

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140722.2 + phase: 0
(555 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
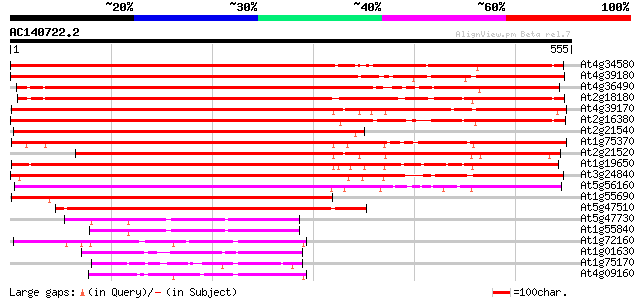
Score E
Sequences producing significant alignments: (bits) Value
At4g34580 putative protein 636 0.0
At4g39180 SEC14 - like protein 635 0.0
At4g36490 hypothetical protein 627 e-180
At2g18180 putative phosphatidylinositol/phophatidylcholine trans... 613 e-175
At4g39170 unknown protein 568 e-162
At2g16380 putative phosphatidylinositol/phosphatidylcholine tran... 556 e-159
At2g21540 putative phosphatidylinositol/phophatidylcholine trans... 556 e-158
At1g75370 sec14 cytosolic factor, putative 507 e-144
At2g21520 putative phosphatidylinositol/phosphatidylcholine tran... 497 e-141
At1g19650 sec14 cytosolic factor- like protein 493 e-139
At3g24840 phosphatidylinositol transfer protein like 439 e-123
At5g56160 putative protein 377 e-105
At1g55690 Phosphatidylinositol Transfer Protein like 371 e-103
At5g47510 putative protein 302 4e-82
At5g47730 putative protein 92 8e-19
At1g55840 polyphosphoinositide binding protein, putative 85 1e-16
At1g72160 cytosolic factor, putative 84 2e-16
At1g01630 polyphosphoinositide binding protein, putative 76 4e-14
At1g75170 unknown protein 70 4e-12
At4g09160 putative protein 68 2e-11
>At4g34580 putative protein
Length = 560
Score = 636 bits (1641), Expect = 0.0
Identities = 340/558 (60%), Positives = 423/558 (74%), Gaps = 24/558 (4%)
Query: 1 MEHSEDEKK-KKVGSIKKVALSASSKFKNSFTKKGRKHS-RVMSICIEDSFDAEELQAVD 58
+E SE+E+K K+ S+KK A++AS++FKNSF KKGR+ S RVMS+ IED DAE+LQA+D
Sbjct: 8 IEMSEEERKIVKISSLKKKAINASNRFKNSFKKKGRRSSSRVMSVPIEDDIDAEDLQALD 67
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
A RQ LIL+ELLPSK DD HMMLRFLRARK+DIEK KQMW+DM++WRK+FGADTI+EDF+
Sbjct: 68 AFRQALILDELLPSKLDDLHMMLRFLRARKFDIEKAKQMWSDMIQWRKDFGADTIIEDFD 127
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFA 178
FEE+DEV+K YPQG+HGVDK+GRPVYIERLGQ+D NKLLQVT+++RY+KYHV+EFE+ F
Sbjct: 128 FEEIDEVMKHYPQGYHGVDKEGRPVYIERLGQIDANKLLQVTTMDRYVKYHVKEFEKTFK 187
Query: 179 VKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFII 238
VK P+CS+AA KHIDQSTTILDVQGVGL++ +K+AR+LLQRL KID +NYPE+LNRMFII
Sbjct: 188 VKFPSCSVAANKHIDQSTTILDVQGVGLKNFSKSARELLQRLCKIDNENYPETLNRMFII 247
Query: 239 NAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGG 298
NAGSGFRLLW+TVKSFLDPKTT+KIHVLGNKY SKLLEVIDASELPEF GG CTC DKGG
Sbjct: 248 NAGSGFRLLWSTVKSFLDPKTTAKIHVLGNKYHSKLLEVIDASELPEFFGGACTCEDKGG 307
Query: 299 CMLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDVH 358
CM SDKGPWNDPE+LK+A N + +S E K + Q + GF E+L+
Sbjct: 308 CMRSDKGPWNDPEVLKIAINREAK--CSPISEDEHKHVDQGRST--SGF-----ESLE-- 356
Query: 359 CLDQPKSYGVYQYDSFVPVLDKAVDSSW-KKTIQNDKYALSKDCFSNNNGMNSSGFSKQF 417
+ K+ Y+ + +DK++D +W KT + + + +SK G G
Sbjct: 357 -RIKKKTDEDNVYEKQIATIDKSMDMAWLAKTQKAENFPISKGITKLMKGAPKKG-DGLL 414
Query: 418 VGGIMALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMIK------AAAISNNEYM 471
VGG+MA VMGIV ++R++ +PRK+TEAALYG + Y + + K AA +S++EYM
Sbjct: 415 VGGVMAFVMGIVAMVRLSKDVPRKLTEAALYGNSVCYEESTKSKQNQGQFAAPVSSSEYM 474
Query: 472 AMMKRMAELEEKVTVLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQV 530
M+KRMAELE+K L +KP + EKEE L AL RV LEQEL TKKALE+AL Q
Sbjct: 475 LMVKRMAELEDKCMFLDLKPAHVESEKEEKLQAALNRVQVLEQELTETKKALEEALVSQK 534
Query: 531 ELEGQIDKKKKKKKKLVR 548
E+ I+ KKKKKKKLVR
Sbjct: 535 EILAYIE-KKKKKKKLVR 551
>At4g39180 SEC14 - like protein
Length = 554
Score = 635 bits (1637), Expect = 0.0
Identities = 326/554 (58%), Positives = 416/554 (74%), Gaps = 21/554 (3%)
Query: 1 MEHSEDEKK-KKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDA 59
+E SED+K+ K+ S+KK A++A++KFK+S TKKGR+HSRV + I D D EELQAVDA
Sbjct: 17 VEISEDDKRLTKLCSLKKKAINATNKFKHSMTKKGRRHSRVACVSIVDEIDTEELQAVDA 76
Query: 60 LRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEF 119
RQ LIL+ELLPSKHDD HMMLRFLRARK+D+EK KQMW+DML WRKE+GADTIMEDF+F
Sbjct: 77 FRQALILDELLPSKHDDHHMMLRFLRARKFDLEKAKQMWSDMLNWRKEYGADTIMEDFDF 136
Query: 120 EELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAV 179
+E++EV+K YPQG+HGVDK+GRP+YIERLGQVD KL++VT+++RY+KYHV+EFE+ F V
Sbjct: 137 KEIEEVVKYYPQGYHGVDKEGRPIYIERLGQVDATKLMKVTTIDRYVKYHVKEFEKTFNV 196
Query: 180 KLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIIN 239
K PACSIAAK+HIDQSTTILDVQGVGL + NKAA+DLLQ +QKID DNYPE+LNRMFIIN
Sbjct: 197 KFPACSIAAKRHIDQSTTILDVQGVGLSNFNKAAKDLLQSIQKIDNDNYPETLNRMFIIN 256
Query: 240 AGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGC 299
AG GFRLLWNTVKSFLDPKTT+KIHVLGNKYQ+KLLE+IDA+ELPEFLGG CTCADKGGC
Sbjct: 257 AGCGFRLLWNTVKSFLDPKTTAKIHVLGNKYQTKLLEIIDANELPEFLGGKCTCADKGGC 316
Query: 300 MLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDVHC 359
M SDKGPWNDPEI K+ QNG GR ++LSG+EEKTI + +K K ET
Sbjct: 317 MRSDKGPWNDPEIFKLVQNGEGRCLRRSLSGIEEKTIFEYNNETKK--KCEPEETHKQSA 374
Query: 360 LDQPKSYGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALS--KDCFSNNNGMNSSGFSKQF 417
+ K + D+ V D A + W + + + KD +S N + G+
Sbjct: 375 AEMEKKF----IDTNV---DAAAAADWPTKLNKAEKNPTDLKDVYSAVNPLERKGY---L 424
Query: 418 VGGIMALVMGIVTIIRMTSSMPRKITEAALYG--GNSVYYDGSMIKAAAISNNEYMAMMK 475
G +MAL+MGIV ++R+T +MPR++TEA +Y G++VY DG +S EY+AM+K
Sbjct: 425 YGSVMALLMGIVGVMRLTKNMPRRLTEANVYSREGSAVYQDG----VTVMSKQEYIAMVK 480
Query: 476 RMAELEEKVTVLSVKPVMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQ 535
++ +LEEK + + E+E+ L+ AL R+ LE +L T KAL++ +TRQ E+
Sbjct: 481 KITDLEEKCKSMEAQAAFYMEREKTLDAALRRIDQLELQLSETNKALDETMTRQHEIMAF 540
Query: 536 IDKKKKKKKKLVRY 549
I+KKKKKK+K + +
Sbjct: 541 IEKKKKKKRKFLLF 554
>At4g36490 hypothetical protein
Length = 543
Score = 627 bits (1616), Expect = e-180
Identities = 336/550 (61%), Positives = 414/550 (75%), Gaps = 31/550 (5%)
Query: 7 EKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSI-CIEDSFDAEELQAVDALRQTLI 65
E K ++GS KK S+S + S TK+ R+ S+VMS+ IED DAEEL+AVDA RQ+LI
Sbjct: 8 ELKPRMGSFKK--RSSSKNLRYSMTKR-RRSSKVMSVEIIEDVHDAEELKAVDAFRQSLI 64
Query: 66 LEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEV 125
L+ELLP KHDD HMMLRFL+ARK+D+EKTKQMWT+ML+WRKEFGADT+ME+F+F+E+DEV
Sbjct: 65 LDELLPEKHDDYHMMLRFLKARKFDLEKTKQMWTEMLRWRKEFGADTVMEEFDFKEIDEV 124
Query: 126 LKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACS 185
LK YPQGHHGVDK+GRPVYIERLG VD KL+QVT+++RY+ YHV EFER F VK PACS
Sbjct: 125 LKYYPQGHHGVDKEGRPVYIERLGLVDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACS 184
Query: 186 IAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFR 245
IAAKKHIDQSTTILDVQGVGL++ NKAARDL+ RLQK+DGDNYPE+LNRMFIINAGSGFR
Sbjct: 185 IAAKKHIDQSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGSGFR 244
Query: 246 LLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKG 305
+LWNTVKSFLDPKTT+KIHVLGNKYQSKLLE+ID SELPEFLGG+CTCAD GGCM SDKG
Sbjct: 245 MLWNTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDESELPEFLGGSCTCADNGGCMRSDKG 304
Query: 306 PWNDPEILKMAQNGVGRYTI-KALSGVEEKTIKQEETAYQKGFKDSFPETLDVHCLDQPK 364
PW +PEI+K NG + + EKTI +E+ + + + + +V + P
Sbjct: 305 PWKNPEIMKRVHNGDHKCSKGSQAENSGEKTIPEEDDSTTEPASEEEKASKEVEIV--PA 362
Query: 365 SYGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMAL 424
++ + + +A S K + YA+ + C NN G S F G+MAL
Sbjct: 363 AHPAWN-------MPEAHKFSLSK---KEVYAIQEAC---NNATTEGGRSPIFT-GVMAL 408
Query: 425 VMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMIKAA---------AISNNEYMAMMK 475
VMG+VT+I++T ++PRK+TE+ LY D SM K+A AIS ++MA+MK
Sbjct: 409 VMGVVTMIKVTKNVPRKLTESTLYSSPVYCDDASMNKSAMQSEKMTVPAISGEDFMAIMK 468
Query: 476 RMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELEG 534
RMAELE+KVTVLS +P VMPP+KEEMLN A++R + LEQEL ATKKAL+D+L RQ EL
Sbjct: 469 RMAELEQKVTVLSAQPTVMPPDKEEMLNAAISRSNVLEQELAATKKALDDSLGRQEELVA 528
Query: 535 QIDKKKKKKK 544
I+KKKKKKK
Sbjct: 529 YIEKKKKKKK 538
>At2g18180 putative phosphatidylinositol/phophatidylcholine transfer
protein
Length = 558
Score = 613 bits (1580), Expect = e-175
Identities = 331/556 (59%), Positives = 404/556 (72%), Gaps = 37/556 (6%)
Query: 8 KKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICI-EDSFDAEELQAVDALRQTLIL 66
++ V S KK + SK S TKK R+ S+VMS+ I ED DAEEL+ VDA RQ LIL
Sbjct: 13 ERPNVCSFKK---RSCSKLSCSLTKK-RRSSKVMSVEIFEDEHDAEELKVVDAFRQVLIL 68
Query: 67 EELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVL 126
+ELLP KHDD HMMLRFL+ARK+D+EKT QMW+DML+WRKEFGADT+MEDFEF+E+DEVL
Sbjct: 69 DELLPDKHDDYHMMLRFLKARKFDLEKTNQMWSDMLRWRKEFGADTVMEDFEFKEIDEVL 128
Query: 127 KCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSI 186
K YPQGHHGVDK+GRPVYIERLGQVD KL+QVT+++RY+ YHV EFER F VK PACSI
Sbjct: 129 KYYPQGHHGVDKEGRPVYIERLGQVDSTKLMQVTTMDRYVNYHVMEFERTFNVKFPACSI 188
Query: 187 AAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRL 246
AAKKHIDQSTTILDVQGVGL++ NKAARDL+ RLQK+DGDNYPE+LNRMFIINAGSGFR+
Sbjct: 189 AAKKHIDQSTTILDVQGVGLKNFNKAARDLITRLQKVDGDNYPETLNRMFIINAGSGFRM 248
Query: 247 LWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKGP 306
LWNTVKSFLDPKTT+KIHVLGNKYQSKLLE+IDASELPEFLGG+CTCAD GGCM SDKGP
Sbjct: 249 LWNTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDASELPEFLGGSCTCADNGGCMRSDKGP 308
Query: 307 WNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQKGFKDSFPETLDV-HCLDQPKS 365
WN+P+I+K NG + + E +G + E + P
Sbjct: 309 WNNPDIMKRVNNG------DHICSKRSQADNAGENIISQGNNSAVEEAPETDQSQPSPCQ 362
Query: 366 YGVYQYDSF-VPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMAL 424
V + ++ +P K S + D YA+ + C + N S F+ G+MA
Sbjct: 363 NVVVAHPAWNIPEAHKFSLS------KRDVYAIQEACKATNESGRSPIFT-----GVMAF 411
Query: 425 VMGIVTIIRMTSSMPRKITEAALYGGNSVYYD----------GSMIKAAAISNNEYMAMM 474
VMG+VT+IR+T ++PRK+TE+ +Y + VY D G + IS ++MA+M
Sbjct: 412 VMGVVTMIRVTKNVPRKLTESTIY-SSPVYCDENSMNKSSMHGKKMATTTISGEDFMAVM 470
Query: 475 KRMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVELE 533
KRMAELE+KVT LS +P MPPEKEEMLN A++R LEQEL ATKKAL+D+LTRQ +L
Sbjct: 471 KRMAELEQKVTNLSAQPATMPPEKEEMLNAAISRADFLEQELAATKKALDDSLTRQEDLV 530
Query: 534 GQIDKKKKKKKKLVRY 549
++ +KKKKKKLVR+
Sbjct: 531 AYVE-RKKKKKKLVRF 545
>At4g39170 unknown protein
Length = 614
Score = 568 bits (1463), Expect = e-162
Identities = 317/590 (53%), Positives = 405/590 (67%), Gaps = 46/590 (7%)
Query: 2 EHSEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHS-RVMSICIEDSFDAEELQAVDAL 60
E+SEDE++ ++GS+KK A++AS+KFK+S KK RK RV S+ IED D EELQAVD
Sbjct: 30 ENSEDERRTRIGSLKKKAINASTKFKHSLKKKRRKSDVRVSSVSIEDVRDVEELQAVDEF 89
Query: 61 RQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFE 120
RQ L++EELLP KHDD HMMLRFL+ARK+DIEK K MW DM++WRKEFG DTI++DF+FE
Sbjct: 90 RQALVMEELLPHKHDDYHMMLRFLKARKFDIEKAKHMWADMIQWRKEFGTDTIIQDFQFE 149
Query: 121 ELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVK 180
E+DEVLK YP G+H VDK+GRPVYIERLG+VD NKL+QVT+++RY++YHV+EFER+F +K
Sbjct: 150 EIDEVLKYYPHGYHSVDKEGRPVYIERLGKVDPNKLMQVTTLDRYIRYHVKEFERSFMLK 209
Query: 181 LPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINA 240
PAC+IAAKK+ID STTILDVQGVGL++ K+AR+L+ RLQKIDGDNYPE+L++MFIINA
Sbjct: 210 FPACTIAAKKYIDSSTTILDVQGVGLKNFTKSARELITRLQKIDGDNYPETLHQMFIINA 269
Query: 241 GSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCM 300
G GFRLLW+TVKSFLDPKTTSKIHVLG KYQSKLLE+ID+SELPEFLGG CTCAD+GGCM
Sbjct: 270 GPGFRLLWSTVKSFLDPKTTSKIHVLGCKYQSKLLEIIDSSELPEFLGGACTCADQGGCM 329
Query: 301 LSDKGPWNDPEILKMAQNG---VGRYTIKALSGVEEKTIKQEETAYQ--KGFKDSFPETL 355
LSDKGPW +PEI+KM +G + +K L+ + K I + +Y KG S E+
Sbjct: 330 LSDKGPWKNPEIVKMVLHGGAHRAKQVVKVLNS-DGKVIAYAKPSYPWIKGSDTSTAESG 388
Query: 356 D--VHCLDQPKSYGVYQ--------------------------YDSFVPVLDKAVDSSWK 387
+ PK+ Y YD +VP++DKAVD++WK
Sbjct: 389 SEAEDIVVSPKAVKSYSHLRLTPVREEAKVGSGETSFAGSFAGYDEYVPMVDKAVDATWK 448
Query: 388 KTIQNDKYALSKDC-FSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAA 446
A SK N + FS + + MA VM I+T R S+ R +T+
Sbjct: 449 VKPTAINRAPSKGAHMPPNVPKDHESFSARVLVTFMAFVMAILTFFRTVSN--RVVTKQL 506
Query: 447 LYGGNSVYYDGSMIKAAAISNNEYMAMMKRMAELEEKVTVLSVKP-VMPPEKEEMLNNAL 505
+ +GS AAA + +++K++ ELEEK+ L KP MP EKEE+LN A+
Sbjct: 507 PPPPSQPQIEGS---AAAEEADLLNSVLKKLTELEEKIGALQSKPSEMPYEKEELLNAAV 563
Query: 506 TRVSTLEQELGATKKALEDALTRQVELEGQIDKKK----KKKKKLVRYIC 551
RV LE EL ATKKAL +AL RQ EL ID+++ +KK K + C
Sbjct: 564 CRVDALEAELIATKKALYEALMRQEELLAYIDRQEAAQHQKKNKRKQMFC 613
>At2g16380 putative phosphatidylinositol/phosphatidylcholine
transfer protein
Length = 582
Score = 556 bits (1434), Expect = e-159
Identities = 304/585 (51%), Positives = 391/585 (65%), Gaps = 59/585 (10%)
Query: 1 MEHSEDEKKK-KVGSIKKVALSASSKFKNSFTKKGRK-HSRVMSICIEDSFDAEELQAVD 58
ME+SED +K K+ S+K+ A+SAS++FKNSF KK R+ S+++S+ D + ++ +V+
Sbjct: 6 MENSEDGRKLVKMSSLKQKAISASNRFKNSFKKKTRRTSSKIVSVANTDDINGDDYLSVE 65
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
A RQ L+L++LLP KHDD HMMLRFLRARK+D EK KQMW+DML+WR +FG DTI+EDFE
Sbjct: 66 AFRQVLVLDDLLPPKHDDLHMMLRFLRARKFDKEKAKQMWSDMLQWRMDFGVDTIIEDFE 125
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFA 178
FEE+D+VLK YPQG+HGVDK+GRPVYIERLGQ+D NKLLQ T+++RY KYHV+EFE+ F
Sbjct: 126 FEEIDQVLKHYPQGYHGVDKEGRPVYIERLGQIDANKLLQATTMDRYEKYHVKEFEKMFK 185
Query: 179 VKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFII 238
+K P+CS AAKKHIDQSTTI DVQGVGL++ NK+AR+LLQRL KID DNYPE+LNRMFII
Sbjct: 186 IKFPSCSAAAKKHIDQSTTIFDVQGVGLKNFNKSARELLQRLLKIDNDNYPETLNRMFII 245
Query: 239 NAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGG 298
NAG GFRLLW +K FLDPKTTSKIHVLGNKYQ KLLE IDASELP F GG CTCADKGG
Sbjct: 246 NAGPGFRLLWAPIKKFLDPKTTSKIHVLGNKYQPKLLEAIDASELPYFFGGLCTCADKGG 305
Query: 299 CMLSDKGPWNDPEILKMAQNGVGRYTI------------------------KALSGVEEK 334
C+ SDKGPWNDPE+LK+A+N R++ + +EE
Sbjct: 306 CLRSDKGPWNDPELLKIARNPEARFSTISEEDYLLVEEGTSMSMVFEPLERNKMKTIEEN 365
Query: 335 TIKQEETAYQKGFKDSFPETLDVHCLDQPKSYGVYQYDSFVPVLDKAVDSSWKKTIQNDK 394
++ A K S P + L + Y Y S +P+ + + K Q
Sbjct: 366 VSEKHIDAVDKFMALSLPPKPHLKTL---RKVCRYMYCSKIPICLRLLHCYVGKEPQK-- 420
Query: 395 YALSKDCFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALYGGNSVY 454
D F VGG++A VMGIV ++R++ ++PRK+T+ AL NSVY
Sbjct: 421 ---KDDSF--------------LVGGVIAFVMGIVAMLRLSKAVPRKLTDVALL-TNSVY 462
Query: 455 YDGSM--------IKAAAISNNEYMAMMKRMAELEEKVTVLSVKPVMPP-EKEEMLNNAL 505
Y+ + + A +S++EY+ M+KRMAELEEK L K EK++ L AL
Sbjct: 463 YEEAKMSKPNQDEVSAPPVSSSEYVIMVKRMAELEEKYKSLDSKSADEALEKDDKLQAAL 522
Query: 506 TRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKKKKLVRYI 550
RV LE EL TKKAL++ + Q + I+ KK KKK++VR I
Sbjct: 523 NRVQVLEHELSETKKALDETMVNQQGILAYIE-KKNKKKRMVRQI 566
>At2g21540 putative phosphatidylinositol/phophatidylcholine transfer
protein
Length = 371
Score = 556 bits (1432), Expect = e-158
Identities = 268/350 (76%), Positives = 309/350 (87%), Gaps = 2/350 (0%)
Query: 4 SEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEELQAVDALRQT 63
SEDEKK K+ S+KK A++AS+KFK+SFTK+ R++SRVMS+ I D D EELQAVDA RQ
Sbjct: 20 SEDEKKTKLCSLKKKAINASNKFKHSFTKRTRRNSRVMSVSIVDDIDLEELQAVDAFRQA 79
Query: 64 LILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELD 123
LIL+ELLPSKHDD HMMLRFLRARK+D+EK KQMWTDM+ WRKEFG DTIMEDF+F+E+D
Sbjct: 80 LILDELLPSKHDDHHMMLRFLRARKFDLEKAKQMWTDMIHWRKEFGVDTIMEDFDFKEID 139
Query: 124 EVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPA 183
EVLK YPQG+HGVDKDGRPVYIERLGQVD KL+QVT+++RY+KYHVREFE+ F +KLPA
Sbjct: 140 EVLKYYPQGYHGVDKDGRPVYIERLGQVDATKLMQVTTIDRYVKYHVREFEKTFNIKLPA 199
Query: 184 CSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSG 243
CSIAAKKHIDQSTTILDVQGVGL+S +KAARDLLQR+QKID DNYPE+LNRMFIINAGSG
Sbjct: 200 CSIAAKKHIDQSTTILDVQGVGLKSFSKAARDLLQRIQKIDSDNYPETLNRMFIINAGSG 259
Query: 244 FRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSD 303
FRLLW+TVKSFLDPKTT+KIHVLGNKYQSKLLE+ID++ELPEFLGG CTCADKGGCM SD
Sbjct: 260 FRLLWSTVKSFLDPKTTAKIHVLGNKYQSKLLEIIDSNELPEFLGGNCTCADKGGCMRSD 319
Query: 304 KGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEE--TAYQKGFKDSF 351
KGPWNDP+I KM QNG G+ K LS +EEKTI +E T + +K+ F
Sbjct: 320 KGPWNDPDIFKMVQNGEGKCPRKTLSNIEEKTISVDENTTMVKAKYKNIF 369
>At1g75370 sec14 cytosolic factor, putative
Length = 612
Score = 507 bits (1306), Expect = e-144
Identities = 296/593 (49%), Positives = 379/593 (62%), Gaps = 54/593 (9%)
Query: 2 EHSEDEKKKKVGSI--KKVALSASSKFKNSFTKKG----RKHSRVMSICIEDSFDAEELQ 55
E SEDEKK ++G+ KK A ASSK ++S KKG R R S+ IED D EEL+
Sbjct: 30 EVSEDEKKTRIGNFNFKKKAAKASSKLRHSLKKKGSSRRRSSDRTFSLTIEDIHDVEELR 89
Query: 56 AVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIME 115
AVD R L+ E LLP DD H+MLRFL+ARK+DI KTK MW++M+KWRK+FG DTI E
Sbjct: 90 AVDEFRNLLVSENLLPPTLDDYHIMLRFLKARKFDIGKTKLMWSNMIKWRKDFGTDTIFE 149
Query: 116 DFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFER 175
DFEFEE DEVLK YP G+HGVDK+GRPVYIERLG VD KL+QVT+VER+++YHVREFE+
Sbjct: 150 DFEFEEFDEVLKYYPHGYHGVDKEGRPVYIERLGLVDPAKLMQVTTVERFIRYHVREFEK 209
Query: 176 AFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRM 235
+KLPAC IAAK+HID STTILDVQGVG ++ +K ARDL+ +LQKID DNYPE+L+RM
Sbjct: 210 TVNIKLPACCIAAKRHIDSSTTILDVQGVGFKNFSKPARDLIIQLQKIDNDNYPETLHRM 269
Query: 236 FIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCAD 295
FIIN GSGF+L+W TVK FLDPKT +KIHV+GNKYQ+KLLE+IDAS+LP+FLGGTCTCAD
Sbjct: 270 FIINGGSGFKLVWATVKQFLDPKTVTKIHVIGNKYQNKLLEIIDASQLPDFLGGTCTCAD 329
Query: 296 KGGCMLSDKGPWNDPEILKMAQNG---------------VGRYTIKALSGVE-------E 333
+GGCM SDKGPWNDPEILKM Q+G V + SG++ E
Sbjct: 330 RGGCMRSDKGPWNDPEILKMLQSGGPLCRHNSALNSFSRVSSCDKPSFSGIKASDTSTAE 389
Query: 334 KTIKQEETAYQKGFKDSFPETLDVHCLDQPKSYGVY-----QYDSFVPVLDKAVDSSWKK 388
+ EE A K ++ L C D + Y +YDS P++DK VD +W
Sbjct: 390 SGSEVEEMASPKVNRELRVPKLTPVCEDIRGTAISYPTDSSEYDS--PMVDKVVDVAW-- 445
Query: 389 TIQNDKYALSKDCFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALY 448
+ ++K SK + SG + L+M V + + S+ E
Sbjct: 446 -MAHEKPKASK----GSEDTPDSGKIRTVTYIWRWLMMFFVNLFTLLISLALPQREGHSQ 500
Query: 449 GGNSVYYDG---------SMIKAAAISNNEYMAMMKRMAELEEKV-TVLSVKPVMPPEKE 498
+SV DG S A N + +++ R+ +LE++V T+ S + MP EKE
Sbjct: 501 SESSV--DGPNARESRPPSPAFATIAERNVFSSVVNRLGDLEKQVETLHSKRHEMPREKE 558
Query: 499 EMLNNALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKKKKLVRYIC 551
E+LN A+ RV LE EL ATKKAL +AL RQ +L ID+++ +K + +C
Sbjct: 559 ELLNTAVYRVDALEAELIATKKALHEALMRQDDLLAYIDREEDEKYHKKKKVC 611
>At2g21520 putative phosphatidylinositol/phosphatidylcholine
transfer protein
Length = 531
Score = 497 bits (1280), Expect = e-141
Identities = 274/527 (51%), Positives = 355/527 (66%), Gaps = 48/527 (9%)
Query: 66 LEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEV 125
++ELLP +HDD HMMLRFL+ARK+D+EK KQMW DM++WRKEFG DTI++DF+FEE++EV
Sbjct: 1 MDELLPDRHDDYHMMLRFLKARKFDVEKAKQMWADMIQWRKEFGTDTIIQDFDFEEINEV 60
Query: 126 LKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACS 185
LK YPQ +HGVDK+GRP+YIERLG+VD N+L+QVTS++RY++YHV+EFER+F +K P+C+
Sbjct: 61 LKHYPQCYHGVDKEGRPIYIERLGKVDPNRLMQVTSMDRYVRYHVKEFERSFMIKFPSCT 120
Query: 186 IAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFR 245
I+AK+HID STTILDVQGVGL++ NK+ARDL+ RLQKIDGDNYPE+L++MFIINAG GFR
Sbjct: 121 ISAKRHIDSSTTILDVQGVGLKNFNKSARDLITRLQKIDGDNYPETLHQMFIINAGPGFR 180
Query: 246 LLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLSDKG 305
LLWNTVKSFLDPKT++KIHVLG KY SKLLEVID +ELPEFLGG CTCAD+GGCMLSDKG
Sbjct: 181 LLWNTVKSFLDPKTSAKIHVLGYKYLSKLLEVIDVNELPEFLGGACTCADQGGCMLSDKG 240
Query: 306 PWNDPEILKMAQNG---VGRYTIKALSGVEEKTIKQEETAYQ--KGFKDSFPET-LDVHC 359
PW +PEI+KM +G R +K L+ E K I + +Y KG S E+ D
Sbjct: 241 PWKNPEIVKMVLHGGAHRARQVVKVLNS-EGKVIAYAKPSYTWIKGSDTSTAESGSDAED 299
Query: 360 LDQPKSYGVYQ-------------------YDSFVPVLDKAVDSSWKKTIQNDKYALSKD 400
+ PK+ + YD +VP++DKAVD++WK + A
Sbjct: 300 IGSPKAIKSFSHLRLTPVPGETSLAGSFPGYDEYVPMVDKAVDATWKVKPAIQRVASRGA 359
Query: 401 CFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIR-MTSSMPRKITEA-ALYGGNSVYY--D 456
S + G + + MA +M + T R +T +P T + A GN++ +
Sbjct: 360 LMSPTVPKDHEGIKARVLVMFMAFLMAVFTFFRTVTKKLPATTTSSPAETQGNAIELGSN 419
Query: 457 GSMIKAAA--------ISNNEYM-AMMKRMAELEEKVTVLSVKP-VMPPEKEEMLNNALT 506
G +K ++ + + + K++ ELE K+ L KP MP EKEE+LN A+
Sbjct: 420 GEGVKEECRPPSPVPDLTETDLLNCVTKKLTELEGKIGTLQSKPNEMPYEKEELLNAAVC 479
Query: 507 RVSTLEQELGATKKALEDALTRQVEL--------EGQIDKKKKKKKK 545
RV LE EL ATKKAL +AL RQ EL E Q K KKKKKK
Sbjct: 480 RVDALEAELIATKKALYEALMRQEELLAYIDRQEEAQFQKMKKKKKK 526
>At1g19650 sec14 cytosolic factor- like protein
Length = 608
Score = 493 bits (1268), Expect = e-139
Identities = 281/581 (48%), Positives = 383/581 (65%), Gaps = 46/581 (7%)
Query: 2 EHSEDEKKKKVGSIKKVALSASSKFKNSFTKKG-RKHSRVMSICIEDSFDAEELQAVDAL 60
E SEDEKK ++G I K S+ SKF++S ++G R R +S+ ED DAEEL+ V
Sbjct: 27 EVSEDEKKTRIGGILKKK-SSKSKFRHSLKRRGSRSIDRTLSLTFEDIHDAEELRYVSEF 85
Query: 61 RQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFE 120
RQ+LI + LLP DD H+MLRFL ARK+D+ K K MWT+M++WR++FG DTI+EDFEF
Sbjct: 86 RQSLISDHLLPPNLDDYHIMLRFLFARKFDLGKAKLMWTNMIQWRRDFGTDTILEDFEFP 145
Query: 121 ELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVK 180
ELDEVL+ YPQG+HGVDK+GRPVYIERLG+VD +KL+QVT++ERYL+YHV+EFE+ VK
Sbjct: 146 ELDEVLRYYPQGYHGVDKEGRPVYIERLGKVDASKLMQVTTLERYLRYHVKEFEKTITVK 205
Query: 181 LPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINA 240
PAC IAAK+HID STTILDVQG+GL++ K ARDL+ +LQKID DNYPE+L+RMFIINA
Sbjct: 206 FPACCIAAKRHIDSSTTILDVQGLGLKNFTKTARDLIIQLQKIDSDNYPETLHRMFIINA 265
Query: 241 GSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCM 300
GSGF+LLW TVKSFLDPKT SKIHVLGNKYQ+KLLE+IDAS+LP+F GGTCTCAD+GGCM
Sbjct: 266 GSGFKLLWGTVKSFLDPKTVSKIHVLGNKYQNKLLEMIDASQLPDFFGGTCTCADQGGCM 325
Query: 301 LSDKGPWNDPEILKMAQNG------VGRY--TIKALSGVEEKT--IKQEETAYQKGFKD- 349
SDKGPW D EILKM ++G G + + +S ++ T +K +T+ K +
Sbjct: 326 RSDKGPWKDSEILKMGRSGGTFCRHAGAFLSSDSQISSSDKPTYSLKVSDTSTAKSGSEL 385
Query: 350 ---SFPETLDVHCLDQPKSYGVY-----------QYDSFVPVLDKAVDSSWKKTIQNDKY 395
+ P+T + + + Y +Y+ VP++DK VD +W+ +Q
Sbjct: 386 EEMASPKTNTNNHVPKLTPVSEYANGNISPTVLSEYEECVPMVDKVVDVAWQ--LQEMPN 443
Query: 396 ALSKDCFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALYGGNSVYY 455
A ++++ G G + + A + T++ + ++P+ + L+ +SV
Sbjct: 444 ASEGPQYTSSLG--KIGSVRHIWSWLTAFFISFFTLL-ASLALPQTKEHSQLH-SSSVRA 499
Query: 456 D------------GSMIKAAAISNNEYMAMMKRMAELEEKVTVL-SVKPVMPPEKEEMLN 502
+ S ++ +++ R+ +LE+++ L S K MP EKEE+LN
Sbjct: 500 ELCDERIARESRPPSPPRSTITERVIISSVLSRLGDLEKQIENLHSRKSEMPHEKEELLN 559
Query: 503 NALTRVSTLEQELGATKKALEDALTRQVELEGQIDKKKKKK 543
A+ RV LE EL TKKAL +AL RQ EL G ID++K+ K
Sbjct: 560 AAVYRVDALEAELITTKKALHEALIRQEELLGYIDRQKEAK 600
>At3g24840 phosphatidylinositol transfer protein like
Length = 579
Score = 439 bits (1128), Expect = e-123
Identities = 252/569 (44%), Positives = 359/569 (62%), Gaps = 46/569 (8%)
Query: 1 MEHSEDEK--KKKVGSIKKVALSASSKFKNSFTKKGRKHS-RVMSICIEDSFDAEELQAV 57
+E SEDEK + + S+KK A+ AS+K +S K+G++ + + I IED D EE +AV
Sbjct: 22 IEVSEDEKITRTRSRSLKKKAIKASNKLTHSLRKRGKRVADQYAPIVIEDVRDEEEEKAV 81
Query: 58 DALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDF 117
+ R+ L+ +LLP +HDD H MLRFL+AR++D+EKT QMW +MLKWRKE G DTI++DF
Sbjct: 82 NVFRKALVSLDLLPPRHDDYHTMLRFLKARRFDLEKTVQMWEEMLKWRKENGVDTIIQDF 141
Query: 118 EFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAF 177
++E +EV + YP G+HGVD++GRPVYIERLG++D KL++VT++ER+L+YHV+ FE+ F
Sbjct: 142 VYDEYEEVQQYYPHGYHGVDREGRPVYIERLGKIDPGKLMKVTTLERFLRYHVQGFEKTF 201
Query: 178 AVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFI 237
+ K PACSIAAK+HI+ STTI+DV GV S K A+DL+ R+QKIDGDNYPE+LN+M+I
Sbjct: 202 SEKFPACSIAAKRHINSSTTIIDVHGVSWMSFRKLAQDLVMRMQKIDGDNYPETLNQMYI 261
Query: 238 INAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKG 297
INAG+GF+L+WNTVK FLDPKTTSKIHVLGNKY+S LLE+ID SELPEFLGG C CA +G
Sbjct: 262 INAGNGFKLVWNTVKGFLDPKTTSKIHVLGNKYRSHLLEIIDPSELPEFLGGNCKCAHEG 321
Query: 298 GCMLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEE---------KTIKQEETAYQKGF- 347
GCM +KGPWNDPEI+K+ ++ Y K + +E + + + ++ G
Sbjct: 322 GCMRFNKGPWNDPEIMKLVRSRDAMYKPKEMGLLENGEVAKLFSLRHVNTDMSSPDGGHV 381
Query: 348 --KDSFPE-TLDVHCLDQPKSYGV---YQYDSFVPVLDK-AVDSSWKKTIQNDKYALSKD 400
++S PE +Q ++ GV Q DS P+ + AV+ S ++Q
Sbjct: 382 RERESHPEHDKRAQLSNQAEAVGVGRMEQSDSTSPLPNNLAVERSLTTSLQ--------- 432
Query: 401 CFSNNNGMNSSGFSKQFVGGIMALVMGIVTIIRMTSSMPRKITEAALYGGNSVYYDGSMI 460
+ F +F I+ L+ + + R+ + K E L SV +
Sbjct: 433 --------KVASFLARF---ILQLLGSLCLMFRILGRLVNKQPENQLRPELSV-----SV 476
Query: 461 KAAAISNNEYMAMMKRMAELEEKVTVLSVKP-VMPPEKEEMLNNALTRVSTLEQELGATK 519
+ + R+ LE VTVL KP +P EKE++L ++L R+ ++EQ+L TK
Sbjct: 477 SQQQVPPPQVHPCWLRLQNLETMVTVLCDKPSSIPQEKEDILRDSLDRIKSIEQDLQKTK 536
Query: 520 KALEDALTRQVELEGQIDKKKKKKKKLVR 548
KAL ++Q+EL + K+ +R
Sbjct: 537 KALFLTASKQIELAECFENLKESSSTGMR 565
>At5g56160 putative protein
Length = 592
Score = 377 bits (969), Expect = e-105
Identities = 217/565 (38%), Positives = 340/565 (59%), Gaps = 32/565 (5%)
Query: 5 EDEKKKKVGSIKKVALSASSKFKNSFT-KKGRKHSRVMSICIEDSFDAEELQAVDALRQT 63
E+ ++ ++G++KK A S S+K + +KG++ IED D +E + V LRQ
Sbjct: 34 EEPRRSRIGNLKKKAFSCSTKLTHPLKMRKGKRKIDFQIPLIEDVRDEKEEKLVSKLRQQ 93
Query: 64 LILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELD 123
L+ ++LLP HDD HM+LRFL+ ++ IEKT W +MLKWRKEFG D I++DF F+ELD
Sbjct: 94 LLQKDLLPPVHDDYHMLLRFLKTMEFKIEKTVTAWEEMLKWRKEFGTDRIIQDFNFKELD 153
Query: 124 EVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPA 183
EV + YPQG+HGVDKDGRP+YIERLG+ KL++VT++ERYLKYHV+EFER KLPA
Sbjct: 154 EVTRHYPQGYHGVDKDGRPIYIERLGKAHPGKLMEVTTIERYLKYHVQEFERTLQEKLPA 213
Query: 184 CSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSG 243
CS+AAK+ + +TTILDV+G+G+++ A +LL + K+D + YPE+L+RMFI+NAG G
Sbjct: 214 CSVAAKRRVTTTTTILDVEGLGMKNFTPTAANLLATIAKVDCNYYPETLHRMFIVNAGIG 273
Query: 244 FR-LLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTCADKGGCMLS 302
FR LW + LDP T +KI VL + SKLLE ID+S+LPEFLGG C C ++GGC+ S
Sbjct: 274 FRSFLWPAAQKLLDPMTIAKIQVLEPRSLSKLLEAIDSSQLPEFLGGLCKCPNEGGCLRS 333
Query: 303 DKGPWNDPEILKMAQ----NGVGRYTIKALS---------GVEEKTIKQEETAYQKGFKD 349
+KGPWNDPEI+++ N V + T L + K +EE ++ +
Sbjct: 334 NKGPWNDPEIVELVHHMEVNNVPQTTTAPLHVRDYDSTTCTISPKETLKEEPEPEEYYSS 393
Query: 350 SFPETLDVHCLDQPKS--YGVYQYDSFVPVLDKAVDSSWKKTIQNDKYALSKDCFSNNNG 407
+ + C+ P S D F+ ++ +++S+ + + D +++ F+N +
Sbjct: 394 TGSRSSMHTCIVPPLSDKASTSDGDKFITTVE-SIESAQSQLLDAD----TENTFANTS- 447
Query: 408 MNSSGFSKQFVGGIMALVMG--IVTIIRMTSSMPRKITEAALYGGNSVYY---DGSMIKA 462
+ G +F G + + I ++++ P K+ L+G Y +++
Sbjct: 448 VREGGQILRF-GALREKINSENIFHLVKILLVFPLKLF--VLFGFLLPGYWQRQNTVVVP 504
Query: 463 AAISNNEYMAMMKRMAELEEKVTVLSVKPVMPPE-KEEMLNNALTRVSTLEQELGATKKA 521
+ +NN+ + R+ ++E++ T +S K V PE E++L +L R+ +LE +L TK
Sbjct: 505 DSSTNNKVLECFDRLKKMEKEFTEISRKQVKIPEANEKLLAESLERIKSLELDLDKTKSV 564
Query: 522 LEDALTRQVELEGQIDKKKKKKKKL 546
L LT+Q+++ Q++ + ++ L
Sbjct: 565 LHITLTKQLQITEQLESQDEEVSPL 589
>At1g55690 Phosphatidylinositol Transfer Protein like
Length = 625
Score = 371 bits (953), Expect = e-103
Identities = 180/323 (55%), Positives = 250/323 (76%), Gaps = 5/323 (1%)
Query: 2 EHSEDEKKK-KVGSIKKVALSASSKFKNSFTKKGRKHS--RVMSICIEDSFDAEELQAVD 58
E SEDE+++ K+G++KK A++AS+KF +S K+G++ RV ++ IED D +E V
Sbjct: 21 EISEDERRRSKIGNLKKKAINASTKFTHSLKKRGKRKIDYRVPAVSIEDVRDEKEESVVL 80
Query: 59 ALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFE 118
R+ L+ +LLP +HD+ H +LRFL+AR +IEKT Q+W +ML+WRKE+G DTI+EDF+
Sbjct: 81 EFRRKLLERDLLPPRHDEYHTLLRFLKARDLNIEKTTQLWEEMLRWRKEYGTDTILEDFD 140
Query: 119 FEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFA 178
FEEL+EVL+ YPQG+HGVDK+GRPVYIERLG+ +KL+++T+++RYLKYHV+EFERA
Sbjct: 141 FEELEEVLQYYPQGYHGVDKEGRPVYIERLGKAHPSKLMRITTIDRYLKYHVQEFERALQ 200
Query: 179 VKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFII 238
K PACSIAAK+ I +TTILDVQG+G+++ A +L+ + KID YPE+L+RM+I+
Sbjct: 201 EKFPACSIAAKRRICSTTTILDVQGLGIKNFTPTAANLVAAMSKIDNSYYPETLHRMYIV 260
Query: 239 NAGSGF-RLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPEFLGGTCTC-ADK 296
NAG+GF ++LW + FLD KT +KIHVL K KL EVID+S+LPEFLGG+C+C D
Sbjct: 261 NAGTGFKKMLWPAAQKFLDAKTIAKIHVLEPKSLFKLHEVIDSSQLPEFLGGSCSCFGDG 320
Query: 297 GGCMLSDKGPWNDPEILKMAQNG 319
GGC+ S+KGPWNDPEI+K+ +G
Sbjct: 321 GGCLRSNKGPWNDPEIMKLIYHG 343
Score = 41.2 bits (95), Expect = 0.002
Identities = 23/80 (28%), Positives = 45/80 (55%), Gaps = 1/80 (1%)
Query: 474 MKRMAELEEKVTVLSVKPV-MPPEKEEMLNNALTRVSTLEQELGATKKALEDALTRQVEL 532
++R+ +LE+ + KPV +P EKE ML ++L R+ ++E +L TK+ L + +Q+E+
Sbjct: 546 LERIQKLEKSYEDIRNKPVAIPVEKERMLMDSLDRIKSVEFDLDKTKRLLHATVMKQMEI 605
Query: 533 EGQIDKKKKKKKKLVRYICC 552
+ + + R + C
Sbjct: 606 TEMLQNIRDSQLHRRRRLFC 625
>At5g47510 putative protein
Length = 403
Score = 302 bits (773), Expect = 4e-82
Identities = 146/308 (47%), Positives = 204/308 (65%), Gaps = 5/308 (1%)
Query: 46 EDSFDAEELQAVDALRQTLILEELLPSKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWR 105
E S + EE+ V+A R L+L LP KH D + + RFL+ R +D+EK+K+ + + +KWR
Sbjct: 19 EQSPNNEEM--VEAFRNLLLLHGHLPDKHGDHNTLRRFLKMRDFDLEKSKEAFLNYMKWR 76
Query: 106 KEFGADTIMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERY 165
++ D I + F+FEE EV K YP G H VDK GRP+YIERLG D N L+ T++ERY
Sbjct: 77 VDYKVDLISQKFKFEEYGEVKKHYPHGFHKVDKTGRPIYIERLGMTDLNAFLKATTIERY 136
Query: 166 LKYHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDG 225
+ YH++E E+ +++ PACSIA+ KH+ +TTILDV GVG+ + +K AR L +QKID
Sbjct: 137 VNYHIKEQEKTMSLRYPACSIASDKHVSSTTTILDVSGVGMSNFSKPARSLFMEIQKIDS 196
Query: 226 DNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPE 285
+ YPE+L+R+F++NA SGFR+LW +K+FLD +T +K+ VLG Y +LLE I+ S LP
Sbjct: 197 NYYPETLHRLFVVNASSGFRMLWLALKTFLDARTLAKVQVLGPNYLGELLEAIEPSNLPT 256
Query: 286 FLGGTCTCADKGGCMLSDKGPWNDPEILKMAQNGVGRYTIKALSGVEEKTIKQEETAYQK 345
FLGG CTC+D GGC+ SD+GPWNDP I + + TI+ + + A QK
Sbjct: 257 FLGGNCTCSDHGGCLFSDEGPWNDPGIKEKIEE---PSTIEDAHSETMDKVSENAPANQK 313
Query: 346 GFKDSFPE 353
KDS E
Sbjct: 314 VHKDSVNE 321
>At5g47730 putative protein
Length = 341
Score = 92.0 bits (227), Expect = 8e-19
Identities = 68/241 (28%), Positives = 117/241 (48%), Gaps = 15/241 (6%)
Query: 55 QAVDALRQTLI-LEELLPSKHDDPHM------MLRFLRARKYDIEKTKQMWTDMLKWRKE 107
+A+D ++ + +EE L ++ H + RFL+AR +++ K M + L+WR +
Sbjct: 7 EAIDEFQELMDQVEEPLKKTYERVHQGYLRENLGRFLKARDWNVCKAHTMLVECLRWRVD 66
Query: 108 FGADTIMED--FEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERY 165
D+I+ E +V G G K+G PV+ +G +K SV Y
Sbjct: 67 NEIDSILSKPIVPTELYRDVRDSQLIGMSGYTKEGLPVFAIGVGLSTFDK----ASVHYY 122
Query: 166 LKYHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDG 225
++ H++ E V LP+ S + I +LD+ G+ L ++++ L+ + ID
Sbjct: 123 VQSHIQINEYRDRVLLPSISKKNGRPITTCVKVLDMTGLKLSALSQIK--LVTIISTIDD 180
Query: 226 DNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLGNKYQSKLLEVIDASELPE 285
NYPE N +++NA F W VK L +T K+HVL + +LL+++D + LP
Sbjct: 181 LNYPEKTNTYYVVNAPYIFSACWKVVKPLLQERTRKKVHVLSGCGRDELLKIMDFTSLPH 240
Query: 286 F 286
F
Sbjct: 241 F 241
>At1g55840 polyphosphoinositide binding protein, putative
Length = 325
Score = 84.7 bits (208), Expect = 1e-16
Identities = 60/209 (28%), Positives = 99/209 (46%), Gaps = 8/209 (3%)
Query: 80 MLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMED--FEFEELDEVLKCYPQGHHGVD 137
+LRFL+AR +++K +M + L+WR + D I+ + + G G
Sbjct: 39 LLRFLKARDGNVQKAHKMLLECLEWRTQNEIDKILTKPIVPVDLYRGIRDTQLVGVSGYS 98
Query: 138 KDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKKHIDQSTT 197
K+G PV +G +K SV Y++ H++ E V LP+ S + I
Sbjct: 99 KEGLPVIAIGVGLSTYDK----ASVHYYVQSHIQMNEYRDRVVLPSASKKQGRPICTCLK 154
Query: 198 ILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKSFLDP 257
ILD+ G+ L ++++ L+ + ID NYPE +++N F W T+K L
Sbjct: 155 ILDMSGLKLSALSQIK--LMTAITTIDDLNYPEKTETYYVVNVPYIFSACWKTIKPLLQE 212
Query: 258 KTTSKIHVLGNKYQSKLLEVIDASELPEF 286
+T KI VL + +LL+++D LP F
Sbjct: 213 RTKKKIQVLKGCGKDELLKIMDYESLPHF 241
>At1g72160 cytosolic factor, putative
Length = 490
Score = 84.3 bits (207), Expect = 2e-16
Identities = 75/311 (24%), Positives = 143/311 (45%), Gaps = 32/311 (10%)
Query: 4 SEDEKKKKVGSIKKVALSASSKFKNSFTKKGRKHSRVMSICIEDSFDAEEL-----QAVD 58
+E ++ K S K+VA S + + S++ + + +EL +A+D
Sbjct: 80 TEKQEVKDEASQKEVAEEKKSMIPQNLGSFKEESSKLSDLSNSEKKSLDELKHLVREALD 139
Query: 59 ALRQTLILEEL----LPSKHDDPH--MMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADT 112
+ T EE+ +P DD ++L+FLRAR++ ++ + M + +KWRKEF D
Sbjct: 140 NHQFTNTPEEVKIWGIPLLEDDRSDVVLLKFLRAREFKVKDSFAMLKNTIKWRKEFKIDE 199
Query: 113 IMEDFEFEELDEVLKCYPQGHHGVDKDGRPVYIERLGQVDCNKLLQVT-----SVERYLK 167
++E+ ++LD+V+ HG D++G PV G+ +L T + +L+
Sbjct: 200 LVEEDLVDDLDKVVFM-----HGHDREGHPVCYNVYGEFQNKELYNKTFSDEEKRKHFLR 254
Query: 168 YHVREFERAFAVKLPACSIAAKKHIDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDN 227
++ ER +++ S I Q + + G+G + + A + ++ LQ DN
Sbjct: 255 TRIQFLER--SIRKLDFSSGGVSTIFQVNDMKNSPGLGKKELRSATKQAVELLQ----DN 308
Query: 228 YPESLNRMFIINAGSGFRLLWNTVKSFLDPKTTSKIHVLG-NKYQSKLLEVIDASELPEF 286
YPE + + IN + + + + F+ P++ SK+ G ++ L + I ++P
Sbjct: 309 YPEFVFKQAFINVPWWYLVFYTVIGPFMTPRSKSKLVFAGPSRSAETLFKYISPEQVPVQ 368
Query: 287 LGG----TCTC 293
GG C C
Sbjct: 369 YGGLSVDPCDC 379
>At1g01630 polyphosphoinositide binding protein, putative
Length = 255
Score = 76.3 bits (186), Expect = 4e-14
Identities = 65/219 (29%), Positives = 96/219 (43%), Gaps = 19/219 (8%)
Query: 72 SKHDDPHMMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQ 131
+K D M+ RFLRAR DIEK M+ + L W++ + + E K Q
Sbjct: 45 TKEVDDLMIRRFLRARDLDIEKASTMFLNYLTWKRSMLPKGHIPEAEIANDLSHNKMCMQ 104
Query: 132 GHHGVDKDGRPVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKKH 191
GH DK GRP+ + + + K + EF+R L +
Sbjct: 105 GH---DKMGRPIAVA------------IGNRHNPSKGNPDEFKRFVVYTLEKICARMPRG 149
Query: 192 IDQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTV 251
++ I D+QG G + + R L L + D YPE L +++I++A F W +
Sbjct: 150 QEKFVAIGDLQGWGYSNCD--IRGYLAALSTLQ-DCYPERLGKLYIVHAPYIFMTAWKVI 206
Query: 252 KSFLDPKTTSKIHVLGN-KYQSKLLEVIDASELPEFLGG 289
F+D T KI + N K LLE ID S+LP+ GG
Sbjct: 207 YPFIDANTKKKIVFVENKKLTPTLLEDIDESQLPDIYGG 245
>At1g75170 unknown protein
Length = 296
Score = 69.7 bits (169), Expect = 4e-12
Identities = 66/214 (30%), Positives = 99/214 (45%), Gaps = 28/214 (13%)
Query: 82 RFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGHHGVDKDGR 141
R+L AR +++ K K+M + LKWR F + I + E E K Y G H D+ GR
Sbjct: 49 RYLEARNWNVGKAKKMLEETLKWRSSFKPEEIRWN-EVSGEGETGKVYKAGFH--DRHGR 105
Query: 142 PVYIERLGQVDCNKLLQVTSVERYLKYHVREFERAFAVKLPACSIAAKKHIDQSTTILDV 201
V I R G L S+E +K+ V E A + LP + +Q + ++D
Sbjct: 106 TVLILRPG------LQNTKSLENQMKHLVYLIENAI-LNLP-------EDQEQMSWLIDF 151
Query: 202 QGVGLRSM--NKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVKSFLDPKT 259
G + + K+AR+ + LQ ++YPE L F+ N F W VK F+D KT
Sbjct: 152 TGWSMSTSVPIKSARETINILQ----NHYPERLAVAFLYNPPRLFEAFWKIVKYFIDAKT 207
Query: 260 TSKIHVLGNKYQSKLLEVI----DASELPEFLGG 289
K+ + K S+ +E++ D LP GG
Sbjct: 208 FVKVKFVYPK-NSESVELMSTFFDEENLPTEFGG 240
>At4g09160 putative protein
Length = 723
Score = 67.8 bits (164), Expect = 2e-11
Identities = 56/226 (24%), Positives = 104/226 (45%), Gaps = 23/226 (10%)
Query: 79 MMLRFLRARKYDIEKTKQMWTDMLKWRKEFGADTIMEDFEFEELDEVLKCYPQGHHGVDK 138
++L+FLRAR + ++ M L+WR +F + ++++ ++LD+V+ + QG DK
Sbjct: 341 VLLKFLRARDFKPQEAYSMLNKTLQWRIDFNIEELLDENLGDDLDKVV--FMQGQ---DK 395
Query: 139 DGRPVYIERLGQVDCNKLLQVT-----SVERYLKYHVREFERAFA-VKLPACSIAAKKHI 192
+ PV G+ L Q T ER+L++ ++ E++ + A ++ I
Sbjct: 396 ENHPVCYNVYGEFQNKDLYQKTFSDEEKRERFLRWRIQFLEKSIRNLDFVAGGVST---I 452
Query: 193 DQSTTILDVQGVGLRSMNKAARDLLQRLQKIDGDNYPESLNRMFIINAGSGFRLLWNTVK 252
Q + + G G + A + L LQ DNYPE +++ IN + + +
Sbjct: 453 CQVNDLKNSPGPGKTELRLATKQALHLLQ----DNYPEFVSKQIFINVPWWYLAFYRIIS 508
Query: 253 SFLDPKTTSKIHVLG-NKYQSKLLEVIDASELPEFLGG----TCTC 293
F+ ++ SK+ G ++ LL+ I +P GG C C
Sbjct: 509 PFMSQRSKSKLVFAGPSRSAETLLKYISPEHVPVQYGGLSVDNCEC 554
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,478,657
Number of Sequences: 26719
Number of extensions: 547926
Number of successful extensions: 2139
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 2025
Number of HSP's gapped (non-prelim): 97
length of query: 555
length of database: 11,318,596
effective HSP length: 104
effective length of query: 451
effective length of database: 8,539,820
effective search space: 3851458820
effective search space used: 3851458820
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140722.2


