

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140774.7 + phase: 0 /pseudo
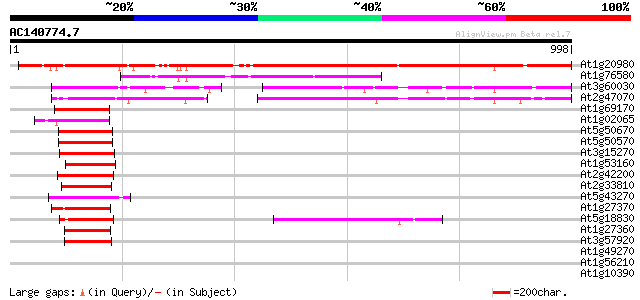
(998 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20980 putative SPL1-related protein 908 0.0
At1g76580 unknown protein 357 2e-98
At3g60030 squamosa promoter binding protein-like 12 289 6e-78
At2g47070 squamosa promoter binding protein-like 1 287 2e-77
At1g69170 squamosa promoter binding protein-like 6 (spl6) 129 1e-29
At1g02065 squamosa promoter binding protein-like 8 (SPL8) 126 7e-29
At5g50670 putative protein 125 1e-28
At5g50570 putative protein 125 1e-28
At3g15270 squamosa promoter binding protein-like 5 122 8e-28
At1g53160 squamosa promoter binding protein-like 4 (spl4) 122 1e-27
At2g42200 squamosa promoter binding protein-like 9 121 2e-27
At2g33810 squamosa-promoter binding protein-like 3 120 3e-27
At5g43270 squamosa promoter binding protein-like 2 (emb|CAB56576.1) 119 1e-26
At1g27370 squamosa-promoter binding protein-like 10 117 2e-26
At5g18830 squamosa promoter binding protein-like 7 115 1e-25
At1g27360 squamosa-promoter binding protein-like 11 113 5e-25
At3g57920 squamosa promoter-binding protein homolog 110 3e-24
At1g49270 hypothetical protein 39 0.015
At1g56210 unknown protein 37 0.056
At1g10390 unknown protein 35 0.16
>At1g20980 putative SPL1-related protein
Length = 1035
Score = 908 bits (2346), Expect = 0.0
Identities = 532/1060 (50%), Positives = 682/1060 (64%), Gaps = 110/1060 (10%)
Query: 16 LSSHQFYDSSNTKKRDL---LSSYDVVHIPN--DNWNPKEWNWDSIRFMTAKSTTVEPQQ 70
+++ F S +KRDL +S+ V P D WN K W+WDS RF AK VE Q+
Sbjct: 8 VAAPMFIHQSLGRKRDLYYPMSNRLVQSQPQRRDEWNSKMWDWDSRRF-EAKPVDVEVQE 66
Query: 71 V----------EESLNLNLGS-----------TGLVRPNKRIRSGSPTSASYPMCQVDNC 109
E L+LNLGS T VRPNK++RSGSP +YPMCQVDNC
Sbjct: 67 FDLTLRNRSGEERGLDLNLGSGLTAVEETTTTTQNVRPNKKVRSGSP-GGNYPMCQVDNC 125
Query: 110 KEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLA 169
EDLS AKDYHRRHKVCE HSKA+KAL+G QMQRFCQQCSRFH L EFDEGKRSCRRRLA
Sbjct: 126 TEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHLLSEFDEGKRSCRRRLA 185
Query: 170 GHNRRRRKT-QPDEVAVGGSPPLNQ------VAANLEIFNLLTAIADGSQGKFEER---- 218
GHNRRRRKT QP+EVA G P N AN+++ LLTA+A +QGK +
Sbjct: 186 GHNRRRRKTTQPEEVASGVVVPGNHDTTNNTANANMDLMALLTALAC-AQGKNAVKPPVG 244
Query: 219 RSQVPDKEQLVQILNRI---PLPADLTAKLLDVGNNLNAKNDNVQMETSPSYHHRDDQLN 275
VPD+EQL+QILN+I PLP DL +KL ++G+ D+ P+ + ++D +N
Sbjct: 245 SPAVPDREQLLQILNKINALPLPMDLVSKLNNIGSLARKNMDH------PTVNPQND-MN 297
Query: 276 NAPPAPLTKDFLAVLSTT--PSTP-----ARNGG---------------NGSTSSADHMR 313
A P+ T D LAVLSTT S+P GG NG T++ +
Sbjct: 298 GASPS--TMDLLAVLSTTLGSSSPDALAILSQGGFGNKDSEKTKLSSYENGVTTNLEKRT 355
Query: 314 --------ERSSGSSQSPNDDSDCQ-EDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSE 364
ERSS S+QSP+ DSD + +D R L LQLF SSPE++S + SSRKY+SS
Sbjct: 356 FGFSSVGGERSSSSNQSPSQDSDSRGQDTRSSLSLQLFTSSPEDESRPTVASSRKYYSSA 415
Query: 365 SSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCISTGFGGNANKETSQSHSCTTIPLDL 424
SSNPV++R+PSSSP + E+ F LQ S +K +S C +PL+L
Sbjct: 416 SSNPVEDRSPSSSPVMQEL-FPLQASPETMRSK---------NHKNSSPRTGC--LPLEL 463
Query: 425 FKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHF 484
F S N + + ++GYASSGSDYSPPSLNSD QDRTG+I+FKL DK PS
Sbjct: 464 FGAS---NRGAADPNFKGFGQQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQL 520
Query: 485 PGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSD 544
PGTLR++IYNWLS PS++ESYIRPGCVVLS+Y +MS AAW QLE+ LQR+ L+ NS
Sbjct: 521 PGTLRSEIYNWLSNIPSEMESYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNSP 580
Query: 545 SDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWRSPELISVSPLAIVGGQETSISLKGR 604
SDFWRN RF+V +G QLASHK+G++R K W TW SPELISVSP+A+V G+ETS+ ++GR
Sbjct: 581 SDFWRNARFIVNTGRQLASHKNGKVRCSKSWRTWNSPELISVSPVAVVAGEETSLVVRGR 640
Query: 605 NLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEIKLSGFEVQNTSPSVLGRCFIEVENG 664
+L+ G I CT Y + EV + ++DE+ ++ F+VQN P LGRCFIEVENG
Sbjct: 641 SLTNDGISIRCTHMGSYMAMEVTRAVCRQTIFDELNVNSFKVQNVHPGFLGRCFIEVENG 700
Query: 665 FKGNSFPVIIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNEL 724
F+G+SFP+IIANASICKEL L EF K D E+ + P SR+E L FLNEL
Sbjct: 701 FRGDSFPLIIANASICKELNRLGEEF--HPKSQDMTEEQAQSSNRGPTSREEVLCFLNEL 758
Query: 725 GWLFQRERFSNVHEVPDYSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGS 784
GWLFQ+ + S + E D+SL RFKF+L SVER+ C L++TLLDMLV+++ + L+ +
Sbjct: 759 GWLFQKNQTSELREQSDFSLARFKFLLVCSVERDYCALIRTLLDMLVERNLVNDELNREA 818
Query: 785 VEMLKAIQLLNRAVKRKCTSMVDLLINYSITSKN-DTSKKYVFPPNLEGPGGITPLHLAA 843
++ML IQLLNRAVKRK T MV+LLI+Y + +S+K+VF PN+ GPGGITPLHLAA
Sbjct: 819 LDMLAEIQLLNRAVKRKSTKMVELLIHYLVNPLTLSSSRKFVFLPNITGPGGITPLHLAA 878
Query: 844 STTDSEGVIDSLTNDPQE----CWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQR 899
T+ S+ +ID LTNDPQE W TL D GQTP++YA +RNNH+YN LVARK +D++
Sbjct: 879 CTSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAAIRNNHNYNSLVARKLADKRN 938
Query: 900 SEVSVRIDNEIEHPSLGIELMQKRIN-QVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGP 958
+VS+ I++E+ + + KR++ ++ + SC+ CA ++ +RR SGS+ P
Sbjct: 939 KQVSLNIEHEVVDQT----GLSKRLSLEMNKSSSSCASCATVALKYQRRVSGSQRLFPTP 994
Query: 959 FIHSMLAVAAVCVCVCVLFRGTPYVGSVSPFRWENLNYGT 998
IHSMLAVA VCVCVCV P V S F W L+YG+
Sbjct: 995 IIHSMLAVATVCVCVCVFMHAFPIVRQGSHFSWGGLDYGS 1034
>At1g76580 unknown protein
Length = 488
Score = 357 bits (915), Expect = 2e-98
Identities = 221/501 (44%), Positives = 296/501 (58%), Gaps = 59/501 (11%)
Query: 198 LEIFNLLTAI--ADGSQGKFEERRSQVPDKEQLVQILNRI---PLPADLTAKLLDVGNNL 252
+++ LLTA+ A G VP +EQL+QILN+I PLP +LT+KL ++G L
Sbjct: 1 MDVMALLTALVCAQGRNEATTNGSPGVPQREQLLQILNKIKALPLPMNLTSKLNNIGI-L 59
Query: 253 NAKNDNVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPA-------RNGGNGS 305
KN PS + + +N A +P T D LA LS + + A GG G+
Sbjct: 60 ARKNPE-----QPSPMNPQNSMNGAS-SPSTMDLLAALSASLGSSAPEAIAFLSQGGFGN 113
Query: 306 TSSADHMR-------------------------ERSSGSSQSPNDDSDCQ-EDVRVKLPL 339
S D + ER+S ++ SP+ SD + +D R L L
Sbjct: 114 KESNDRTKLTSSDHSATTSLEKKTLEFPSFGGGERTSSTNHSPSQYSDSRGQDTRSSLSL 173
Query: 340 QLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFNSNCI 399
QLF SSPE +S K+ SS KY+SS SSNPV++R+PSSSP + E+ F +
Sbjct: 174 QLFTSSPEEESRPKVASSTKYYSSASSNPVEDRSPSSSPVMQEL----------FPLHTS 223
Query: 400 STGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSPP 459
N K+TS S + +PL+LF SN + + + ++GYASSGSDYSPP
Sbjct: 224 PETRRYNNYKDTSTSPRTSCLPLELF--GASNRGATANPNYNVLRHQSGYASSGSDYSPP 281
Query: 460 SLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYAS 519
SLNS+ Q+RTG+I FKLF+K PS P TLRT+I+ WLS+ PSD+ES+IRPGCV+LS+Y +
Sbjct: 282 SLNSNAQERTGKISFKLFEKDPSQLPNTLRTEIFRWLSSFPSDMESFIRPGCVILSVYVA 341
Query: 520 MSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWR 579
MS++AW QLEEN LQRV SL+ DS+FW N RFLV +G QLASHK GRIR+ K W T
Sbjct: 342 MSASAWEQLEENLLQRVRSLV--QDSEFWSNSRFLVNAGRQLASHKHGRIRLSKSWRTLN 399
Query: 580 SPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVIGSGDPGMVYDEI 639
PELI+VSPLA+V G+ET++ ++GRNL+ G ++ C Y S EV G DE+
Sbjct: 400 LPELITVSPLAVVAGEETALIVRGRNLTNDGMRLRCAHMGNYASMEVTGREHRLTKVDEL 459
Query: 640 KLSGFEVQNTSPSVLGRCFIE 660
+S F+VQ+ S LGRCFIE
Sbjct: 460 NVSSFQVQSASSVSLGRCFIE 480
>At3g60030 squamosa promoter binding protein-like 12
Length = 927
Score = 289 bits (739), Expect = 6e-78
Identities = 199/573 (34%), Positives = 296/573 (50%), Gaps = 53/573 (9%)
Query: 450 ASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRP 509
+ S SD SP S + D Q RT RI+FKLF K P+ FP LR QI NWL+ P+D+ESYIRP
Sbjct: 383 SDSASDQSPSSSSGDAQSRTDRIVFKLFGKEPNDFPVALRGQILNWLAHTPTDMESYIRP 442
Query: 510 GCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRI 569
GC+VL+IY A+W +L + + L+ SD W +G + +QLA +G++
Sbjct: 443 GCIVLTIYLRQDEASWEELCCDLSFSLRRLLDLSDDPLWTDGWLYLRVQNQLAFAFNGQV 502
Query: 570 RM--CKPWGTWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGADCYTSSEVI 627
+ P + ++I+V PLA+ ++ ++KG NL PGT++ CT + E
Sbjct: 503 VLDTSLPLRSHDYSQIITVRPLAVT--KKAQFTVKGINLRRPGTRLLCTVEGTHLVQEAT 560
Query: 628 GSG----DPGMVYDEIKLSGFEVQNTSPSVLGRCFIEVEN--GFKGNSFPVIIA-NASIC 680
G D +EI F + P GR F+E+E+ G + FP I++ + IC
Sbjct: 561 QGGMEERDDLKENNEIDFVNFSCE--MPIASGRGFMEIEDQGGLSSSFFPFIVSEDEDIC 618
Query: 681 KELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHEVP 740
E+R LES + F S +A+ F++E+GWL R +
Sbjct: 619 SEIRRLESTLE----------------FTGTDSAMQAMDFIHEIGWLLHRSELKSRLAAS 662
Query: 741 D------YSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAIQLL 794
D +SL RFKF++ FS++R C ++K LL++L FE + L + LL
Sbjct: 663 DHNPEDLFSLIRFKFLIEFSMDREWCCVMKKLLNIL----FEEGTVDPSPDAALSELCLL 718
Query: 795 NRAVKRKCTSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGVIDS 854
+RAV++ MV++L+ +S KN T +F P+ GPGG+TPLH+AA SE V+D+
Sbjct: 719 HRAVRKNSKPMVEMLLRFSPKKKNQTLAG-LFRPDAAGPGGLTPLHIAAGKDGSEDVLDA 777
Query: 855 LTNDPQ----ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDR--QRSEVSVRIDN 908
LT DP + W+ D G TP YA +R + SY LV RK S + + V V I
Sbjct: 778 LTEDPGMTGIQAWKNSRDNTGFTPEDYARLRGHFSYIHLVQRKLSRKPIAKEHVVVNIPE 837
Query: 909 --EIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFIHSMLAV 966
IEH M ++ ++ ++C + + + + +S + P + SM+A+
Sbjct: 838 SFNIEHKQEKRSPMDSSSLEITQI----NQCKLCDHKRVFVTTHHKSVAYRPAMLSMVAI 893
Query: 967 AAVCVCVCVLFRGTPYVGSV-SPFRWENLNYGT 998
AAVCVCV +LF+ P V V PFRWE L YGT
Sbjct: 894 AAVCVCVALLFKSCPEVLYVFQPFRWELLEYGT 926
Score = 149 bits (376), Expect = 8e-36
Identities = 114/321 (35%), Positives = 151/321 (46%), Gaps = 41/321 (12%)
Query: 75 LNLNLGSTGLVRPN-KRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKAS 133
L LNLG + K+ + G + CQVDNC DLSK KDYHRRHKVCE HSKA+
Sbjct: 97 LTLNLGGNNIEGNGVKKTKLGGGIPSRAICCQVDNCGADLSKVKDYHRRHKVCEIHSKAT 156
Query: 134 KALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQ 193
AL+G MQRFCQQCSRFH L EFDEGKRSCRRRLAGHN+RRRK PD + G S +Q
Sbjct: 157 TALVGGIMQRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANPDTIGNGTSMSDDQ 216
Query: 194 VAANLEIFNLLTAIADGSQGKFEERRSQVPDKEQLVQILNRIPLPAD-----LTAKLLDV 248
+N + LL +++ + Q D++ L +L + A LL
Sbjct: 217 -TSNYMLITLLKILSN----IHSNQSDQTGDQDLLSHLLKSLVSQAGEHIGRNLVGLLQG 271
Query: 249 GNNLNAKND--NVQMETSPSYHHRDDQLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGST 306
G L A + N+ S R+D +++ +S TP
Sbjct: 272 GGGLQASQNIGNLSALLSLEQAPREDIKHHS------------VSETPWQEVYANSAQER 319
Query: 307 SSADHMRERSSGSSQSPND---DSDCQEDVRVKLPLQLFGSSPENDSPSKL-------PS 356
+ D ++ + ND DSD D+ P P N + S L S
Sbjct: 320 VAPDRSEKQVKVNDFDLNDIYIDSDDTTDIERSSP------PPTNPATSSLDYHQDSRQS 373
Query: 357 SRKYFSSESSNPVDERTPSSS 377
S S +S+ +++PSSS
Sbjct: 374 SPPQTSRRNSDSASDQSPSSS 394
Score = 29.6 bits (65), Expect = 8.9
Identities = 24/85 (28%), Positives = 36/85 (42%), Gaps = 12/85 (14%)
Query: 271 DDQLNNAPPA-PLTKDFLAVLSTTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDC 329
D + ++ PP P T + S+P + S S++D SSG +QS D
Sbjct: 348 DIERSSPPPTNPATSSLDYHQDSRQSSPPQTSRRNSDSASDQSPSSSSGDAQSRTD---- 403
Query: 330 QEDVRVKLPLQLFGSSPENDSPSKL 354
++ +LFG P ND P L
Sbjct: 404 ------RIVFKLFGKEP-NDFPVAL 421
>At2g47070 squamosa promoter binding protein-like 1
Length = 881
Score = 287 bits (734), Expect = 2e-77
Identities = 198/579 (34%), Positives = 296/579 (50%), Gaps = 54/579 (9%)
Query: 442 SVPFKAGYASSGSDYSPPSLNSDTQDRTGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPS 501
S P + + S SD SP S + D Q RTGRI+FKLF K P+ FP LR QI +WLS P+
Sbjct: 334 SPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPT 393
Query: 502 DLESYIRPGCVVLSIYASMSSAAWVQLEENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQL 561
D+ESYIRPGC+VL+IY + AW +L ++ + L+ SD W G V +QL
Sbjct: 394 DMESYIRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQL 453
Query: 562 ASHKDGRIRMCKPWG--TWRSPELISVSPLAIVGGQETSISLKGRNLSAPGTKIHCTGAD 619
A +G++ + + +ISV PLAI ++ ++KG NL GT++ C+
Sbjct: 454 AFVYNGQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRQRGTRLLCSVEG 513
Query: 620 CYTSSEVIGSGDPGMVYDEIKLSGFEVQNTS-----PSVLGRCFIEVEN-GFKGNSFP-V 672
Y E D+ K + V+ + P + GR F+E+E+ G + FP +
Sbjct: 514 KYLIQETTHDSTT-REDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFL 572
Query: 673 IIANASICKELRPLESEFDEEEKMCDAISEEHEHHFGRPKSRDEALHFLNELGWLFQRER 732
++ + +C E+R LE+ + F S +A+ F++E+GWL R +
Sbjct: 573 VVEDDDVCSEIRILETTLE----------------FTGTDSAKQAMDFIHEIGWLLHRSK 616
Query: 733 FSNVHEVPD-YSLDRFKFVLTFSVERNCCMLVKTLLDMLVDKHFEGEGLSTGSVEMLKAI 791
P + L RF++++ FS++R C +++ LL+M D GE S+ S L +
Sbjct: 617 LGESDPNPGVFPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAV-GE-FSSSSNATLSEL 674
Query: 792 QLLNRAVKRKCTSMVDLLINYSITSKNDTSKKYVFPPNLEGPGGITPLHLAASTTDSEGV 851
LL+RAV++ MV++L+ Y + ++ +F P+ GP G+TPLH+AA SE V
Sbjct: 675 CLLHRAVRKNSKPMVEMLLRYIPKQQRNS----LFRPDAAGPAGLTPLHIAAGKDGSEDV 730
Query: 852 IDSLTNDPQ----ECWETLADENGQTPHAYAMMRNNHSYNMLVARKCSDRQRSEVSVRI- 906
+D+LT DP E W+T D G TP YA +R + SY L+ RK + + +E V +
Sbjct: 731 LDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHVVVN 790
Query: 907 ------DNEIEHPSLGIELMQKRINQVKRVGDSCSKCAIAEVRAKRRFSGSRSWLHGPFI 960
D E + P G I Q+ C C V R RS + P +
Sbjct: 791 IPVSFSDREQKEPKSGPMASALEITQI-----PCKLCDHKLVYGTTR----RSVAYRPAM 841
Query: 961 HSMLAVAAVCVCVCVLFRGTPYVGSV-SPFRWENLNYGT 998
SM+A+AAVCVCV +LF+ P V V PFRWE L+YGT
Sbjct: 842 LSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGT 880
Score = 155 bits (392), Expect = 1e-37
Identities = 113/305 (37%), Positives = 153/305 (50%), Gaps = 38/305 (12%)
Query: 74 SLNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKAS 133
+LNLN S GL P K+ +SG+ +CQV+NC+ DLSK KDYHRRHKVCE HSKA+
Sbjct: 84 TLNLNGESDGLF-PAKKTKSGA-------VCQVENCEADLSKVKDYHRRHKVCEMHSKAT 135
Query: 134 KALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVGGSPPLNQ 193
A +G +QRFCQQCSRFH L EFDEGKRSCRRRLAGHN+RRRKT P+ A G P +
Sbjct: 136 SATVGGILQRFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGN--PSDD 193
Query: 194 VAANLEIFNLLTAIAD-----GSQGKFEERRSQVPDK--EQLVQILNRIPLPADLTAKLL 246
++N + LL +++ G Q + EQL + L + L + L
Sbjct: 194 HSSNYLLITLLKILSNMHNHTGDQDLMSHLLKSLVSHAGEQLGKNLVELLLQGGGSQGSL 253
Query: 247 DVGNNLNAKNDNVQMETSPSYHHRDDQL---NNAPPAPLTKDF--LAVLSTTPSTPARNG 301
++GN+ + E + R D N + DF + + T
Sbjct: 254 NIGNSALLGIEQAPQEELKQFSARQDGTATENRSEKQVKMNDFDLNDIYIDSDDTDVERS 313
Query: 302 ---GNGSTSSADH------------MRERSSGSSQSPNDDSDCQEDVRVKLPLQLFGSSP 346
N +TSS D+ R S S QSP+ S+ + ++ +LFG P
Sbjct: 314 PPPTNPATSSLDYPSWIHQSSPPQTSRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEP 373
Query: 347 ENDSP 351
N+ P
Sbjct: 374 -NEFP 377
>At1g69170 squamosa promoter binding protein-like 6 (spl6)
Length = 405
Score = 129 bits (323), Expect = 1e-29
Identities = 60/97 (61%), Positives = 73/97 (74%), Gaps = 1/97 (1%)
Query: 81 STGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQ 140
S G P+K+ R+ + S + P+CQV C +DLS +KDYH+RH+VCEAHSK S ++
Sbjct: 102 SRGFALPSKKSRASNLCSQN-PLCQVYGCSKDLSSSKDYHKRHRVCEAHSKTSVVIVNGL 160
Query: 141 MQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRK 177
QRFCQQCSRFH L EFD+GKRSCRRRLAGHN RRRK
Sbjct: 161 EQRFCQQCSRFHFLSEFDDGKRSCRRRLAGHNERRRK 197
>At1g02065 squamosa promoter binding protein-like 8 (SPL8)
Length = 333
Score = 126 bits (316), Expect = 7e-29
Identities = 70/146 (47%), Positives = 80/146 (53%), Gaps = 15/146 (10%)
Query: 44 DNWNPKEWNWDSIRFMTAKSTTVEPQQVEESLNLNLGS------------TGLVRPNKRI 91
D P + S F+ K+ E + LNLG + L R ++
Sbjct: 119 DQTGPGSGSGSSYNFLIPKT---EVDFTSNRIGLNLGGRTYFSAADDDFVSRLYRRSRPG 175
Query: 92 RSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRF 151
SG S S P CQ + C DLS AK YHRRHKVCE HSKAS + QRFCQQCSRF
Sbjct: 176 ESGMANSLSTPRCQAEGCNADLSHAKHYHRRHKVCEFHSKASTVVAAGLSQRFCQQCSRF 235
Query: 152 HPLVEFDEGKRSCRRRLAGHNRRRRK 177
H L EFD GKRSCR+RLA HNRRRRK
Sbjct: 236 HLLSEFDNGKRSCRKRLADHNRRRRK 261
>At5g50670 putative protein
Length = 359
Score = 125 bits (314), Expect = 1e-28
Identities = 54/96 (56%), Positives = 65/96 (67%)
Query: 88 NKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQ 147
+KR R + P+C VD C D S ++YH+RHKVC+ HSK + QRFCQQ
Sbjct: 85 SKRTRGNGVGTNQMPICLVDGCDSDFSNCREYHKRHKVCDVHSKTPVVTINGHKQRFCQQ 144
Query: 148 CSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEV 183
CSRFH L EFDEGKRSCR+RL GHNRRRRK QP+ +
Sbjct: 145 CSRFHALEEFDEGKRSCRKRLDGHNRRRRKPQPEHI 180
>At5g50570 putative protein
Length = 359
Score = 125 bits (314), Expect = 1e-28
Identities = 54/96 (56%), Positives = 65/96 (67%)
Query: 88 NKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQ 147
+KR R + P+C VD C D S ++YH+RHKVC+ HSK + QRFCQQ
Sbjct: 85 SKRTRGNGVGTNQMPICLVDGCDSDFSNCREYHKRHKVCDVHSKTPVVTINGHKQRFCQQ 144
Query: 148 CSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEV 183
CSRFH L EFDEGKRSCR+RL GHNRRRRK QP+ +
Sbjct: 145 CSRFHALEEFDEGKRSCRKRLDGHNRRRRKPQPEHI 180
>At3g15270 squamosa promoter binding protein-like 5
Length = 181
Score = 122 bits (307), Expect = 8e-28
Identities = 57/98 (58%), Positives = 67/98 (68%)
Query: 89 KRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQC 148
+R+R S +CQVD C +L++AK Y+RRH+VCE H+KAS A + QRFCQQC
Sbjct: 48 ERVRGPSTDRVPSRLCQVDRCTVNLTEAKQYYRRHRVCEVHAKASAATVAGVRQRFCQQC 107
Query: 149 SRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVG 186
SRFH L EFDE KRSCRRRLAGHN RRRK D G
Sbjct: 108 SRFHELPEFDEAKRSCRRRLAGHNERRRKISGDSFGEG 145
>At1g53160 squamosa promoter binding protein-like 4 (spl4)
Length = 174
Score = 122 bits (306), Expect = 1e-27
Identities = 57/89 (64%), Positives = 63/89 (70%)
Query: 100 SYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEFDE 159
S +CQVD C D+ +AK YHRRHKVCE H+KAS L QRFCQQCSRFH L EFDE
Sbjct: 50 SLRLCQVDRCTADMKEAKLYHRRHKVCEVHAKASSVFLSGLNQRFCQQCSRFHDLQEFDE 109
Query: 160 GKRSCRRRLAGHNRRRRKTQPDEVAVGGS 188
KRSCRRRLAGHN RRRK+ + GS
Sbjct: 110 AKRSCRRRLAGHNERRRKSSGESTYGEGS 138
>At2g42200 squamosa promoter binding protein-like 9
Length = 375
Score = 121 bits (304), Expect = 2e-27
Identities = 59/101 (58%), Positives = 68/101 (66%), Gaps = 1/101 (0%)
Query: 86 RPNKRIRSG-SPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRF 144
R N+R+R G S S P CQV+ C DL+ AK Y+ RH+VC HSK K + QRF
Sbjct: 55 RSNRRVRGGGSGQSGQIPRCQVEGCGMDLTNAKGYYSRHRVCGVHSKTPKVTVAGIEQRF 114
Query: 145 CQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAV 185
CQQCSRFH L EFD KRSCRRRLAGHN RRRK QP ++V
Sbjct: 115 CQQCSRFHQLPEFDLEKRSCRRRLAGHNERRRKPQPASLSV 155
>At2g33810 squamosa-promoter binding protein-like 3
Length = 131
Score = 120 bits (302), Expect = 3e-27
Identities = 56/90 (62%), Positives = 67/90 (74%), Gaps = 1/90 (1%)
Query: 92 RSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRF 151
+ G TS+S +CQV++C D+SKAK YH+RHKVC+ H+KA + QRFCQQCSRF
Sbjct: 43 QKGKATSSS-GVCQVESCTADMSKAKQYHKRHKVCQFHAKAPHVRISGLHQRFCQQCSRF 101
Query: 152 HPLVEFDEGKRSCRRRLAGHNRRRRKTQPD 181
H L EFDE KRSCRRRLAGHN RRRK+ D
Sbjct: 102 HALSEFDEAKRSCRRRLAGHNERRRKSTTD 131
>At5g43270 squamosa promoter binding protein-like 2 (emb|CAB56576.1)
Length = 419
Score = 119 bits (297), Expect = 1e-26
Identities = 69/149 (46%), Positives = 85/149 (56%), Gaps = 11/149 (7%)
Query: 70 QVEESLNLNLGST---GLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVC 126
+VE + L L T V P K+ +S P P CQV+ C DLS AKDYHR+H++C
Sbjct: 133 EVENAKGLGLPVTLASSSVSPVKKSKS-IPQRLQTPHCQVEGCNLDLSSAKDYHRKHRIC 191
Query: 127 EAHSKASKALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAVG 186
E HSK K ++ +RFCQQCSRFH L EFDE KRSCRRRL+ HN RRRK P
Sbjct: 192 ENHSKFPKVVVSGVERRFCQQCSRFHCLSEFDEKKRSCRRRLSDHNARRRKPNPGR-TYD 250
Query: 187 GSPPLNQVAANLEIFNLLTAIADGSQGKF 215
G P ++ V +N I S+ KF
Sbjct: 251 GKPQVDFV------WNRFALIHPRSEEKF 273
>At1g27370 squamosa-promoter binding protein-like 10
Length = 396
Score = 117 bits (294), Expect = 2e-26
Identities = 55/106 (51%), Positives = 72/106 (67%), Gaps = 2/106 (1%)
Query: 74 SLNLNLGSTGLVRPNKRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKAS 133
++++NL + +V K G S P CQ+D C+ DLS +KDYHR+H+VCE HSK
Sbjct: 148 AVSMNLLTPSVVARKKTKSCGQ--SMQVPRCQIDGCELDLSSSKDYHRKHRVCETHSKCP 205
Query: 134 KALLGNQMQRFCQQCSRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQ 179
K ++ +RFCQQCSRFH + EFDE KRSCR+RL+ HN RRRK Q
Sbjct: 206 KVVVSGLERRFCQQCSRFHAVSEFDEKKRSCRKRLSHHNARRRKPQ 251
>At5g18830 squamosa promoter binding protein-like 7
Length = 801
Score = 115 bits (288), Expect = 1e-25
Identities = 84/314 (26%), Positives = 143/314 (44%), Gaps = 15/314 (4%)
Query: 469 TGRIMFKLFDKHPSHFPGTLRTQIYNWLSTRPSDLESYIRPGCVVLSIYASMSSAAWVQL 528
TGRI FKL+D +P+ FP LR QI+ WL+ P +LE YIRPGC +L+++ +M W +L
Sbjct: 320 TGRISFKLYDWNPAEFPRRLRHQIFQWLANMPVELEGYIRPGCTILTVFIAMPEIMWAKL 379
Query: 529 EENFLQRVDSLIHNSDSDFWRNGRFLVYSGSQLASHKDGRIRMCKPWGTWRSPELISVSP 588
++ + +D I + G VY + + G + + SP+L V P
Sbjct: 380 SKDPVAYLDEFILKPGKMLFGRGSMTVYLNNMIFRLIKGGTTLKRVDVKLESPKLQFVYP 439
Query: 589 LAIVGGQETSISLKGRNLSAPGTKIHCTGADCY---TSSEVIGSGDPGMVYDEIKLSGFE 645
G+ + + G+NL P + + + Y S V G K
Sbjct: 440 TCFEAGKPIELVVCGQNLLQPKCRFLVSFSGKYLPHNYSVVPAPDQDGKRSCNNKFYKIN 499
Query: 646 VQNTSPSVLGRCFIEVEN-GFKGNSFPVIIANASICKELRPLESEFD-------EEEKMC 697
+ N+ PS+ G F+EVEN N P+II +A++C E++ +E +F+ +E C
Sbjct: 500 IVNSDPSLFGPAFVEVENESGLSNFIPLIIGDAAVCSEMKLIEQKFNATLFPEGQEVTAC 559
Query: 698 DAISEEHEHHFGRPKSRDEALHFLNELGWLFQRERFSNVHE-VPDYSLDRFKFVLTFSVE 756
+++ FG +S L L ++ W + + V + R+ VL + ++
Sbjct: 560 SSLT-CCCRDFGERQSTFSGL--LLDIAWSVKVPSAERTEQPVNRCQIKRYNRVLNYLIQ 616
Query: 757 RNCCMLVKTLLDML 770
N ++ +L L
Sbjct: 617 NNSASILGNVLHNL 630
Score = 99.4 bits (246), Expect = 9e-21
Identities = 46/97 (47%), Positives = 65/97 (66%), Gaps = 4/97 (4%)
Query: 89 KRIRSGSPTSASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQC 148
KR+R GS + CQV +C+ D+S+ K YH+RH+VC + AS +L + +R+CQQC
Sbjct: 127 KRVRGGSGVAR----CQVPDCEADISELKGYHKRHRVCLRCATASFVVLDGENKRYCQQC 182
Query: 149 SRFHPLVEFDEGKRSCRRRLAGHNRRRRKTQPDEVAV 185
+FH L +FDEGKRSCRR+L HN RR++ D+ V
Sbjct: 183 GKFHLLPDFDEGKRSCRRKLERHNNRRKRKPVDKGGV 219
>At1g27360 squamosa-promoter binding protein-like 11
Length = 393
Score = 113 bits (283), Expect = 5e-25
Identities = 51/82 (62%), Positives = 59/82 (71%)
Query: 98 SASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEF 157
S P CQ+D C+ DLS AK YHR+HKVCE HSK K + +RFCQQCSRFH + EF
Sbjct: 169 SMPVPRCQIDGCELDLSSAKGYHRKHKVCEKHSKCPKVSVSGLERRFCQQCSRFHAVSEF 228
Query: 158 DEGKRSCRRRLAGHNRRRRKTQ 179
DE KRSCR+RL+ HN RRRK Q
Sbjct: 229 DEKKRSCRKRLSHHNARRRKPQ 250
>At3g57920 squamosa promoter-binding protein homolog
Length = 354
Score = 110 bits (276), Expect = 3e-24
Identities = 52/83 (62%), Positives = 60/83 (71%)
Query: 98 SASYPMCQVDNCKEDLSKAKDYHRRHKVCEAHSKASKALLGNQMQRFCQQCSRFHPLVEF 157
S++ CQV+ C+ DLS K Y+ RHKVC HSK+SK ++ QRFCQQCSRFH L EF
Sbjct: 53 SSTTARCQVEGCRMDLSNVKAYYSRHKVCCIHSKSSKVIVSGLHQRFCQQCSRFHQLSEF 112
Query: 158 DEGKRSCRRRLAGHNRRRRKTQP 180
D KRSCRRRLA HN RRRK QP
Sbjct: 113 DLEKRSCRRRLACHNERRRKPQP 135
>At1g49270 hypothetical protein
Length = 699
Score = 38.9 bits (89), Expect = 0.015
Identities = 40/134 (29%), Positives = 53/134 (38%), Gaps = 12/134 (8%)
Query: 276 NAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSSADHMRERSSGSSQSPNDDSDCQEDVRV 335
N+PPAP + PS P+ N ++ +E +S S D + + +
Sbjct: 9 NSPPAPPPP-------SPPSPPSSNDQQTTSPPPSDNQETTSPPPPSSPDIAPPPQQQQE 61
Query: 336 KLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVDERTPSSSPPVVEMNFGLQGGIRGFN 395
P L PEN S SS S SS PS+SPP N G +G N
Sbjct: 62 SPPPPL----PENSSDGSSSSSPPPPSDSSSQSQSPPPPSTSPPQQSDNNGNKGNNNENN 117
Query: 396 -SNCISTGFGGNAN 408
N S+G GGN N
Sbjct: 118 KGNDGSSGDGGNKN 131
>At1g56210 unknown protein
Length = 364
Score = 37.0 bits (84), Expect = 0.056
Identities = 41/176 (23%), Positives = 66/176 (37%), Gaps = 25/176 (14%)
Query: 213 GKFEERRSQVPDKEQLVQILNRIPLPADLTAKLLDVGNNLNAKNDNVQMETSPSYHHRDD 272
GK E+ ++PD ++ P P D K N K + VQ+ + D+
Sbjct: 98 GKNAEQLPEIPDP------VDNKPKPVDPKEK-----NKKKKKEEKVQITNEATSSGIDN 146
Query: 273 QLNNAPPAPLTKDFLAVLSTTPSTPARNGGNGSTSSA-------DHMRERSSGSSQSPND 325
P P + + S P G+G +S D ++E+ SG +SP+
Sbjct: 147 -----PEKPGSGECDKPESEKPVDEKCLSGDGGETSGPVKEEKKDVLKEKDSGKEESPSP 201
Query: 326 DSDCQEDVRVKLPLQLFGSSPENDSPSKLPSSRKYFSSESSNPVD--ERTPSSSPP 379
+D K G+ P+N K + + ++NP D RT S PP
Sbjct: 202 PADSSAPAAEKKAEDTGGAVPDNGKVGKKKKKKGQSLATTNNPTDGPARTQSLPPP 257
>At1g10390 unknown protein
Length = 1041
Score = 35.4 bits (80), Expect = 0.16
Identities = 39/133 (29%), Positives = 54/133 (40%), Gaps = 6/133 (4%)
Query: 342 FGSSPENDSPSKLPSSRKYFSSESSNPVDERTP---SSSPPVVEMNFGLQGGIRGFNSNC 398
F SP S P++ + SS S+NP +TP SSS NFG +SN
Sbjct: 421 FSPSPAFGQTSANPTN-PFSSSTSTNPFAPQTPTIASSSFGTATSNFGSSPFGVTSSSNL 479
Query: 399 ISTGFGGNANKETSQSHSCTTIPLDLFKGSKSNNMIQQSSSVQSVPFKAGYASSGSDYSP 458
+G + S S TT P LF S + SS S P + + G+ S
Sbjct: 480 FGSGSSTTTSVFGSSSAFGTTTPSPLFGSSSTPGFGSSSSIFGSAPGQGATPAFGN--SQ 537
Query: 459 PSLNSDTQDRTGR 471
PS ++ TG+
Sbjct: 538 PSTLFNSTPSTGQ 550
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,183,994
Number of Sequences: 26719
Number of extensions: 1115123
Number of successful extensions: 3624
Number of sequences better than 10.0: 75
Number of HSP's better than 10.0 without gapping: 22
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 3505
Number of HSP's gapped (non-prelim): 116
length of query: 998
length of database: 11,318,596
effective HSP length: 109
effective length of query: 889
effective length of database: 8,406,225
effective search space: 7473134025
effective search space used: 7473134025
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC140774.7


