

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140848.2 + phase: 0 /pseudo
(573 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
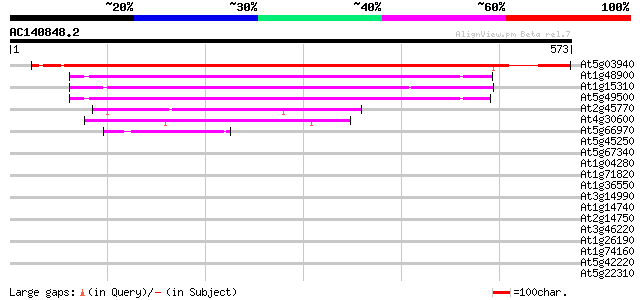
Score E
Sequences producing significant alignments: (bits) Value
At5g03940 signal recognition particle 54CP protein precursor 760 0.0
At1g48900 unknown protein (At1g48900) 211 1e-54
At1g15310 putative signal recognition particle 54 kDa subunit 192 4e-49
At5g49500 SRP54 (signal recognition particle 54 KDa) protein 188 6e-48
At2g45770 putative signal recognition particle receptor (alpha s... 155 7e-38
At4g30600 signal recognition particle receptor-like protein 126 3e-29
At5g66970 putative protein 52 1e-06
At5g45250 disease resistance protein RPS4 38 0.017
At5g67340 unknown proteins 34 0.25
At1g04280 unknown protein 33 0.33
At1g71820 unknown protein (At1g71820) 33 0.43
At1g36550 hypothetical protein 32 0.73
At3g14990 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate bios... 32 0.96
At1g14740 unknown protein 32 0.96
At2g14750 putative adenosine phosphosulfate kinase 32 1.2
At3g46220 unknown protein 31 1.6
At1g26190 unknown protein (At1g26190) 31 1.6
At1g74160 unknown protein 31 2.1
At5g42220 unknown protein 30 2.8
At5g22310 unknown protein 30 2.8
>At5g03940 signal recognition particle 54CP protein precursor
Length = 564
Score = 760 bits (1962), Expect = 0.0
Identities = 410/559 (73%), Positives = 457/559 (81%), Gaps = 43/559 (7%)
Query: 23 SSSKLCSSLSYRNSFAKEIWGFVQSKSVTMRRMDMIRPVVVRAEMFGQLTTGLESAWNKL 82
+S+ L SS RN +EIW +V+SK+V R VRAEMFGQLT GLE+AW+KL
Sbjct: 38 NSASLSSS---RNLSTREIWSWVKSKTVVGH--GRYRRSQVRAEMFGQLTGGLEAAWSKL 92
Query: 83 KGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLVK 142
KGEEVLTKDNI EPMRDIRRALLEADVSLPVVRRFVQSV+DQAVG+GV RGVKPDQQLVK
Sbjct: 93 KGEEVLTKDNIAEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGMGVIRGVKPDQQLVK 152
Query: 143 IVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAG 202
IVHDELV+LMGGEVSEL FAK+GPTVILLAGLQGVGKTTVCAKLA Y KKQGKSCML+AG
Sbjct: 153 IVHDELVKLMGGEVSELQFAKSGPTVILLAGLQGVGKTTVCAKLACYLKKQGKSCMLIAG 212
Query: 203 DVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKIDVVIVDTAGRLQI 262
DVYRPAAIDQL ILG+QV VPVYTAGTDVKP+DIAKQG +EAKK +DVVI+DTAGRLQI
Sbjct: 213 DVYRPAAIDQLVILGEQVGVPVYTAGTDVKPADIAKQGLKEAKKNNVDVVIMDTAGRLQI 272
Query: 263 DKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTKLDGDSRGG 322
DK MMDELKDVK+ LNPTEVLLVVDAMTGQEAAALVTTFN+EIGITGAILTKLDGDSRGG
Sbjct: 273 DKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAAALVTTFNVEIGITGAILTKLDGDSRGG 332
Query: 323 AALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQEDAE 382
AALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKA EVM+QEDAE
Sbjct: 333 AALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKATEVMRQEDAE 392
Query: 383 DLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGMAKVTPAQIREAERNLEIMEVI 442
DLQKKIMSAKFDFNDFLKQTRAVA+MGS++RV+GMIPGM KV+PAQIREAE+NL +ME +
Sbjct: 393 DLQKKIMSAKFDFNDFLKQTRAVAKMGSMTRVLGMIPGMGKVSPAQIREAEKNLLVMEAM 452
Query: 443 IKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILSNFEKYTTIM--------SG 494
I+ MTPEERE+PELLAESP RRKR+A+DSGKTEQQ A+++ + M G
Sbjct: 453 IEVMTPEERERPELLAESPERRKRIAKDSGKTEQQVSALVAQIFQMRVKMKNLMGVMEGG 512
Query: 495 KPTCSSTISNACSDEEPNGCNGRWIDANT**S*GST*NRGKVCLYQAPPGTARRRKKAES 554
S + +A E+ +APPGTARR++KA+S
Sbjct: 513 SIPALSGLEDALKAEQ-----------------------------KAPPGTARRKRKADS 543
Query: 555 RRLFADSTLRQP-PRGFGS 572
R+ F +S +P PRGFGS
Sbjct: 544 RKKFVESASSKPGPRGFGS 562
>At1g48900 unknown protein (At1g48900)
Length = 495
Score = 211 bits (536), Expect = 1e-54
Identities = 125/436 (28%), Positives = 226/436 (51%), Gaps = 10/436 (2%)
Query: 62 VVRAEMFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSV 121
+V AE+ G++T ++ ++ ++ + + E + +I RALL++DVS P+V+ ++
Sbjct: 1 MVLAELGGRITRAIQ----QMSNVTIIDEKALNECLNEITRALLQSDVSFPLVKEMQSNI 56
Query: 122 TDQAVGVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTT 181
+ G + + + + EL +++ K +V++ GLQG GKTT
Sbjct: 57 KKIVNLEDLAAGHNKRRIIEQAIFSELCKMLDPGKPAFAPKKAKASVVMFVGLQGAGKTT 116
Query: 182 VCAKLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGF 241
C K A Y +K+G LV D +R A DQL + +P Y + T+ P IA +G
Sbjct: 117 TCTKYAYYHQKKGYKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVEGV 176
Query: 242 EEAKKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTF 301
+ KK+ D++IVDT+GR + + ++ +E++ V P V+ V+D+ GQ A F
Sbjct: 177 DTFKKENCDLIIVDTSGRHKQEASLFEEMRQVAEATKPDLVIFVMDSSIGQAAFDQAQAF 236
Query: 302 NLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILG 361
+ + I+TK+DG ++GG ALS + P+ +G GE M++ E F R+LG
Sbjct: 237 KQSVAVGAVIITKMDGHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLG 296
Query: 362 MGDVLSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGM 421
MGD FV+K QEV+ ++ +L +K+ F Q + + MG + V M+PG+
Sbjct: 297 MGDWSGFVDKLQEVVPKDQQPELLEKLSQGNFTLRIMYDQFQNILNMGPLKEVFSMLPGI 356
Query: 422 -AKVTP-AQIREAERNLEIMEVIIKAMTPEERE--KPELLAESPVRRKRVAQDSGKTEQQ 477
A++ P +E++ ++ ++ +MT +E + P++ ES R R+A+ SG+ ++
Sbjct: 357 SAEMMPKGHEKESQAKIKRYMTMMDSMTNDELDSSNPKVFNES--RMMRIARGSGRQVRE 414
Query: 478 TDAILSNFEKYTTIMS 493
+L +++ I S
Sbjct: 415 VMEMLEEYKRLAKIWS 430
>At1g15310 putative signal recognition particle 54 kDa subunit
Length = 479
Score = 192 bits (488), Expect = 4e-49
Identities = 122/435 (28%), Positives = 216/435 (49%), Gaps = 7/435 (1%)
Query: 62 VVRAEMFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSV 121
+V AE+ G++T ++ N +E + D + E I RALL++DVS +V + ++
Sbjct: 1 MVLAELGGRITRAIQQMNNVTIIDEKVLNDFLNE----ITRALLQSDVSFGLVEKMQTNI 56
Query: 122 TDQAVGVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTT 181
+ G + + + EL +++ K P+V++ GLQG GKTT
Sbjct: 57 KKIVNLDDLAAGHNKRLIIEQAIFKELCRMLDPGKPAFAPKKAKPSVVMFVGLQGAGKTT 116
Query: 182 VCAKLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGF 241
C K A Y +K+G LV D +R A DQL + +P Y + T+ P IA +G
Sbjct: 117 TCTKYAYYHQKKGYKAALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVEGV 176
Query: 242 EEAKKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTF 301
+ KK+K D++IVDT+GR + ++ +E++ V P V+ V+D+ GQ A F
Sbjct: 177 DRFKKEKCDLIIVDTSGRHKQAASLFEEMRQVAEATEPDLVIFVMDSSIGQAAFEQAEAF 236
Query: 302 NLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILG 361
+ + I+TK+DG ++GG ALS + P+ +G GE M++ E F R+LG
Sbjct: 237 KETVSVGAVIITKMDGHAKGGGALSAVAATKSPVIFIGTGEHMDEFEVFDVKPFVSRLLG 296
Query: 362 MGDVLSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIPGM 421
GD V+K QEV+ ++ +L + + F Q + ++ ++++ M+PG+
Sbjct: 297 KGDWSGLVDKLQEVVPKDLQNELVENLSQGNFTLRSMYDQFQCSLRI-PLNQLFSMLPGI 355
Query: 422 -AKVTPAQIREAER-NLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTD 479
A++ P E R ++ ++ +MT +E + P + R R+A+ SG+ ++
Sbjct: 356 SAEMMPKGHGEESRVKMKRYMTMMDSMTNKELDSPNPKIFNESRIMRIARGSGRLVREVM 415
Query: 480 AILSNFEKYTTIMSG 494
+L +++ M G
Sbjct: 416 EMLEEYKRIAKTMKG 430
>At5g49500 SRP54 (signal recognition particle 54 KDa) protein
Length = 497
Score = 188 bits (478), Expect = 6e-48
Identities = 117/436 (26%), Positives = 221/436 (49%), Gaps = 12/436 (2%)
Query: 62 VVRAEMFGQLTTGLESAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSV 121
+V AE+ G++ + ++ K+ ++ + + + + +I RALL++DVS P+V+ ++
Sbjct: 1 MVLAELGGRIMSAIQ----KMNNVTIIDEKALNDCLNEITRALLQSDVSFPLVKEMQTNI 56
Query: 122 TDQAVGVGVTRGVKPDQQLVKIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQG--VGK 179
+ G + + + + EL +++ S K P+V++ GLQG + K
Sbjct: 57 KKIVNLEDLAAGHNKRRIIEQAIFSELCKMLDPGKSAFAPKKAKPSVVMFVGLQGEVLEK 116
Query: 180 TTVCAKLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQ 239
+ + +++G LV D +R A DQL + +P Y + T P IA +
Sbjct: 117 PQLVPSMLIIIRRKGYKPALVCADTFRAGAFDQLKQNATKSKIPYYGSYTGSDPVKIAVE 176
Query: 240 GFEEAKKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVT 299
G + KK+ D++IVDT+GR + ++ +E++ + P V+ V+D+ GQ A
Sbjct: 177 GVDRFKKENCDLIIVDTSGRHKQQASLFEEMRQISEATKPDLVIFVMDSSIGQTAFEQAR 236
Query: 300 TFNLEIGITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRI 359
F + + I+TK+DG ++GG LS + P+ +G GE M++ E F R+
Sbjct: 237 AFKQTVAVGAVIITKMDGHAKGGGTLSAVAATKSPVIFIGTGEHMDEFEVFDAKPFVSRL 296
Query: 360 LGMGDVLSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQMGSVSRVIGMIP 419
LG GD+ FV K QEV+ ++ +L + + KF Q + + MG + V M+P
Sbjct: 297 LGNGDMSGFVNKLQEVVPKDQQPELLEMLSHGKFTLRIMYDQFQNMLNMGPLKEVFSMLP 356
Query: 420 GM-AKVTP-AQIREAERNLEIMEVIIKAMTPEERE--KPELLAESPVRRKRVAQDSGKTE 475
GM A++ P +E++ ++ ++ +MT EE + P++ ES R R+A+ SG+
Sbjct: 357 GMRAEMMPEGHEKESQAKIKRYMTMMDSMTNEELDSSNPKVFNES--RIMRIARGSGRIV 414
Query: 476 QQTDAILSNFEKYTTI 491
++ +L +++ TT+
Sbjct: 415 REVMEMLEEYKRLTTM 430
>At2g45770 putative signal recognition particle receptor (alpha
subunit)
Length = 366
Score = 155 bits (391), Expect = 7e-38
Identities = 98/285 (34%), Positives = 150/285 (52%), Gaps = 12/285 (4%)
Query: 85 EEVLTKDNIVEPMR---DIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLV 141
+E+L N+ E R ++ ALL +D + R V+ + + + + G + L
Sbjct: 82 DELLLFWNLAETDRVLDELEEALLVSDFGPKITVRIVERLREDIMSGKLKSGSEIKDALK 141
Query: 142 KIVHDELVQLMGGEVSELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVA 201
+ V + L + +L F K P VI++ G+ G GKTT KLA+ K +G ++ A
Sbjct: 142 ESVLEMLAKKNSKTELQLGFRK--PAVIMIVGVNGGGKTTSLGKLAHRLKNEGTKVLMAA 199
Query: 202 GDVYRPAAIDQLTILGKQVDVPVYTA-GTDVKPSDIAKQGFEEAKKKKIDVVIVDTAGRL 260
GD +R AA DQL I ++ + A G K + + + + K++ DVV+ DT+GRL
Sbjct: 200 GDTFRAAASDQLEIWAERTGCEIVVAEGDKAKAATVLSKAVKRGKEEGYDVVLCDTSGRL 259
Query: 261 QIDKAMMDELKDVKRVLN------PTEVLLVVDAMTGQEAAALVTTFNLEIGITGAILTK 314
+ ++M+EL K+ + P E+LLV+D TG FN +GITG ILTK
Sbjct: 260 HTNYSLMEELIACKKAVGKIVSGAPNEILLVLDGNTGLNMLPQAREFNEVVGITGLILTK 319
Query: 315 LDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRI 359
LDG +RGG +SV E G P+K +G GE +EDL+PF P+ I
Sbjct: 320 LDGSARGGCVVSVVEELGIPVKFIGVGEAVEDLQPFDPEAFVNAI 364
>At4g30600 signal recognition particle receptor-like protein
Length = 634
Score = 126 bits (317), Expect = 3e-29
Identities = 81/290 (27%), Positives = 152/290 (51%), Gaps = 18/290 (6%)
Query: 77 SAWNKLKGEEVLTKDNIVEPMRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKP 136
S + + G+ L + ++ ++ ++ L+ +V+ + + +SV G ++ +
Sbjct: 332 SVFQSITGKANLERTDLGPALKALKERLMTKNVAEEIAEKLCESVEASLEGKKLSSFTRI 391
Query: 137 DQQLVKIVHDELVQLMGGEVS-----ELTFAKTG--PTVILLAGLQGVGKTTVCAKLANY 189
+ + D LV+++ S ++ AK P V++ G+ GVGK+T AK+A +
Sbjct: 392 SSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYW 451
Query: 190 FKKQGKSCMLVAGDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQGFEEAKKKKI 249
++ S M+ A D +R A++QL +++ +P++ G + P+ +AK+ +EA +
Sbjct: 452 LQQHKVSVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEATRNGS 511
Query: 250 DVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAALVTTFNLEIG--- 306
DVV+VDTAGR+Q ++ +M L + + P VL V +A+ G +A ++ FN ++
Sbjct: 512 DVVLVDTAGRMQDNEPLMRALSKLINLNQPDLVLFVGEALVGNDAVDQLSKFNQKLSDLS 571
Query: 307 -------ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMEDLE 348
I G +LTK D D + GAALS+ +SG P+ VG G+ DL+
Sbjct: 572 TSGNPRLIDGILLTKFDTIDDKVGAALSMVYISGSPVMFVGCGQSYTDLK 621
>At5g66970 putative protein
Length = 171
Score = 51.6 bits (122), Expect = 1e-06
Identities = 37/129 (28%), Positives = 61/129 (46%), Gaps = 7/129 (5%)
Query: 97 MRDIRRALLEADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLVKIVHDELVQLMGGEV 156
+ +I ALL+AD+ V + + + K ++ +++ EL +
Sbjct: 32 LNEITCALLDADIPRLAVDEI------ETKSLEIINLPKAPKKKGALIYMELSSKLDPGK 85
Query: 157 SELTFAKTGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGDVYRPAAIDQLTIL 216
S L K P++++ GL+GV KT CAK A Y +K+G LV D ++ A +L
Sbjct: 86 SALIRGKLEPSIVMFIGLRGVDKTKTCAKYARYHRKKGFRPALVCADTFKFDAFVRLNKA 145
Query: 217 GKQVDVPVY 225
K +VPVY
Sbjct: 146 AKD-EVPVY 153
>At5g45250 disease resistance protein RPS4
Length = 1217
Score = 37.7 bits (86), Expect = 0.017
Identities = 35/119 (29%), Positives = 59/119 (49%), Gaps = 19/119 (15%)
Query: 165 GPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGDVYRPAAIDQLTILGKQVD--- 221
G +I + G+ G+GKTT+ +L Y QGK R A IDQ+ + K ++
Sbjct: 228 GTRIIGVVGMPGIGKTTLLKEL--YKTWQGK--------FSRHALIDQIRVKSKHLELDR 277
Query: 222 VPVYTAGTDVKPS----DIAKQGFEEAKKKKIDVVIVDTAGRLQID--KAMMDELKDVK 274
+P G K + D K + + ++K+ VV+ D + R QID + ++D +K+ K
Sbjct: 278 LPQMLLGELSKLNHPHVDNLKDPYSQLHERKVLVVLDDVSKREQIDALREILDWIKEGK 336
>At5g67340 unknown proteins
Length = 707
Score = 33.9 bits (76), Expect = 0.25
Identities = 26/88 (29%), Positives = 41/88 (46%), Gaps = 14/88 (15%)
Query: 233 PSDIAKQGFEEAKKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPT-EVLLVVDAMTG 291
P+D+ FEE + +V R +I + LKD K+ + PT EVL+ + TG
Sbjct: 126 PADLISPSFEEC------IELVKLVARDEISYTIDQALKDQKKGVGPTSEVLVKIAESTG 179
Query: 292 QEAAALVTTFNLEIGITGAILTKLDGDS 319
+ N EI + G +LT + D+
Sbjct: 180 LRS-------NQEILVEGVVLTNMKEDA 200
>At1g04280 unknown protein
Length = 480
Score = 33.5 bits (75), Expect = 0.33
Identities = 24/110 (21%), Positives = 51/110 (45%), Gaps = 9/110 (8%)
Query: 106 EADVSLPVVRRFVQSVTDQAVGVGVTRGVKPDQQLVKIVHDELVQLMGGE-------VSE 158
E+D +P ++ FV + T + VT+ +K + + +V + V G +S
Sbjct: 110 ESDQKVPKLKDFVMAATRKQRFERVTKDLKVKRVISTLVEEMRVIGSGSSEPHCTEVMSP 169
Query: 159 LTFAKTGPTVILLAGLQGVGKTTVCAK--LANYFKKQGKSCMLVAGDVYR 206
+ K P ++L+ G G GK+TV L +++ + +++ D ++
Sbjct: 170 VAHNKRSPVLLLMGGGMGAGKSTVLKDIFLESFWSEAQADAVVIEADAFK 219
>At1g71820 unknown protein (At1g71820)
Length = 752
Score = 33.1 bits (74), Expect = 0.43
Identities = 22/55 (40%), Positives = 30/55 (54%), Gaps = 7/55 (12%)
Query: 202 GDVYRPAAIDQLTILGKQVDVPVYTAGTDVKPSDIAKQ------GFEEAKKKKID 250
G +Y PAA+D ILG+QV + V TDV IA F+ A+KK++D
Sbjct: 429 GKLYTPAAVDLFRILGEQVQI-VRDNSTDVMLYRIALAIIQVMIDFQAAEKKRVD 482
>At1g36550 hypothetical protein
Length = 353
Score = 32.3 bits (72), Expect = 0.73
Identities = 24/91 (26%), Positives = 44/91 (47%), Gaps = 6/91 (6%)
Query: 431 EAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDA---ILSN--- 484
E + +EI E++ KA+ E++ K + + S VA+ + + E++T + ILS+
Sbjct: 189 ETQYYVEIEEMLHKAILFEQQVKRKSSSRSSYGSGTVAKPTYQREERTSSYKPILSSRAE 248
Query: 485 FEKYTTIMSGKPTCSSTISNACSDEEPNGCN 515
+ Y + K + S C PNGC+
Sbjct: 249 SKTYAAVQDHKGKAEISTSRVCDVVNPNGCD 279
>At3g14990 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate
biosynthesis protein, putative
Length = 392
Score = 32.0 bits (71), Expect = 0.96
Identities = 25/107 (23%), Positives = 47/107 (43%), Gaps = 13/107 (12%)
Query: 105 LEADVSLPVVRRFVQSVTDQAVGVGV-TRGVKPDQQLVKIVHDELVQLMGGEVSELTFAK 163
+EA + ++RR +V AVG + G + + + +++ DE+ + K
Sbjct: 225 IEAIALVDILRRAKANVVIAAVGNSLEVEGSRKAKLVAEVLLDEVAE------------K 272
Query: 164 TGPTVILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGDVYRPAAI 210
+ ++L GL G + C KL N +KQ ++ G PA +
Sbjct: 273 SFDLIVLPGGLNGAQRFASCEKLVNMLRKQAEANKPYGGICASPAYV 319
>At1g14740 unknown protein
Length = 733
Score = 32.0 bits (71), Expect = 0.96
Identities = 47/194 (24%), Positives = 78/194 (39%), Gaps = 37/194 (19%)
Query: 306 GITGAILTKLDGDSRGGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDV 365
G G++ ++ GG AL+ VSGKP F+P + R
Sbjct: 231 GTNGSVHSRFRPIGDGGVALANNPVSGKPSSSADYS--------FFPSELPAR------- 275
Query: 366 LSFVEKAQEVMQQEDAEDLQKKIMSAKFDFNDFLKQTRAVAQM--GSVSRVIGMIPGMAK 423
EV D+ +KK+ A + ND ++ R + + S+S V +I GMA
Sbjct: 276 -----PGNEVTISGDS---RKKV--ANLEDNDAVRSERVLYDIVSKSISSVALIIQGMAD 325
Query: 424 VTPAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQD--SGKTEQQTDAI 481
T +E RNL +PE++EK L +R ++++ S + Q D +
Sbjct: 326 ETLESAKEYLRNL--------IDSPEKKEKLVNLQNQIDKRSDLSKETLSKCVKDQLDIL 377
Query: 482 LSNFEKYTTIMSGK 495
++ +SGK
Sbjct: 378 VAVRTGLKYFLSGK 391
>At2g14750 putative adenosine phosphosulfate kinase
Length = 276
Score = 31.6 bits (70), Expect = 1.2
Identities = 15/39 (38%), Positives = 22/39 (55%)
Query: 168 VILLAGLQGVGKTTVCAKLANYFKKQGKSCMLVAGDVYR 206
VI + GL G GK+T+ L ++GK C ++ GD R
Sbjct: 103 VIWVTGLSGSGKSTLACALNQMLYQKGKLCYILDGDNVR 141
>At3g46220 unknown protein
Length = 804
Score = 31.2 bits (69), Expect = 1.6
Identities = 21/79 (26%), Positives = 37/79 (46%), Gaps = 8/79 (10%)
Query: 238 KQGFEEAKKKKIDVVIVDTAGRLQIDKAMMDELKDVKRVLNPTEVLLVVDAMTGQEAAAL 297
K G E + K D+V++D++ R + K + L + L +V+A+ G+
Sbjct: 607 KNGTEVEESKTQDLVLLDSSERTALAKNLNGSLSK--------KALALVEALEGKRVDTF 658
Query: 298 VTTFNLEIGITGAILTKLD 316
+ TF +G +L KLD
Sbjct: 659 MVTFRDLAEESGLVLKKLD 677
>At1g26190 unknown protein (At1g26190)
Length = 674
Score = 31.2 bits (69), Expect = 1.6
Identities = 23/77 (29%), Positives = 33/77 (41%), Gaps = 16/77 (20%)
Query: 165 GPTVILLAGLQGVGKTTVCAKLANY--------FKKQGKSCMLVAGDVYRPAAIDQLTIL 216
G ++ +AG G GKT K+ N+ S +V G+ P D T+L
Sbjct: 64 GIILVGVAGPSGAGKTVFTEKILNFLPSVAVISMDNYNDSSRIVDGNFDDPRLTDYDTLL 123
Query: 217 --------GKQVDVPVY 225
GKQV+VP+Y
Sbjct: 124 KNLEDLKEGKQVEVPIY 140
>At1g74160 unknown protein
Length = 1028
Score = 30.8 bits (68), Expect = 2.1
Identities = 31/137 (22%), Positives = 57/137 (40%), Gaps = 12/137 (8%)
Query: 363 GDVLSFVEKAQEVMQQEDAEDLQKKIMSAKFDFN-DFLKQTRAVAQ--MGSVSRVIGMIP 419
G L +++ E MQ + D +K+ S F D+ ++ A + M S +RV
Sbjct: 420 GKDLRALKQILESMQSKGFLDTEKQQQSTNFAVQRDYERENSATSNHAMSSRTRVQSSSS 479
Query: 420 GMAKVTPAQIREAERNLEIMEVIIKAMTPEE---------REKPELLAESPVRRKRVAQD 470
+P I + + +E + ++ P REKP+ S KRV +D
Sbjct: 480 NQVYQSPIVIMKPAKLVEKAGIPASSLIPIHSLTGIKKIRREKPDDKGTSASNSKRVTKD 539
Query: 471 SGKTEQQTDAILSNFEK 487
++ ++ S+F+K
Sbjct: 540 CSPGNRRAESCTSSFDK 556
>At5g42220 unknown protein
Length = 879
Score = 30.4 bits (67), Expect = 2.8
Identities = 29/120 (24%), Positives = 52/120 (43%), Gaps = 19/120 (15%)
Query: 373 QEVMQQEDAEDLQKKIMSAK------FDFNDFLKQTRAVAQMGSVSRVI---GMIPGMAK 423
Q++ QQ D ++++ + FDF+ ++Q M VSR G +P A
Sbjct: 749 QQIAQQVDGQEIENMMSGGAQGEGGGFDFSRMVQQ-----MMPLVSRAFSQGGPLPHPAT 803
Query: 424 VTPAQIREAERNLEIMEVIIKAMTPEEREKPELLAESPVRRKRVAQDSGKTEQQTDAILS 483
+ P + ++ N++ M +I E + PE + + V ++QD E D LS
Sbjct: 804 IQPDDRQPSQVNVQSMAQMI-----EHSDPPEDVFRAMVENAAISQDELVNELCCDEALS 858
>At5g22310 unknown protein
Length = 481
Score = 30.4 bits (67), Expect = 2.8
Identities = 24/80 (30%), Positives = 37/80 (46%), Gaps = 6/80 (7%)
Query: 321 GGAALSVKEVSGKPIKLVGRGERMEDLEPFYPDRMAGRILGMGDVLSFVEKAQEVMQQED 380
G KE K + + R +R +D+ D + G+GD +EK +E+M D
Sbjct: 271 GRELTEAKETERKMKEEMKREKRAKDVLEEVCDELTK---GIGDDKKEMEKEREMMHIAD 327
Query: 381 A---EDLQKKIMSAKFDFND 397
E +Q K+ AKF+F D
Sbjct: 328 VLREERVQMKLTEAKFEFED 347
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,668,330
Number of Sequences: 26719
Number of extensions: 476564
Number of successful extensions: 1773
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 20
Number of HSP's that attempted gapping in prelim test: 1737
Number of HSP's gapped (non-prelim): 41
length of query: 573
length of database: 11,318,596
effective HSP length: 105
effective length of query: 468
effective length of database: 8,513,101
effective search space: 3984131268
effective search space used: 3984131268
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140848.2


