + HL
Sbjct: 619 TSLPAGQLGAPPATPSMVVSPSTSPPAGHLGSPSDTPSSLVSPSTSPPAGHL 670
Score = 27.3 bits (59), Expect = 8.2
Identities = 19/60 (31%), Positives = 28/60 (46%)
Query: 28 PINLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSG 87
P + PS T +S SAS + S P+A+ AS + S PS+ S + S+ G
Sbjct: 673 PSDTPSSVVTPSASPSTSPSASPSVSPSAFPSASPSASPSASPSVSPSASPSASPQSSIG 732
>At3g57120 unknown protein
Length = 456
Score = 36.2 bits (82), Expect = 0.018
Identities = 29/84 (34%), Positives = 39/84 (45%), Gaps = 7/84 (8%)
Query: 64 ASSPSHLSARPSSGGSGTRPSTSGSDMASEL-----TTTSAWGSSSRPSSASGPPTSNQT 118
AS P +RPS G PST S S TTTS + SS+SG +S+
Sbjct: 27 ASEPKRTRSRPSQS-QGRHPSTRRSPSTSNTGTTTTTTTSNSNKTGASSSSSGAASSSVA 85
Query: 119 SQTSLRPRSAETRPGSSHLSRFAE 142
S+TSL E+ P + H+ +E
Sbjct: 86 SRTSL-ASLRESLPENPHIYNVSE 108
>At1g78580 trehalose-6-phosphate synthase
Length = 942
Score = 36.2 bits (82), Expect = 0.018
Identities = 27/86 (31%), Positives = 45/86 (51%), Gaps = 5/86 (5%)
Query: 67 PSHLSARPSSGGSGTRPSTSGSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPR 126
P+ +RPSS SG + S+SG +T + S S+ SS+S +N++SQ SL+
Sbjct: 817 PAIARSRPSSD-SGAK-SSSGDRRPPSKSTHNNNKSGSKSSSSSNSNNNNKSSQRSLQ-- 872
Query: 127 SAETRPGSSHLSRFAEHVGENSVAWN 152
+E + GS+H + ++WN
Sbjct: 873 -SERKSGSNHSLGNSRRPSPEKISWN 897
>At3g57340 dnaJ-like protein
Length = 367
Score = 35.4 bits (80), Expect = 0.030
Identities = 21/64 (32%), Positives = 32/64 (49%), Gaps = 2/64 (3%)
Query: 83 PSTSGSDMASELTTTSAW--GSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRF 140
P+ D+ SEL + GS+ P SA+ +SN + + SLR R + T SS +S
Sbjct: 39 PNLPIDDLVSELNNNKSDEPGSAKSPGSAAAKDSSNSSDRPSLRQRGSSTTSSSSSMSYT 98
Query: 141 AEHV 144
E +
Sbjct: 99 EEQI 102
>At1g27850 transposon protein, putative
Length = 1148
Score = 35.0 bits (79), Expect = 0.039
Identities = 38/135 (28%), Positives = 59/135 (43%), Gaps = 4/135 (2%)
Query: 35 RGTLGWGSKSSSSASNAWGSSLSPNANAG-ASSPSHLSARPSSGGSGTRPSTSGSDMASE 93
RG+ + S S+SP+A+ +SS SH R SS G+ ++SG D
Sbjct: 274 RGSSPASRNGRDAVSTRSRKSVSPSASRSVSSSHSHERDRFSSQSKGS-VASSGDDDLHS 332
Query: 94 LTTTSAWGSSSRPSSASGPPTSNQTSQTS--LRPRSAETRPGSSHLSRFAEHVGENSVAW 151
L + GS S + +++TS++S L P SA RP S L + +S+
Sbjct: 333 LQSIPVGGSERAVSKRASLSPNSRTSRSSKLLSPGSAPRRPFESALRQMEHPKSHHSMFR 392
Query: 152 NGARTTETLGITQRK 166
A + + GI K
Sbjct: 393 PLASSLPSTGIYSGK 407
>At1g06930 hypothetical protein
Length = 169
Score = 34.7 bits (78), Expect = 0.051
Identities = 38/112 (33%), Positives = 50/112 (43%), Gaps = 4/112 (3%)
Query: 28 PINLPSQRGTLGWGSK-SSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTS 86
P SQ GT SK SSSS N+ P + PS+ A PSS + T+
Sbjct: 33 PFKWESQPGTPRRLSKRSSSSGFNSDSDFNFPVSAPLTPPPSYFYASPSSTKHVSPKKTN 92
Query: 87 ---GSDMASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSS 135
S ++ + S+ SSS SS+S P + +TS S R RS GSS
Sbjct: 93 TLFSSLLSKNRSVPSSPASSSSSSSSSVPSSPLRTSDLSNRRRSMWFESGSS 144
>At4g15410 phosphatase like protein
Length = 421
Score = 34.3 bits (77), Expect = 0.067
Identities = 31/113 (27%), Positives = 48/113 (42%), Gaps = 6/113 (5%)
Query: 30 NLPSQRGTLGWGSKSSSSASNAWGSSLSPNANAGASSPSHLSARPSSGGSGTRPSTSGSD 89
NL + T ++++ N G + AGA SPS S P S T PS S S
Sbjct: 34 NLDAAVSTFLDNDAAAAAEPNPTGPPPPSSTIAGAQSPSQ-SHSPDYTPSETSPSPSRSR 92
Query: 90 MASELTTTSAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGSSHLSRFAE 142
AS + + +G SR + T N + RS+ +R + ++ FA+
Sbjct: 93 SASPSSRAAPYGLRSRGGAGENKETENPSG-----IRSSRSRQHAGNIRTFAD 140
>At3g55270 MAP kinase phosphatase (MKP1)
Length = 732
Score = 34.3 bits (77), Expect = 0.067
Identities = 31/97 (31%), Positives = 43/97 (43%), Gaps = 1/97 (1%)
Query: 39 GWGSKSSSSASNAWGSSLSPNANAGASSPSHL-SARPSSGGSGTRPSTSGSDMASELTTT 97
G S ++SS+S A LSP++ +S + L S SSG S RPS S + ++
Sbjct: 439 GSPSSTTSSSSTASPPFLSPDSVCSTNSGNSLKSFSQSSGRSSLRPSIPPSLTLPKFSSL 498
Query: 98 SAWGSSSRPSSASGPPTSNQTSQTSLRPRSAETRPGS 134
S S + P + G T Q S S R GS
Sbjct: 499 SLLPSQTSPKESRGVNTFLQPSPNRKASPSLAERRGS 535
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.125 0.374
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,927,807
Number of Sequences: 26719
Number of extensions: 259470
Number of successful extensions: 1668
Number of sequences better than 10.0: 200
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 143
Number of HSP's that attempted gapping in prelim test: 1227
Number of HSP's gapped (non-prelim): 423
length of query: 256
length of database: 11,318,596
effective HSP length: 97
effective length of query: 159
effective length of database: 8,726,853
effective search space: 1387569627
effective search space used: 1387569627
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140849.12


