

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140773.3 + phase: 0 /partial
(140 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
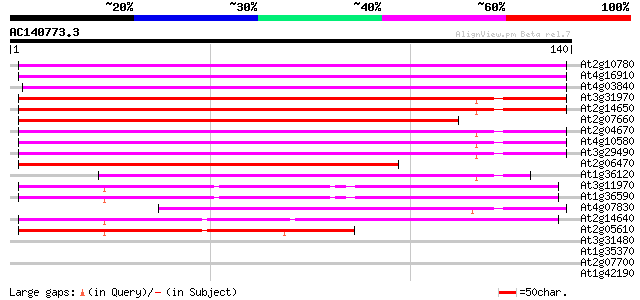
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 118 1e-27
At4g16910 retrotransposon like protein 115 1e-26
At4g03840 putative transposon protein 110 3e-25
At3g31970 hypothetical protein 104 1e-23
At2g14650 putative retroelement pol polyprotein 102 9e-23
At2g07660 putative retroelement pol polyprotein 100 3e-22
At2g04670 putative retroelement pol polyprotein 99 1e-21
At4g10580 putative reverse-transcriptase -like protein 97 2e-21
At3g29490 hypothetical protein 94 3e-20
At2g06470 putative retroelement pol polyprotein 87 3e-18
At1g36120 putative reverse transcriptase gb|AAD22339.1 77 4e-15
At3g11970 hypothetical protein 73 6e-14
At1g36590 hypothetical protein 73 6e-14
At4g07830 putative reverse transcriptase 70 5e-13
At2g14640 putative retroelement pol polyprotein 60 4e-10
At2g05610 putative retroelement pol polyprotein 56 5e-09
At3g31480 hypothetical protein 36 0.008
At1g35370 hypothetical protein 36 0.008
At2g07700 putative retroelement pol polyprotein 33 0.064
At1g42190 hypothetical protein 33 0.064
>At2g10780 pseudogene
Length = 1611
Score = 118 bits (295), Expect = 1e-27
Identities = 58/137 (42%), Positives = 82/137 (59%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ + G GR KKL+ R++GPY++ +RVG V Y++ L P L+ H+VFHVS
Sbjct: 1446 GDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVFHVS 1505
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGES 122
QLRK + D ++ +++N+TVE PVRI DR K RGK L+K +W E
Sbjct: 1506 QLRKCLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMTKGTRGKARDLLKVLWNCRGREE 1565
Query: 123 LTWELESKKLESYPELF 139
TWE E+K ++PE F
Sbjct: 1566 YTWETENKMKANFPEWF 1582
>At4g16910 retrotransposon like protein
Length = 687
Score = 115 bits (287), Expect = 1e-26
Identities = 56/137 (40%), Positives = 81/137 (58%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ + G GR KKL R++GPY++ +RVG V Y++ L P L H+VFHVS
Sbjct: 530 GDLVYLKAMTYKGAGRFTSRKKLRPRYVGPYKVIERVGAVAYKLDLPPKLDAFHNVFHVS 589
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGES 122
QLRK + + ++ +++N+TVE PVRI D+ K RGK + L+K +W E
Sbjct: 590 QLRKCLSEQEESMEDVPPGLKENMTVEAWPVRIMDQMKKGTRGKSMDLLKILWNCGGREE 649
Query: 123 LTWELESKKLESYPELF 139
TWE E+K ++PE F
Sbjct: 650 YTWETETKMKANFPEWF 666
>At4g03840 putative transposon protein
Length = 973
Score = 110 bits (274), Expect = 3e-25
Identities = 54/136 (39%), Positives = 80/136 (58%)
Query: 4 DHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVSQ 63
D V+ + G GR KKL+ R++GPY++ +RVG V Y++ L P L+ H+VFHVSQ
Sbjct: 809 DLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVFHVSQ 868
Query: 64 LRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGESL 123
LRK + + ++ +++N+TVE PV+I DR K RGK L+K +W E
Sbjct: 869 LRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTKGTRGKSRDLLKVLWNCGGREQY 928
Query: 124 TWELESKKLESYPELF 139
TWE E+K ++ E F
Sbjct: 929 TWETENKMKANFSEWF 944
>At3g31970 hypothetical protein
Length = 1329
Score = 104 bits (260), Expect = 1e-23
Identities = 55/139 (39%), Positives = 85/139 (60%), Gaps = 4/139 (2%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ ++ + G R++ KLT R++GP++I +RVG V YR+ L + H VFHVS
Sbjct: 1187 GDRVYLKMAMLRGPNRSISETKLTPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVS 1246
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVW--EGATG 120
LRK + V+ ++ N+T+E PVRI +R++K LR K+IPL+K +W +G T
Sbjct: 1247 MLRKCLHKDDEVLAKILEDLQPNMTLEARPVRILERRIKELRRKKIPLIKVLWNCDGVTE 1306
Query: 121 ESLTWELESKKLESYPELF 139
E TWE E++ S+ + F
Sbjct: 1307 E--TWEPEARMKASFKKWF 1323
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 102 bits (253), Expect = 9e-23
Identities = 52/139 (37%), Positives = 84/139 (60%), Gaps = 4/139 (2%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ ++ + G R++ KL+ R++GP++I +RVG V YR+ L + H VFHVS
Sbjct: 1189 GDRVYLKMAMLRGPNRSISETKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVS 1248
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVW--EGATG 120
LRK + V+ ++ N+T+E PVR+ +R++K LR K+IPL+K +W +G T
Sbjct: 1249 MLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLWDCDGVTK 1308
Query: 121 ESLTWELESKKLESYPELF 139
E TWE E++ + + F
Sbjct: 1309 E--TWEPEARMKARFKKWF 1325
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 100 bits (248), Expect = 3e-22
Identities = 49/110 (44%), Positives = 69/110 (62%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ + G GR KKL+ R++GPY++ +RVG V Y++ L P L+ H+VFHVS
Sbjct: 838 GDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNVFHNVFHVS 897
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVK 112
QLRKY+ D ++ +++N+TVE PVRI DR K RGK L+K
Sbjct: 898 QLRKYLSDQEESVEDIPPGLKENMTVEAWPVRIMDRMSKGTRGKSRDLLK 947
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 98.6 bits (244), Expect = 1e-21
Identities = 51/139 (36%), Positives = 83/139 (59%), Gaps = 4/139 (2%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ ++ + G R++ KL+ R++GP++I +RVG V YR+ L + H VFHV
Sbjct: 1269 GDRVYLKMAMLRGPNRSILETKLSPRYMGPFRIVERVGPVAYRLELPDVMRAFHKVFHVL 1328
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVW--EGATG 120
LRK + V+ ++ N+T+E PVR+ +R++K LR K+IPL+K +W +G T
Sbjct: 1329 MLRKCLHKDDEVLVKIPEDLQPNMTLEARPVRVLERRIKELRRKKIPLIKVLWDCDGVTE 1388
Query: 121 ESLTWELESKKLESYPELF 139
E TWE E++ + + F
Sbjct: 1389 E--TWEPEARMKARFKKWF 1405
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 97.4 bits (241), Expect = 2e-21
Identities = 50/139 (35%), Positives = 82/139 (58%), Gaps = 4/139 (2%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ ++ + G R++ KL+ R++GP++I +RV V YR+ L + H VFHVS
Sbjct: 1098 GDRVYLKMAMLRGPNRSISETKLSPRYMGPFKIVERVEPVAYRLELPDVMRAFHKVFHVS 1157
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVW--EGATG 120
LRK + + ++ N+T+E PVR+ +R++K LR K+IPL+K +W +G T
Sbjct: 1158 MLRKCLHKDDEALAKIPEDLQPNMTLEARPVRVLERRIKELRQKKIPLIKVLWDCDGVTE 1217
Query: 121 ESLTWELESKKLESYPELF 139
E TWE E++ + + F
Sbjct: 1218 E--TWEPEARMKARFKKWF 1234
>At3g29490 hypothetical protein
Length = 438
Score = 93.6 bits (231), Expect = 3e-20
Identities = 48/139 (34%), Positives = 82/139 (58%), Gaps = 4/139 (2%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ ++ + G R++ KL+LR++GP++I +RVG V Y + L + H VFHVS
Sbjct: 199 GDRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDVMRAFHKVFHVS 258
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVW--EGATG 120
LRK + V+ ++ N+T+E VR+ +R++K L+ K+I L+K +W +G T
Sbjct: 259 MLRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKISLIKVLWDCDGVTE 318
Query: 121 ESLTWELESKKLESYPELF 139
E TW+ E++ + + F
Sbjct: 319 E--TWQPEARMKARFKKWF 335
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 87.0 bits (214), Expect = 3e-18
Identities = 41/95 (43%), Positives = 60/95 (63%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVS 62
GD V+ + G GR KKL+ R++GPY++ +RVG V Y++ L P L+ H+VFHVS
Sbjct: 805 GDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDLPPKLNAFHNVFHVS 864
Query: 63 QLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDD 97
QLRK + D ++ +++N+TVE PVRI D
Sbjct: 865 QLRKCLSDQEESVEDVPPGLKENMTVEAWPVRIMD 899
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 76.6 bits (187), Expect = 4e-15
Identities = 41/110 (37%), Positives = 66/110 (59%), Gaps = 4/110 (3%)
Query: 23 KKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHVSQLRKYIPDPSHVIQSDDVQV 82
++ L F +I +RVG V YR+ L + H+VFHVS LRK + V+ +
Sbjct: 1113 RRRELEFEVGTEIVERVGPVAYRLELPDVMRAFHNVFHVSMLRKCLHKDDEVLAKIPEDL 1172
Query: 83 RDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVW--EGATGESLTWELESK 130
+ N+T+E PVR+ +R++K +R K+IP++K +W +G T E TWE E++
Sbjct: 1173 QPNMTLEARPVRVLERRIKEVRRKKIPMIKVLWDCDGVTEE--TWEPEAR 1220
>At3g11970 hypothetical protein
Length = 1499
Score = 72.8 bits (177), Expect = 6e-14
Identities = 43/136 (31%), Positives = 74/136 (53%), Gaps = 5/136 (3%)
Query: 3 GDHVFFRVTPVTGVGRALKS-KKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD+V+ ++ P +++ +KL+ ++ GPY+I DR G V Y++ L Y S +H VFHV
Sbjct: 1365 GDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSY-SQVHPVFHV 1423
Query: 62 SQLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGE 121
SQL+ + + S + V ++D E +P ++ +RK+ +GK + V W E
Sbjct: 1424 SQLKVLVGNVSTTVHLPSV-MQD--VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLE 1480
Query: 122 SLTWELESKKLESYPE 137
TWE +++PE
Sbjct: 1481 EATWEFLFDLQKTFPE 1496
>At1g36590 hypothetical protein
Length = 1499
Score = 72.8 bits (177), Expect = 6e-14
Identities = 43/136 (31%), Positives = 74/136 (53%), Gaps = 5/136 (3%)
Query: 3 GDHVFFRVTPVTGVGRALKS-KKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD+V+ ++ P +++ +KL+ ++ GPY+I DR G V Y++ L Y S +H VFHV
Sbjct: 1365 GDYVYVKLQPYRQQSVVMRANQKLSPKYFGPYKIIDRCGEVAYKLALPSY-SQVHPVFHV 1423
Query: 62 SQLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGE 121
SQL+ + + S + V ++D E +P ++ +RK+ +GK + V W E
Sbjct: 1424 SQLKVLVGNVSTTVHLPSV-MQD--VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLE 1480
Query: 122 SLTWELESKKLESYPE 137
TWE +++PE
Sbjct: 1481 EATWEFLFDLQKTFPE 1496
>At4g07830 putative reverse transcriptase
Length = 611
Score = 69.7 bits (169), Expect = 5e-13
Identities = 39/104 (37%), Positives = 61/104 (58%), Gaps = 4/104 (3%)
Query: 38 RVGTVVYRVGLAPYLSNLHDVFHVSQLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDD 97
RVG V +R+ L+ + H VFHVS LRK + V+ ++ N+T+E PVR+ +
Sbjct: 468 RVGPVAFRLELSDVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVRVLE 527
Query: 98 RKVKTLRGKEIPLVKFV--WEGATGESLTWELESKKLESYPELF 139
R++K LR K+IPL+K + +G T E TWE E++ + + F
Sbjct: 528 RRIKELRRKKIPLIKVLRNCDGVTEE--TWEPEARLKARFKKWF 569
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 60.1 bits (144), Expect = 4e-10
Identities = 41/136 (30%), Positives = 68/136 (49%), Gaps = 3/136 (2%)
Query: 3 GDHVFFRVTPVTGVGRALKS-KKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD V R+ P +S +KL+ RF GP+Q++ + G V YR+ L P + +H VFHV
Sbjct: 801 GDWVLLRIQPYRQKTLFRRSSQKLSHRFYGPFQVASKHGEVAYRLTL-PEGTRIHPVFHV 859
Query: 62 SQLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGE 121
S L+ ++ D +R+N ++ P + + + ++ K + + WEG E
Sbjct: 860 SLLKPWVGD-GEPDMGQLPPLRNNGELKLQPTAVLEVRWRSQDKKRVADLLVQWEGLHIE 918
Query: 122 SLTWELESKKLESYPE 137
TWE + S+PE
Sbjct: 919 DATWEEYDQLAASFPE 934
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 56.2 bits (134), Expect = 5e-09
Identities = 32/88 (36%), Positives = 54/88 (61%), Gaps = 5/88 (5%)
Query: 3 GDHVFFRVTPVTGVGRALKS-KKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD+VF ++ P ++S +KL+ ++ GPY++ DR G V Y++ L P S +H VFHV
Sbjct: 693 GDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQL-PANSQVHPVFHV 751
Query: 62 SQLRKY---IPDPSHVIQSDDVQVRDNL 86
SQLR + +H+++ + +R+NL
Sbjct: 752 SQLRVLVGTVTTSTHLLRCYLMSLRENL 779
>At3g31480 hypothetical protein
Length = 338
Score = 35.8 bits (81), Expect = 0.008
Identities = 15/43 (34%), Positives = 27/43 (61%)
Query: 3 GDHVFFRVTPVTGVGRALKSKKLTLRFIGPYQISDRVGTVVYR 45
GD V+ ++ + R + KL+ +++GP++I +RVG V YR
Sbjct: 166 GDRVYLKMDMLQSPKRFILETKLSPKYMGPFRIVERVGPVAYR 208
>At1g35370 hypothetical protein
Length = 1447
Score = 35.8 bits (81), Expect = 0.008
Identities = 30/125 (24%), Positives = 53/125 (42%), Gaps = 28/125 (22%)
Query: 3 GDHVFFRVTPVTGVGRALK-SKKLTLRFIGPYQISDRVGTVVYRVGLAPYLSNLHDVFHV 61
GD V+ ++ P L+ ++KL+ ++ GPY+I ++ G V+ + +V
Sbjct: 1336 GDFVYVKLQPYRQQSVVLRVNQKLSPKYFGPYKIIEKCGEVM-----------VGNVTTS 1384
Query: 62 SQLRKYIPDPSHVIQSDDVQVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGE 121
+QL +PD E P I +RK+ +G+ +V W G E
Sbjct: 1385 TQLPSVLPD----------------IFEKAPEYILERKLVKRQGRAATMVLVKWIGEPVE 1428
Query: 122 SLTWE 126
TW+
Sbjct: 1429 EATWK 1433
>At2g07700 putative retroelement pol polyprotein
Length = 543
Score = 32.7 bits (73), Expect = 0.064
Identities = 13/43 (30%), Positives = 26/43 (60%)
Query: 81 QVRDNLTVETLPVRIDDRKVKTLRGKEIPLVKFVWEGATGESL 123
+++ NLT P+ I +R+++ + + IP V+ +WE T +L
Sbjct: 193 ELKTNLTYPEGPLEIGERRIRKFKKRLIPQVQVIWENKTDGNL 235
>At1g42190 hypothetical protein
Length = 64
Score = 32.7 bits (73), Expect = 0.064
Identities = 15/46 (32%), Positives = 24/46 (51%)
Query: 43 VYRVGLAPYLSNLHDVFHVSQLRKYIPDPSHVIQSDDVQVRDNLTV 88
+Y++ L + H VFHVS LRK I +VI + +N+ +
Sbjct: 1 MYKLDLPTLMDAFHKVFHVSMLRKCITHQENVISEPPPDLLENMMI 46
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.143 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,062,129
Number of Sequences: 26719
Number of extensions: 115225
Number of successful extensions: 293
Number of sequences better than 10.0: 27
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 261
Number of HSP's gapped (non-prelim): 27
length of query: 140
length of database: 11,318,596
effective HSP length: 89
effective length of query: 51
effective length of database: 8,940,605
effective search space: 455970855
effective search space used: 455970855
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 55 (25.8 bits)
Medicago: description of AC140773.3


