

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140773.4 + phase: 0 /pseudo
(1194 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
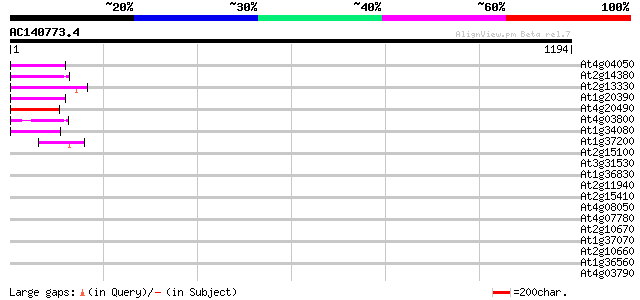
Sequences producing significant alignments: (bits) Value
At4g04050 putative transposon protein 83 1e-15
At2g14380 putative retroelement pol polyprotein 78 3e-14
At2g13330 F14O4.9 76 1e-13
At1g20390 hypothetical protein 76 1e-13
At4g20490 putative protein 75 2e-13
At4g03800 61 4e-09
At1g34080 putative protein 60 1e-08
At1g37200 hypothetical protein 44 6e-04
At2g15100 putative retroelement pol polyprotein 39 0.014
At3g31530 hypothetical protein 39 0.018
At1g36830 hypothetical protein 39 0.018
At2g11940 putative retroelement gag/pol polyprotein 38 0.030
At2g15410 putative retroelement pol polyprotein 37 0.052
At4g08050 37 0.088
At4g07780 putative athila transposon protein 36 0.15
At2g10670 pseudogene 36 0.15
At1g37070 hypothetical protein 36 0.15
At2g10660 putative Athila retroelement ORF1 protein 34 0.44
At1g36560 hypothetical protein 34 0.44
At4g03790 putative athila-like protein 33 0.98
>At4g04050 putative transposon protein
Length = 681
Score = 82.8 bits (203), Expect = 1e-15
Identities = 41/117 (35%), Positives = 69/117 (58%)
Query: 2 LFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLG 61
L I+ + ++ +++D G++V++L ++G DS+L+ LT + G + SLG
Sbjct: 539 LVIRIDVGNYELSHIMIDTGSSVDVLFYDAFKRMGHLDSELQGRKTPLTGFAGDTTFSLG 598
Query: 62 AIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGL 118
I+L + +R+T+F+VV KA FN +LGR W+H + AVPST HQ I + G+
Sbjct: 599 TIQLPTIARGVRRLTSFLVVNKKAPFNAILGRPWLHAMKAVPSTYHQCIKFPSDKGI 655
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 77.8 bits (190), Expect = 3e-14
Identities = 42/127 (33%), Positives = 73/127 (57%), Gaps = 1/127 (0%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL I+ I V ++L+D G++VN++ + L K+ ++D +KP LT ++G++ +
Sbjct: 347 PLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLTGFDGNTMMTN 406
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVE 120
G IKL + +G F+VV +N++LG WIH + A+PS+ HQ I I G +E
Sbjct: 407 GTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCIKIPTSIG-IE 465
Query: 121 NVEADQS 127
+ +Q+
Sbjct: 466 TIRGNQN 472
>At2g13330 F14O4.9
Length = 889
Score = 76.3 bits (186), Expect = 1e-13
Identities = 48/172 (27%), Positives = 84/172 (47%), Gaps = 8/172 (4%)
Query: 2 LFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLG 61
L I+ + ++ ++VD G++V++L + G DS L+ LT + G + S+G
Sbjct: 65 LVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSKLQGRKTPLTGFAGDTTFSIG 124
Query: 62 AIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLV-- 119
I+L + +++T F+VV KA FN +LGR W+H + AVPST HQ I G+
Sbjct: 125 TIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKAVPSTYHQCIKFPSYKGIAVV 184
Query: 120 -ENVEADQSYFLAEVNTITKRN-----FDKQLAHIAPVISPGPKQMASNDEI 165
+ + + ++ I K + + +LA + V S P Q + +
Sbjct: 185 YGSQRSSRKCYMGSYEDIKKADPVVLMIEDELAEMKTVRSLDPSQRGTRKSL 236
>At1g20390 hypothetical protein
Length = 1791
Score = 76.3 bits (186), Expect = 1e-13
Identities = 39/118 (33%), Positives = 67/118 (56%)
Query: 1 PLFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSL 60
PL ++ I+ V +VL+D G++V+L+ + L + + D +KP + L ++G ++
Sbjct: 570 PLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPVSKPLAGFDGDFVMTI 629
Query: 61 GAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGL 118
G IKL + VG F+V+ A +NV+LG WIH + A+PST HQ + +G+
Sbjct: 630 GTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPSTYHQCVKFPTHNGI 687
Score = 42.0 bits (97), Expect = 0.002
Identities = 19/41 (46%), Positives = 25/41 (60%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW+EA + +V +F+ KFII R G+P I TD G
Sbjct: 1534 FTKWVEAESYATIRANDVQNFVWKFIICRHGLPYEIITDNG 1574
>At4g20490 putative protein
Length = 853
Score = 75.1 bits (183), Expect = 2e-13
Identities = 37/104 (35%), Positives = 64/104 (60%)
Query: 2 LFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLG 61
L I+ + V ++L+D G++VNL+ L K+G+ +S + P +T Y+G + S+G
Sbjct: 481 LVIKLVMEDFDVERILIDTGSSVNLMFLKTLFKMGISESKIMPKIRPMTGYDGEAKMSIG 540
Query: 62 AIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPST 105
IKL++ VG + T F+V+ S+ +N +LG WI+ + A+PST
Sbjct: 541 EIKLQVQVGGITQKTKFVVIDSEPIYNAILGSPWIYSMKAIPST 584
>At4g03800
Length = 637
Score = 60.8 bits (146), Expect = 4e-09
Identities = 40/124 (32%), Positives = 64/124 (51%), Gaps = 15/124 (12%)
Query: 2 LFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLG 61
L I + ++++LVD G++V+L+ FLG P + ++ + S SLG
Sbjct: 203 LIITLDVANFKISRILVDTGSSVDLI---FLG----------PPSPLVA-FTSESAMSLG 248
Query: 62 AIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGLVEN 121
IKL ++ G +I F+V A +N++LG WI + AVPST HQ + +G VE
Sbjct: 249 TIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIFQMKAVPSTYHQWLKFPTSNG-VET 307
Query: 122 VEAD 125
+ D
Sbjct: 308 IWGD 311
>At1g34080 putative protein
Length = 811
Score = 59.7 bits (143), Expect = 1e-08
Identities = 32/107 (29%), Positives = 56/107 (51%)
Query: 2 LFIQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLG 61
L I + V ++L+D G++V+L+ L ++G+ D+K L ++ + SLG
Sbjct: 477 LVISLDVGNCEVQRILIDTGSSVDLIFLDTLVRMGISKKDIKGAPSPLVSFTIETSMSLG 536
Query: 62 AIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQ 108
I L + ++ F V A +N++LG W++ + AVPST HQ
Sbjct: 537 TITLPVTAQGVVKMIEFTVFDRPAAYNIILGTPWLYEMKAVPSTYHQ 583
>At1g37200 hypothetical protein
Length = 1564
Score = 43.9 bits (102), Expect = 6e-04
Identities = 34/108 (31%), Positives = 49/108 (44%), Gaps = 11/108 (10%)
Query: 62 AIKLELMVGSTKRITTFMVVPS---KANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGL 118
AI + + VG+ K + S KA FN +LGR W+H + AVPST HQ I E G+
Sbjct: 504 AIVIRIDVGNYKLSRIMIDTGSSVDKAPFNAILGRPWLHAMKAVPSTYHQCIKFPSEKGI 563
Query: 119 VENVEADQ--------SYFLAEVNTITKRNFDKQLAHIAPVISPGPKQ 158
+ + SY L + + + +LA + V S P Q
Sbjct: 564 AVIYGSQRSSRRCYMGSYELIKKAELVVLMIEDKLAEMKTVRSSDPSQ 611
Score = 42.0 bits (97), Expect = 0.002
Identities = 19/40 (47%), Positives = 26/40 (64%)
Query: 1155 TKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
TKWIE + VT+++V F+ + I+YR GIP I TD G
Sbjct: 1396 TKWIEGKAFQQVTEKQVEVFLLENIVYRHGIPYEIVTDNG 1435
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 39.3 bits (90), Expect = 0.014
Identities = 18/50 (36%), Positives = 27/50 (54%)
Query: 59 SLGAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQ 108
SLG + L + ++ F V A +NV+LG W++ + VPST HQ
Sbjct: 352 SLGTVVLPVTAQGVVKMVEFTVFDRPAAYNVILGTPWLYEMKVVPSTYHQ 401
Score = 34.7 bits (78), Expect = 0.34
Identities = 17/40 (42%), Positives = 24/40 (59%)
Query: 1155 TKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
T+ +EA ++T +V +FI K II R G+P I TD G
Sbjct: 1073 TRGVEAAAYSNITHVQVWNFIWKDIICRHGLPYEIVTDNG 1112
>At3g31530 hypothetical protein
Length = 831
Score = 38.9 bits (89), Expect = 0.018
Identities = 18/41 (43%), Positives = 24/41 (57%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
F+KWIEA + +V +F+ K I+ R GIP I TD G
Sbjct: 688 FSKWIEAESYASIKDAQVENFVLKHILCRHGIPYEIVTDNG 728
Score = 35.8 bits (81), Expect = 0.15
Identities = 18/83 (21%), Positives = 38/83 (45%)
Query: 59 SLGAIKLELMVGSTKRITTFMVVPSKANFNVLLGREWIHGVGAVPSTVHQRIAIWKEDGL 118
+LG IKL + +I F + +N ++G W++ AV ST H + D +
Sbjct: 2 TLGTIKLPVRAKGVTKIVDFSITDQPTVYNAIIGTPWLNQFRAVASTYHLCLKFPTSDSV 61
Query: 119 VENVEADQSYFLAEVNTITKRNF 141
E + +++ + N + + ++
Sbjct: 62 WEKLGHERAEAVKAENQLLETSY 84
>At1g36830 hypothetical protein
Length = 243
Score = 38.9 bits (89), Expect = 0.018
Identities = 18/40 (45%), Positives = 26/40 (65%)
Query: 1155 TKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
TKWIEA + VT+++V F+ + I+ + GIP I TD G
Sbjct: 27 TKWIEAKAFQQVTEKQVEDFLWENIVCQHGIPYEIITDNG 66
>At2g11940 putative retroelement gag/pol polyprotein
Length = 1212
Score = 38.1 bits (87), Expect = 0.030
Identities = 19/42 (45%), Positives = 24/42 (56%)
Query: 1153 LFTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
L +W+EA +TQ +V FI K II R G+P I TD G
Sbjct: 1009 LLLEWVEAAAYSYITQVQVRHFIWKEIICRHGLPYEIVTDNG 1050
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 37.4 bits (85), Expect = 0.052
Identities = 18/41 (43%), Positives = 23/41 (55%)
Query: 1154 FTKWIEAIPLKDVTQEEVISFIQKFIIYRFGIPETITTDQG 1194
FTKW EA + +EV +F+ K II G+P I TD G
Sbjct: 1530 FTKWFEAESYARIQSKEVQNFVWKNIICHHGLPYEIVTDNG 1570
>At4g08050
Length = 1428
Score = 36.6 bits (83), Expect = 0.088
Identities = 29/99 (29%), Positives = 49/99 (49%), Gaps = 5/99 (5%)
Query: 17 LVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAIK-LELMVGSTKRI 75
L D GA+VNL+P S ++ KP ++ L + SS G +K L +M+ +
Sbjct: 592 LCDLGASVNLMPLSMARRLEFI--QYKPCDLTLILADRSSRKHFGMLKDLPVMINGVEVP 649
Query: 76 TTFMVVPSKANFN--VLLGREWIHGVGAVPSTVHQRIAI 112
T F+V+ + ++LGR ++ VGAV +I +
Sbjct: 650 TDFVVLDMEVEHKDPLILGRPFLASVGAVIDVKEGKIGL 688
>At4g07780 putative athila transposon protein
Length = 446
Score = 35.8 bits (81), Expect = 0.15
Identities = 33/115 (28%), Positives = 58/115 (49%), Gaps = 17/115 (14%)
Query: 3 FIQAKINGVG------------VNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILT 50
FIQ K++ G N+ L D GA+V+L+P S ++G+ + K N+ L
Sbjct: 276 FIQEKLDDPGSFTLPCSLGPLTFNRCLCDLGASVSLMPLSTAKRLGIV--EYKFCNLALL 333
Query: 51 NYEGSSGNSLGAI-KLELMVGSTKRITTFMVVPS--KANFNVLLGREWIHGVGAV 102
+GS + G I L + +G+ + T F+V+ + + ++LGR ++ GAV
Sbjct: 334 LPDGSVAHPHGLIGNLPVKIGNDEIPTDFVVLDTDEEGKDPLILGRPFLASAGAV 388
>At2g10670 pseudogene
Length = 929
Score = 35.8 bits (81), Expect = 0.15
Identities = 27/92 (29%), Positives = 50/92 (54%), Gaps = 5/92 (5%)
Query: 14 NKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAIK-LELMVGST 72
N L D GA+V+L+P S + K+G KP ++ L + +S G ++ + +M+
Sbjct: 589 NNCLCDLGASVSLMPLSVVKKLGFV--HYKPCDLTLILADRTSKTPFGLLEDVPVMINGV 646
Query: 73 KRITTFMV--VPSKANFNVLLGREWIHGVGAV 102
+ T F+V + ++ ++LGR ++ VGAV
Sbjct: 647 EVPTDFVVLEMDGESKDPLILGRPFLASVGAV 678
>At1g37070 hypothetical protein
Length = 590
Score = 35.8 bits (81), Expect = 0.15
Identities = 29/92 (31%), Positives = 50/92 (53%), Gaps = 5/92 (5%)
Query: 14 NKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAIK-LELMVGST 72
N+ L D GA+V+L+P S ++G+ + K N+ L +GS + G I+ L + + +
Sbjct: 272 NRCLCDLGASVSLMPLSTAKRLGI--MEYKFCNLALLLADGSVAHPHGLIENLPVKIENV 329
Query: 73 KRITTFMV--VPSKANFNVLLGREWIHGVGAV 102
+ T F+V V K ++LGR ++ GAV
Sbjct: 330 EIPTDFVVLDVDEKGKDPLILGRPFLASAGAV 361
>At2g10660 putative Athila retroelement ORF1 protein
Length = 1303
Score = 34.3 bits (77), Expect = 0.44
Identities = 29/99 (29%), Positives = 51/99 (51%), Gaps = 5/99 (5%)
Query: 17 LVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAIK-LELMVGSTKRI 75
L D GA+V+L+P S K+ + +P ++ L + SS G +K L LM+ +
Sbjct: 449 LCDLGASVSLMPLSVARKLEF--TQYRPCDLSLILADRSSRKPFGLLKHLPLMINGVEVP 506
Query: 76 TTFMVVPSKANFN--VLLGREWIHGVGAVPSTVHQRIAI 112
T F+V+ + ++LGR ++ VGA+ RI++
Sbjct: 507 TDFVVLGMEVEPKDPLILGRPFLASVGAMIDVKDGRISL 545
>At1g36560 hypothetical protein
Length = 524
Score = 34.3 bits (77), Expect = 0.44
Identities = 26/102 (25%), Positives = 51/102 (49%), Gaps = 5/102 (4%)
Query: 4 IQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAI 63
+ + + NK L D GA+V+L+P S ++G + K N+ L +GS G +
Sbjct: 127 LPCSLGSLTFNKCLCDLGASVSLMPLSVAKRLGF--NKYKYCNISLILADGSVRQPHGVL 184
Query: 64 K-LELMVGSTKRITTFMV--VPSKANFNVLLGREWIHGVGAV 102
+ L + +G + T F++ + + ++LGR ++ GA+
Sbjct: 185 EDLPIKIGKVEVPTDFIILNMDEEPKDPLILGRPFLATAGAI 226
>At4g03790 putative athila-like protein
Length = 1064
Score = 33.1 bits (74), Expect = 0.98
Identities = 28/102 (27%), Positives = 50/102 (48%), Gaps = 5/102 (4%)
Query: 4 IQAKINGVGVNKVLVDGGATVNLLPQSFLGKIGLYDSDLKPHNVILTNYEGSSGNSLGAI 63
+ I + + L D GA+V+L+P S ++ KP ++ L + SS G +
Sbjct: 416 LPCSIGELAFSDFLCDLGASVSLMPLSMGRRLEFI--HYKPCDLTLILADRSSRKPFGML 473
Query: 64 K-LELMVGSTKRITTFMVVPSKANFN--VLLGREWIHGVGAV 102
K L +M+ + T F+V+ + ++LGR ++ VGAV
Sbjct: 474 KDLPVMINGVEVPTDFVVLNMEVKHKDPLILGRPFLASVGAV 515
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.348 0.153 0.528
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,115,697
Number of Sequences: 26719
Number of extensions: 959487
Number of successful extensions: 3289
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 17
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 3252
Number of HSP's gapped (non-prelim): 50
length of query: 1194
length of database: 11,318,596
effective HSP length: 110
effective length of query: 1084
effective length of database: 8,379,506
effective search space: 9083384504
effective search space used: 9083384504
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 66 (30.0 bits)
Medicago: description of AC140773.4


