

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.8 + phase: 0 /pseudo
(59 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
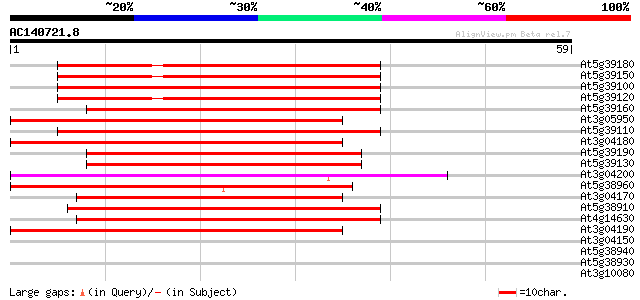
Score E
Sequences producing significant alignments: (bits) Value
At5g39180 germin-like protein 48 8e-07
At5g39150 germin - like protein 48 8e-07
At5g39100 germin-like protein (GLP6) 48 1e-06
At5g39120 germin - like protein 47 1e-06
At5g39160 germin-like protein (GLP2a) copy1 44 1e-05
At3g05950 germin-like protein 44 1e-05
At5g39110 germin -like protein 44 2e-05
At3g04180 germin-like protein 43 3e-05
At5g39190 germin-like protein (GLP2a) copy2 42 4e-05
At5g39130 germin - like protein 42 4e-05
At3g04200 germin-like protein 42 6e-05
At5g38960 germin - like protein 41 1e-04
At3g04170 germin-like protein 41 1e-04
At5g38910 germin - like protein 41 1e-04
At4g14630 germin precursor oxalate oxidase 41 1e-04
At3g04190 germin-like protein 41 1e-04
At3g04150 germin-like protein 37 0.002
At5g38940 germin - like protein 35 0.007
At5g38930 germin - like protein 34 0.016
At3g10080 germin-like protein 30 0.17
>At5g39180 germin-like protein
Length = 221
Score = 48.1 bits (113), Expect = 8e-07
Identities = 23/34 (67%), Positives = 29/34 (84%), Gaps = 1/34 (2%)
Query: 6 LLVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
+L+++ AL + IA AYDPSPLQDFCVAI DPK+G
Sbjct: 8 ILITLSALVT-IAKAYDPSPLQDFCVAIDDPKNG 40
>At5g39150 germin - like protein
Length = 221
Score = 48.1 bits (113), Expect = 8e-07
Identities = 23/34 (67%), Positives = 29/34 (84%), Gaps = 1/34 (2%)
Query: 6 LLVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
+L+++ AL + IA AYDPSPLQDFCVAI DPK+G
Sbjct: 8 ILITLSALVT-IAKAYDPSPLQDFCVAIDDPKNG 40
>At5g39100 germin-like protein (GLP6)
Length = 221
Score = 47.8 bits (112), Expect = 1e-06
Identities = 22/34 (64%), Positives = 27/34 (78%)
Query: 6 LLVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
+L+++ AL S A AYDPSPLQDFCVAI D K+G
Sbjct: 8 ILITLSALVISFAEAYDPSPLQDFCVAIDDLKNG 41
>At5g39120 germin - like protein
Length = 221
Score = 47.4 bits (111), Expect = 1e-06
Identities = 23/34 (67%), Positives = 28/34 (81%), Gaps = 1/34 (2%)
Query: 6 LLVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
+L++ AL + IA AYDPSPLQDFCVAI DPK+G
Sbjct: 8 ILITFWALVT-IAKAYDPSPLQDFCVAIDDPKNG 40
>At5g39160 germin-like protein (GLP2a) copy1
Length = 200
Score = 44.3 bits (103), Expect = 1e-05
Identities = 21/31 (67%), Positives = 23/31 (73%)
Query: 9 SILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
+I+AL S AYDPSPLQDFCVAI D K G
Sbjct: 11 AIIALVLSFVNAYDPSPLQDFCVAIDDLKGG 41
>At3g05950 germin-like protein
Length = 229
Score = 44.3 bits (103), Expect = 1e-05
Identities = 20/35 (57%), Positives = 24/35 (68%)
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVAIKD 35
++ L +LALASS YDPSPLQDFCVA+ D
Sbjct: 5 LRFLVAKAILLALASSFVSCYDPSPLQDFCVAVDD 39
>At5g39110 germin -like protein
Length = 222
Score = 43.9 bits (102), Expect = 2e-05
Identities = 21/34 (61%), Positives = 26/34 (75%)
Query: 6 LLVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
+L+++ AL S A A DPSPLQDFCVAI D K+G
Sbjct: 8 ILITLSALVISFAEANDPSPLQDFCVAIGDLKNG 41
>At3g04180 germin-like protein
Length = 222
Score = 43.1 bits (100), Expect = 3e-05
Identities = 18/35 (51%), Positives = 25/35 (71%)
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVAIKD 35
++ L + +LALASS + YDPSPLQD+CVA +
Sbjct: 5 LQFLLAKIILLALASSFVYCYDPSPLQDYCVATNE 39
>At5g39190 germin-like protein (GLP2a) copy2
Length = 222
Score = 42.4 bits (98), Expect = 4e-05
Identities = 20/29 (68%), Positives = 22/29 (74%)
Query: 9 SILALASSIAFAYDPSPLQDFCVAIKDPK 37
+I+AL S AYDPSPLQDFCVAI D K
Sbjct: 11 AIIALVLSFVNAYDPSPLQDFCVAIDDLK 39
>At5g39130 germin - like protein
Length = 222
Score = 42.4 bits (98), Expect = 4e-05
Identities = 20/29 (68%), Positives = 22/29 (74%)
Query: 9 SILALASSIAFAYDPSPLQDFCVAIKDPK 37
+I+AL S AYDPSPLQDFCVAI D K
Sbjct: 11 AIIALVLSFVNAYDPSPLQDFCVAIDDLK 39
>At3g04200 germin-like protein
Length = 227
Score = 42.0 bits (97), Expect = 6e-05
Identities = 23/59 (38%), Positives = 32/59 (53%), Gaps = 13/59 (22%)
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVA-------------IKDPKDGDYSDLTH 46
+++L V +LALA+S YDP+PLQDFCVA KDPK +D ++
Sbjct: 6 LRLLVTQVILLALATSFVSCYDPNPLQDFCVAASETNRVFVNGKFCKDPKSVTANDFSY 64
>At5g38960 germin - like protein
Length = 221
Score = 41.2 bits (95), Expect = 1e-04
Identities = 20/38 (52%), Positives = 25/38 (65%), Gaps = 2/38 (5%)
Query: 1 MKILYLLVSILALASSIAFAY--DPSPLQDFCVAIKDP 36
MK LYL + L AS++ FA DPSPLQDFC+ + P
Sbjct: 1 MKNLYLAILYLLAASTLPFAIASDPSPLQDFCIGVNTP 38
>At3g04170 germin-like protein
Length = 227
Score = 41.2 bits (95), Expect = 1e-04
Identities = 17/28 (60%), Positives = 22/28 (77%)
Query: 8 VSILALASSIAFAYDPSPLQDFCVAIKD 35
+ +LALASS YDPSPLQD+CVA+ +
Sbjct: 12 IILLALASSFVSCYDPSPLQDYCVAVPE 39
>At5g38910 germin - like protein
Length = 222
Score = 40.8 bits (94), Expect = 1e-04
Identities = 18/33 (54%), Positives = 24/33 (72%)
Query: 7 LVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
++SILA+ S++ A DPS LQDFCV + P DG
Sbjct: 9 VLSILAITLSLSKASDPSSLQDFCVGVNTPADG 41
>At4g14630 germin precursor oxalate oxidase
Length = 222
Score = 40.8 bits (94), Expect = 1e-04
Identities = 17/32 (53%), Positives = 21/32 (65%)
Query: 8 VSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
+S+ AL + A DPSPLQDFCV + P DG
Sbjct: 12 LSLFALTLPLVIASDPSPLQDFCVGVNTPADG 43
>At3g04190 germin-like protein
Length = 222
Score = 40.8 bits (94), Expect = 1e-04
Identities = 17/35 (48%), Positives = 24/35 (68%)
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVAIKD 35
+ L + +LALASS + Y+PSPLQD+CVA +
Sbjct: 5 LHFLLAKIILLALASSFVYCYEPSPLQDYCVATNE 39
>At3g04150 germin-like protein
Length = 229
Score = 37.0 bits (84), Expect = 0.002
Identities = 15/32 (46%), Positives = 22/32 (67%)
Query: 1 MKILYLLVSILALASSIAFAYDPSPLQDFCVA 32
++ L + +L LAS+ YDP+PLQD+CVA
Sbjct: 5 VQFLVAKIILLVLASTFVHCYDPNPLQDYCVA 36
>At5g38940 germin - like protein
Length = 221
Score = 35.0 bits (79), Expect = 0.007
Identities = 16/33 (48%), Positives = 22/33 (66%)
Query: 7 LVSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
++S+LAL +A A DPS LQDFCV+ +G
Sbjct: 9 VLSLLALTLPLAIASDPSQLQDFCVSANTSANG 41
>At5g38930 germin - like protein
Length = 223
Score = 33.9 bits (76), Expect = 0.016
Identities = 19/41 (46%), Positives = 24/41 (58%), Gaps = 2/41 (4%)
Query: 1 MKILYLL--VSILALASSIAFAYDPSPLQDFCVAIKDPKDG 39
MK L L +S+LAL + A DPS LQDFCV+ +G
Sbjct: 3 MKSLSFLAALSLLALTLPLTIASDPSQLQDFCVSANSSANG 43
>At3g10080 germin-like protein
Length = 227
Score = 30.4 bits (67), Expect = 0.17
Identities = 17/42 (40%), Positives = 23/42 (54%), Gaps = 4/42 (9%)
Query: 4 LYLLVSILALASSIAF--AYDPSPLQDFCVAIKDPKDGDYSD 43
L+LL ++ L ++ F A DP P+QDFC I P Y D
Sbjct: 6 LFLLKALCLLCFNVCFTLASDPDPIQDFC--IPKPVTSPYHD 45
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.145 0.443
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,349,851
Number of Sequences: 26719
Number of extensions: 38529
Number of successful extensions: 143
Number of sequences better than 10.0: 28
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 117
Number of HSP's gapped (non-prelim): 28
length of query: 59
length of database: 11,318,596
effective HSP length: 35
effective length of query: 24
effective length of database: 10,383,431
effective search space: 249202344
effective search space used: 249202344
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 52 (24.6 bits)
Medicago: description of AC140721.8


