

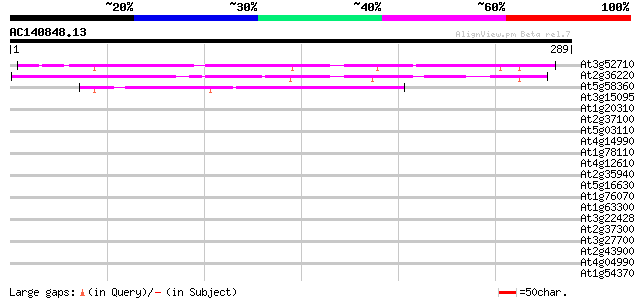
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140848.13 + phase: 0
(289 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g52710 unknown protein 169 1e-42
At2g36220 unknown protein 168 4e-42
At5g58360 similar to unknown protein (gb|AAB63094.1) 41 9e-04
At3g15095 unknown protein, 3' partial 40 0.001
At1g20310 unknown protein 40 0.001
At2g37100 unknown protein 39 0.003
At5g03110 putative protein 38 0.006
At4g14990 unknown protein 37 0.012
At1g78110 unknown protein 36 0.028
At4g12610 putative protein 35 0.061
At2g35940 putative homeodomain transcription factor 35 0.061
At5g16630 unknown protein 33 0.18
At1g76070 unknown protein 33 0.23
At1g63300 hypothetical protein 33 0.23
At3g22428 hypothetical protein 32 0.31
At2g37300 hypothetical protein 32 0.31
At3g27700 unknown protein 32 0.40
At2g43900 putative inositol polyphosphate 5'-phosphatase 32 0.40
At4g04990 32 0.52
At1g54370 Na+/H+ exchanger 5 (NHX5) 32 0.52
>At3g52710 unknown protein
Length = 289
Score = 169 bits (429), Expect = 1e-42
Identities = 125/302 (41%), Positives = 164/302 (53%), Gaps = 41/302 (13%)
Query: 5 ETLVSACAGGSTDRKIACETQADDLKTDPPPESSHHHP------DSPPESFWLSRDEENE 58
+ + SAC GGS DRKI+CET ADD + P +S P D PPES+ LS++ + E
Sbjct: 3 QVVASACTGGS-DRKISCETLADD--NEDSPHNSKIRPVSISAVDFPPESYSLSKEAQLE 59
Query: 59 WLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSF 118
WL+ N +ERKES KGNSS+ NPN+N NS+S S LKSK S+I LPKPQK F
Sbjct: 60 WLNDNAFFERKESQKGNSSAPISNPNTNPNSSSHRIS-----LKSKASIIRLPKPQKTCF 114
Query: 119 -DAKNLRNHKPSNIKLFPKRSASVGKS---FVEPSSPKVSCMGKVRSKRGKTSTVTAAAK 174
+AK RN + + + PKR S KS EP SPKVSC+G+VRSKR + ++
Sbjct: 115 NEAKKRRNCRIARTLMIPKRIGSRLKSDPTLSEPCSPKVSCIGRVRSKRDR-------SR 167
Query: 175 AAVKEKTARTERKQ------SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTN 228
++K+ RT + K FF SFRAIFR+GG K D + + +
Sbjct: 168 RMQRQKSGRTNSFKDKPVPVKKPGFFASFRAIFRTGGGCK-DLSASGAHAPRRDVVVSPP 226
Query: 229 SVSDSKARDSTASLNDVSFVESV-------TRNSVSEGEP--PGLGVMMRFTSGRRSESW 279
VS ++ D L +S +R S+ GEP PGLG M RFTSGRR +
Sbjct: 227 RVSVRRSTDIRGRLPPGDVGKSSPQRNSTGSRRSIDGGEPVLPGLGGMTRFTSGRRPDLL 286
Query: 280 VD 281
V+
Sbjct: 287 VE 288
>At2g36220 unknown protein
Length = 263
Score = 168 bits (425), Expect = 4e-42
Identities = 119/285 (41%), Positives = 157/285 (54%), Gaps = 41/285 (14%)
Query: 2 LQVETLVSA-CAGGSTDRKIACETQADDLKTDPPPESSHHHP-DSPPESFWLSRDEENEW 59
+ +E VS+ C GGS RKI CET ADD P +S P D PPES++LS D + EW
Sbjct: 4 VDIEAFVSSVCIGGSDHRKIVCETLADDSTIPPYYNNSAVSPSDFPPESYFLSNDAQLEW 63
Query: 60 LDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFD 119
L N ++RK+S KGNS N NPNSN +SQRF LKSK S+IGLPKPQK F+
Sbjct: 64 LSDNAFFDRKDSQKGNSGILNSNPNSN------PSSQRFL-LKSKASIIGLPKPQKTCFN 116
Query: 120 AKNLRNHKPSNIKLFPKRSASVGK---SFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAA 176
R H N ++ KR S K S +EPSSPKVSC+G+VRS+R + ++
Sbjct: 117 EAKQRRHAGKN-RVILKRVGSRIKTDISLLEPSSPKVSCIGRVRSRRER-------SRRM 168
Query: 177 VKEKTARTE--RKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSK 234
++K++R E + K F SFRAIFR G K D SA ++ + T+NT+ +
Sbjct: 169 HRQKSSRVEPVNRVKKPGFMASFRAIFRIKGGCK-----DVSARETHTSTRNTHDIRSRL 223
Query: 235 ARDSTASLNDVSFVESVTRNSVSEGEP--PGLGVMMRFTSGRRSE 277
E+ ++ GEP PGLG M RF SGRR++
Sbjct: 224 P------------AEADEKSVFDGGEPVVPGLGGMTRFASGRRAD 256
>At5g58360 similar to unknown protein (gb|AAB63094.1)
Length = 296
Score = 40.8 bits (94), Expect = 9e-04
Identities = 44/177 (24%), Positives = 75/177 (41%), Gaps = 16/177 (9%)
Query: 37 SSHHHP---DSPPESFWLSRDEENEWLDRNFIYERKESTK-GNSSSTNLNPNSNSNSNST 92
S+ HHP +SPP+S R + R +Y+ K SSS N S+S++N+
Sbjct: 62 STAHHPQASNSPPKSSSFKRK-----IKRKTVYKPSSRLKLSTSSSLNHRSKSSSSANAI 116
Query: 93 SNSQRFANLK-----SKTSMIGLPKPQKPSFDAKN-LRNHKPSNIKLFPKRSASVGKSFV 146
S+S + L S + + P+P S D K+ L K ++ P S ++
Sbjct: 117 SDSAVGSFLDRVSSPSDQNFVHDPEPHS-SIDIKDELSVRKLDDVPEDPSVSPNLSPETA 175
Query: 147 EPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRS 203
+ ++ K++ + +S + K K+K RT + K KSF + S
Sbjct: 176 KEPPFEMMTQQKLKKPKAHSSGIKIPTKIVRKKKKERTSQVSKKKGVVKSFAIVLSS 232
>At3g15095 unknown protein, 3' partial
Length = 417
Score = 40.4 bits (93), Expect = 0.001
Identities = 37/121 (30%), Positives = 57/121 (46%), Gaps = 16/121 (13%)
Query: 77 SSTNLNPNSNSNSNSTSNSQRFANLKS-KTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFP 135
SS +++ + S +TS S+R S K + G+ P F A R S +
Sbjct: 44 SSKSIHSPARSACLTTSLSRRLRTSGSLKNASAGVLN--SPMFGANGGRKRSGSGYE--- 98
Query: 136 KRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFK 195
S + + +EPSSPKV+C+G+VR K K K+ AR+ RK ++SF +
Sbjct: 99 -NSNNNNNNNIEPSSPKVTCIGQVRVK---------TRKHVKKKMRARSRRKGGENSFRR 148
Query: 196 S 196
S
Sbjct: 149 S 149
>At1g20310 unknown protein
Length = 240
Score = 40.4 bits (93), Expect = 0.001
Identities = 49/204 (24%), Positives = 82/204 (40%), Gaps = 43/204 (21%)
Query: 87 SNSNSTSNSQRFANLKSKTSMIG-LPKPQKPSFDAKNLRNHKPSNIKLF----------- 134
SNSN S +F+ + + IG P P D R +N +F
Sbjct: 5 SNSNK---STKFSKMLQRAMSIGHSAAPFSPRRDFHQHRTTSTANRGIFFSSPLVPTAAR 61
Query: 135 PKRSASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTE--RKQSKHS 192
+R+ F EP+SPKVSC+G+V+ R K A +K ++ + K+ +
Sbjct: 62 VRRNTKYEAVFAEPTSPKVSCIGQVKLARPKCPEKKNKAPKNLKTASSLSSCVIKEEDNG 121
Query: 193 FFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVT 252
F + IF R+ R ++S+A +++ A + ++ V++VT
Sbjct: 122 SFSKLKRIFSM--RSYPSRKSNSTAFAAAA-----------------AREHPIAEVDAVT 162
Query: 253 RNSVSEGEPPGLGVMMRFTSGRRS 276
P LG M +F S R +
Sbjct: 163 -------AAPSLGAMKKFASSREA 179
>At2g37100 unknown protein
Length = 297
Score = 38.9 bits (89), Expect = 0.003
Identities = 20/53 (37%), Positives = 31/53 (57%), Gaps = 3/53 (5%)
Query: 134 FPKRSASVGKSFVEPSSPKVSCMGKV---RSKRGKTSTVTAAAKAAVKEKTAR 183
F +++AS EP+SPKV+CMG+V RSK+ K T + A + + +R
Sbjct: 43 FRRKNASAAAETQEPTSPKVTCMGQVRINRSKKPKPETARVSGGATERRRQSR 95
>At5g03110 putative protein
Length = 283
Score = 38.1 bits (87), Expect = 0.006
Identities = 18/33 (54%), Positives = 22/33 (66%)
Query: 133 LFPKRSASVGKSFVEPSSPKVSCMGKVRSKRGK 165
LF +R+ S EPSSPKV+CMG+VR R K
Sbjct: 50 LFVRRNKSAAAVAQEPSSPKVTCMGQVRVNRSK 82
>At4g14990 unknown protein
Length = 787
Score = 37.0 bits (84), Expect = 0.012
Identities = 30/115 (26%), Positives = 54/115 (46%), Gaps = 4/115 (3%)
Query: 43 DSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLK 102
+S + W +E WLD++ + E+ + SS +PNSNS ++S Q+ L+
Sbjct: 110 ESSTATDWTQDNEFTSWLDQHTVEEQVQEASW-SSQPQSSPNSNSLYRTSSYPQQQTQLQ 168
Query: 103 SKTS-MIGLPKPQKPSFDAKNLRNHK--PSNIKLFPKRSASVGKSFVEPSSPKVS 154
+S I +P+ SF + R+ + PS+I P +F P++ +S
Sbjct: 169 HYSSEPIIVPESTFTSFPSPGKRSQQSSPSHIHRAPSLPGGSQSNFSAPNASPLS 223
>At1g78110 unknown protein
Length = 342
Score = 35.8 bits (81), Expect = 0.028
Identities = 32/134 (23%), Positives = 56/134 (40%), Gaps = 22/134 (16%)
Query: 82 NPNSNSNSNSTSNSQRFANLKSKTSMIGLPKP----QKPSFDAKNLRNHKPSNIK----- 132
N + +S+S ++ S+T + PKP +PS + N R H +
Sbjct: 5 NNGNRGSSSSGYSADLLVCFPSRTHLALTPKPICSPSRPSDSSTNRRPHHRRQLSKLSGG 64
Query: 133 ---------LFPKRSASV---GKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEK 180
L+ K+++S G EP+SPKV+C G+++ + K +V E+
Sbjct: 65 GGGGHGSPVLWAKQASSKNMGGDEIAEPTSPKVTCAGQIKVRPSKCGG-RGKNWQSVMEE 123
Query: 181 TARTERKQSKHSFF 194
R +S+ FF
Sbjct: 124 IERIHDNRSQSKFF 137
>At4g12610 putative protein
Length = 649
Score = 34.7 bits (78), Expect = 0.061
Identities = 37/151 (24%), Positives = 55/151 (35%), Gaps = 22/151 (14%)
Query: 44 SPPESFWLSRDEENEWLDRNFIYERKESTK-------------------GNSSSTNLNPN 84
+PPE DEENE + KE K + TN
Sbjct: 317 APPEIKQDEDDEENEEEEGGLSKSGKELKKLLGKANGLDESDEDDDDDSDDEEETNYGTV 376
Query: 85 SNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGKS 144
+NS + + N +K + G P+ P+ +K R + K K S+SV K
Sbjct: 377 TNSKQKEAAKEEPVDNAPAKPAPSGPPRGTPPAKPSKGKRKLNDGDSK---KPSSSVQKK 433
Query: 145 FVEPSSPKVSCMGKVRSKRGKTSTVTAAAKA 175
+ PK S + + K++T T A KA
Sbjct: 434 VKTENDPKSSLKEERANTVSKSNTPTKAVKA 464
>At2g35940 putative homeodomain transcription factor
Length = 680
Score = 34.7 bits (78), Expect = 0.061
Identities = 44/219 (20%), Positives = 88/219 (40%), Gaps = 38/219 (17%)
Query: 66 YERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQ-KPSFDAKNLR 124
Y ++++ N++++N + N+N+N+N+ +N+ F L S P+P F L
Sbjct: 34 YTQQDNDSNNNNNSNNSNNNNTNTNTNNNNSSFVFLDSHA-----PQPNASQQFVGIPLS 88
Query: 125 NHKPSNIKL---------FPKR--SASVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAA 173
H+ ++I +P R + G V+P+ + +C R+++G + T+
Sbjct: 89 GHEAASITAADNISVLHGYPPRVQYSLYGSHQVDPTHQQAAC-ETPRAQQGLSLTL---- 143
Query: 174 KAAVKEKTARTERKQSKHSFFKSFRAIFRSG---------GRNKHDRMNDSSALDSSSIT 224
+++ +++Q H + F SG G N + L SS
Sbjct: 144 -------SSQQQQQQQHHQQHQPIHVGFGSGHGEDIRVGSGSTGSGVTNGIANLVSSKYL 196
Query: 225 KNTNSVSDSKARDSTASLNDVSFVESVTRNSVSEGEPPG 263
K + D + +N S + S + S +P G
Sbjct: 197 KAAQELLDEVVNADSDDMNAKSQLFSSKKGSCGNDKPVG 235
>At5g16630 unknown protein
Length = 865
Score = 33.1 bits (74), Expect = 0.18
Identities = 31/153 (20%), Positives = 67/153 (43%), Gaps = 17/153 (11%)
Query: 80 NLNPNSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSA 139
+L P ++ N +S N + + +TS + +PK Q I +PK+S+
Sbjct: 256 SLKPGADRNESSGQNRAKMKHGIFRTSTLMVPKQQA---------------ISSYPKKSS 300
Query: 140 SVGKSFVEPSSPKVSCMGKVRSKRGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRA 199
S K+ P++ + S + + + V ++ +A + K+ T RK + A
Sbjct: 301 SHVKNKSPFEKPQLG--NPLGSDQVQDNAVNSSCEAGMSIKSDGTRRKGDVEFERQIAMA 358
Query: 200 IFRSGGRNKHDRMNDSSALDSSSITKNTNSVSD 232
+ + + ++N++ + + N++SVSD
Sbjct: 359 LSATADNQQSSQVNNTKKVREITKISNSSSVSD 391
>At1g76070 unknown protein
Length = 272
Score = 32.7 bits (73), Expect = 0.23
Identities = 25/88 (28%), Positives = 45/88 (50%), Gaps = 10/88 (11%)
Query: 147 EPSSPKVSCMGKVRSKRGKTST-VTAAAKAAVKEKTART-------ERKQSKHSFFKSFR 198
EP+SPKVSC+G+++ + K T A +++ K ++T E ++ + S KS
Sbjct: 82 EPTSPKVSCIGQIKLGKSKCPTGKKNKAPSSLIPKISKTSTSSLTKEDEKGRLSKIKSIF 141
Query: 199 AIFRSGGRNKHDRMNDS--SALDSSSIT 224
+ + GRN + + + SA D +T
Sbjct: 142 SFSPASGRNTSRKSHPTAVSAADEHPVT 169
>At1g63300 hypothetical protein
Length = 1029
Score = 32.7 bits (73), Expect = 0.23
Identities = 28/110 (25%), Positives = 48/110 (43%), Gaps = 7/110 (6%)
Query: 152 KVSCMGKVRSKRGKTSTVTAAAKAAV-----KEKTARTERKQSKHSFFKSFRAIFRSGGR 206
K + ++R + S T ++ +V E+ A+ R +KH S +++F R
Sbjct: 191 KAARFAELRRRASIESDSTMSSSGSVIEPNTPEEVAKPLRHPTKH--LHSAKSLFEEPSR 248
Query: 207 NKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFVESVTRNSV 256
+ SS SS +TNS +D ARD+ + +D VE + V
Sbjct: 249 ISESEWSGSSDHGISSTDDSTNSSNDIVARDTAINSSDEDEVEKLKNELV 298
>At3g22428 hypothetical protein
Length = 342
Score = 32.3 bits (72), Expect = 0.31
Identities = 29/87 (33%), Positives = 35/87 (39%), Gaps = 11/87 (12%)
Query: 72 TKGNSSSTNLNP--NSNSNSNSTSNSQRFANLKSKTSMIGL-----PKPQKPSFDAKNLR 124
T SSS N P NS +N SN+ +SK MIG+ KP KP+ K
Sbjct: 22 TAAASSSQNHKPRVNSTNNDGGISNNNNNNKKESKPMMIGVQKTADKKPLKPNPSPKLAP 81
Query: 125 NHKPSNIKLFPKRSASVGKSFVEPSSP 151
K SN P G S P +P
Sbjct: 82 IPKQSN----PNHQNQTGPSSSRPMAP 104
>At2g37300 hypothetical protein
Length = 128
Score = 32.3 bits (72), Expect = 0.31
Identities = 21/78 (26%), Positives = 36/78 (45%), Gaps = 1/78 (1%)
Query: 189 SKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKARDSTASLNDVSFV 248
S S ++ S + + + +DS A+D T +V+ ++ DS++ L DVS
Sbjct: 41 SSSSEVSCISSVCTSASKEEEEETHDSGAVDEGKTDHATENVASTEEADSSSKLKDVSSG 100
Query: 249 E-SVTRNSVSEGEPPGLG 265
E SV ++ S E G
Sbjct: 101 EASVVASTSSHKEESQFG 118
>At3g27700 unknown protein
Length = 908
Score = 32.0 bits (71), Expect = 0.40
Identities = 17/66 (25%), Positives = 30/66 (44%)
Query: 84 NSNSNSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGK 143
++N+N+NS S S NL SK + KP+ + K +P N + +++ K
Sbjct: 809 SNNNNNNSNSLSVSRDNLSSKNKCASVSNDPKPAVEVKTSSTEEPENTNVSGDNDSTLDK 868
Query: 144 SFVEPS 149
+ S
Sbjct: 869 QETKES 874
Score = 30.4 bits (67), Expect = 1.2
Identities = 26/107 (24%), Positives = 46/107 (42%), Gaps = 13/107 (12%)
Query: 28 DLKTDPPPESSHHHPD---SPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPN 84
+L P +S PD P E +S +E++ DRN + RKE T+ S + +
Sbjct: 2 ELSVSSPKQSVLSPPDCMSDPEEEHEISEEEDD---DRNHKHRRKEETRSQSLEQDSSDQ 58
Query: 85 SNS-----NSNSTSNSQRFANLKSKTSMIGLPKPQKPSFDAKNLRNH 126
+ S N N F+ + ++ G Q+ FD + +R++
Sbjct: 59 AFSRPYRKNYRHYENGNSFSEHEKRS--FGTGSGQRVQFDNQRMRSN 103
>At2g43900 putative inositol polyphosphate 5'-phosphatase
Length = 1305
Score = 32.0 bits (71), Expect = 0.40
Identities = 38/188 (20%), Positives = 69/188 (36%), Gaps = 2/188 (1%)
Query: 43 DSPPESFWLSRDEENEWLDRNFIYERKESTKGNSSSTNLNPNSNSNSNSTSNSQRFANLK 102
DS P + ++NE + ++ + + SS + +SNS S+ S+ +
Sbjct: 1102 DSNPSNSKSQSLKKNEGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSS 1161
Query: 103 SKTSMIGLPKPQKPSFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSK 162
K+ K K S N ++ K S+ K S S K G SK
Sbjct: 1162 KKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSNSKSSKKSDGDSCSKSQKKSD--GDTNSK 1219
Query: 163 RGKTSTVTAAAKAAVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSS 222
K +++K+ K + + K+ S ++ +S G + S S S
Sbjct: 1220 SQKKGDGDSSSKSHKKNDGDSSSKSHKKNDGDSSSKSHKKSDGDSSSKSHKKSEGDSSLS 1279
Query: 223 ITKNTNSV 230
+T+ T +
Sbjct: 1280 LTRRTMEI 1287
>At4g04990
Length = 303
Score = 31.6 bits (70), Expect = 0.52
Identities = 42/157 (26%), Positives = 60/157 (37%), Gaps = 6/157 (3%)
Query: 117 SFDAKNLRNHKPSNIKLFPKRSASVGKSFVEPSSPKVSCMGKVRSKR-GKTSTVTAAAKA 175
SFD N + H+ N FP RS S G V SSP+V K G T K
Sbjct: 101 SFDQNNNKVHEKEN---FPVRSESGGSYGV--SSPEVRFFPTAPEKPVGLRRPPTVPVKT 155
Query: 176 AVKEKTARTERKQSKHSFFKSFRAIFRSGGRNKHDRMNDSSALDSSSITKNTNSVSDSKA 235
++ T+ E + + + + + D + S+ + S S S+A
Sbjct: 156 FPQDNTSGDESETMEEMWERVKAEKQPKKPNSLQDHVISRGDTKMSTSSWPLPSRSPSRA 215
Query: 236 RDSTASLNDVSFVESVTRNSVSEGEPPGLGVMMRFTS 272
R T SL+ +S S R S PG +M R S
Sbjct: 216 RRPTPSLSSLSPSSSRARRPPSSPARPGKKLMERIPS 252
>At1g54370 Na+/H+ exchanger 5 (NHX5)
Length = 517
Score = 31.6 bits (70), Expect = 0.52
Identities = 26/75 (34%), Positives = 32/75 (42%), Gaps = 3/75 (4%)
Query: 13 GGSTDRKI-ACETQADDLKTDPPP--ESSHHHPDSPPESFWLSRDEENEWLDRNFIYERK 69
GGST + + A E DDL E S H PP S S DE+ F + K
Sbjct: 425 GGSTGKMLEALEVVGDDLDDSMSEGFEESDHQYVPPPFSIGASSDEDTSSSGSRFKMKLK 484
Query: 70 ESTKGNSSSTNLNPN 84
E K +S T L+ N
Sbjct: 485 EFHKTTTSFTALDKN 499
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.307 0.121 0.337
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,871,769
Number of Sequences: 26719
Number of extensions: 302277
Number of successful extensions: 1683
Number of sequences better than 10.0: 119
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 87
Number of HSP's that attempted gapping in prelim test: 1549
Number of HSP's gapped (non-prelim): 165
length of query: 289
length of database: 11,318,596
effective HSP length: 98
effective length of query: 191
effective length of database: 8,700,134
effective search space: 1661725594
effective search space used: 1661725594
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140848.13


