

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.12 - phase: 0
(109 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
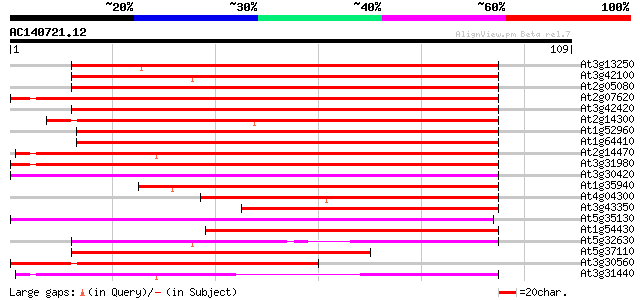
Score E
Sequences producing significant alignments: (bits) Value
At3g13250 hypothetical protein 93 2e-20
At3g42100 putative protein 90 2e-19
At2g05080 putative helicase 89 3e-19
At2g07620 putative helicase 89 5e-19
At3g42420 putative protein 88 8e-19
At2g14300 pseudogene; similar to MURA transposase of maize Muta... 87 1e-18
At1g52960 hypothetical protein 86 3e-18
At1g64410 unknown protein 86 4e-18
At2g14470 pseudogene 85 7e-18
At3g31980 hypothetical protein 84 2e-17
At3g30420 hypothetical protein 72 6e-14
At1g35940 hypothetical protein 72 6e-14
At4g04300 hypothetical protein 66 2e-12
At3g43350 putative protein 65 4e-12
At5g35130 putative protein 57 1e-09
At1g54430 hypothetical protein 57 2e-09
At5g32630 putative protein 55 8e-09
At5g37110 putative helicase 52 5e-08
At3g30560 hypothetical protein 50 1e-07
At3g31440 hypothetical protein 47 2e-06
>At3g13250 hypothetical protein
Length = 1419
Score = 93.2 bits (230), Expect = 2e-20
Identities = 47/84 (55%), Positives = 61/84 (71%), Gaps = 1/84 (1%)
Query: 13 TNKLIVDELNYN-KDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTG 71
+N+LI+DEL YN + +L K +D + LT EQR ++D+I ++ D GG FF+YG+GGTG
Sbjct: 913 SNRLIIDELRYNNQSDLKKKHSDWIQKLTPEQRGIYDQITNAVFNDLGGVFFVYGFGGTG 972
Query: 72 KTFIWKTSLAVVRSKGLIVLNAAS 95
KTFIWKT A VRSKG I LN AS
Sbjct: 973 KTFIWKTLAAAVRSKGQICLNVAS 996
>At3g42100 putative protein
Length = 1752
Score = 89.7 bits (221), Expect = 2e-19
Identities = 47/84 (55%), Positives = 58/84 (68%), Gaps = 1/84 (1%)
Query: 13 TNKLIVDELNYNKDELTKTQTD-MLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTG 71
+N+L+VDEL YN D K + D ML EQR ++D+I + D GG FF+YG+GGTG
Sbjct: 1245 SNRLVVDELRYNIDSNLKEKHDEWFQMLNTEQRGIYDEITGAVFNDLGGVFFIYGFGGTG 1304
Query: 72 KTFIWKTSLAVVRSKGLIVLNAAS 95
KTFIWKT A VRS+G IVLN AS
Sbjct: 1305 KTFIWKTLAAAVRSRGQIVLNVAS 1328
>At2g05080 putative helicase
Length = 1219
Score = 89.4 bits (220), Expect = 3e-19
Identities = 44/83 (53%), Positives = 58/83 (69%)
Query: 13 TNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTGK 72
TN LI+DE Y+++ TK D + LT EQ+ V+D I+ + + GG FF+YG+GGTGK
Sbjct: 721 TNVLILDEKGYDRESETKKHADSIKKLTLEQKSVYDNIIGAVNENVGGVFFVYGFGGTGK 780
Query: 73 TFIWKTSLAVVRSKGLIVLNAAS 95
TF+WKT A +RSKG IVLN AS
Sbjct: 781 TFLWKTLSAALRSKGDIVLNVAS 803
>At2g07620 putative helicase
Length = 1241
Score = 88.6 bits (218), Expect = 5e-19
Identities = 44/95 (46%), Positives = 63/95 (66%), Gaps = 1/95 (1%)
Query: 1 MPRPNTIDMPTFTNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGG 60
MP+P T + +N+ I +E NYN ++L + D +T EQ+ ++D+IM + + GG
Sbjct: 810 MPKP-TKEGTDHSNRFITEEKNYNVEKLKEDHDDWFNKMTSEQKEIYDEIMKAVLENSGG 868
Query: 61 FFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
FF+YG+GGTGKTF+WKT A VR KGLI +N AS
Sbjct: 869 IFFVYGFGGTGKTFMWKTLSAAVRMKGLISVNVAS 903
>At3g42420 putative protein
Length = 1018
Score = 87.8 bits (216), Expect = 8e-19
Identities = 44/83 (53%), Positives = 59/83 (71%)
Query: 13 TNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTGK 72
+N+LIVDE+ Y++ LT + + MLT E+R V+D+I + D GG FF+YG+GGT K
Sbjct: 639 SNRLIVDEMRYDRQYLTGKHAEWIDMLTPEKRGVYDEITGAVFKDLGGVFFVYGFGGTRK 698
Query: 73 TFIWKTSLAVVRSKGLIVLNAAS 95
TFIWKT A +RS+G IVLN AS
Sbjct: 699 TFIWKTLSAAIRSRGDIVLNVAS 721
>At2g14300 pseudogene; similar to MURA transposase of maize Mutator
transposon
Length = 1230
Score = 87.0 bits (214), Expect = 1e-18
Identities = 47/89 (52%), Positives = 56/89 (62%), Gaps = 2/89 (2%)
Query: 8 DMPTFTNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCV-HDKIMDSKGFDDGGFFFLYG 66
D P F N LI+DE NYN++ L K + L LT E + V HD+IMD D GG FFLY
Sbjct: 789 DEPAF-NPLIIDERNYNRESLKKKHDNWLKTLTPEHKKVYHDEIMDDVLNDKGGVFFLYA 847
Query: 67 YGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
+GGTGKTF+WK A +R KG LN AS
Sbjct: 848 FGGTGKTFLWKVLSAAIRCKGDTCLNVAS 876
>At1g52960 hypothetical protein
Length = 924
Score = 85.9 bits (211), Expect = 3e-18
Identities = 41/82 (50%), Positives = 56/82 (68%)
Query: 14 NKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTGKT 73
N I+DE Y++ L + + L M+T EQ+ ++D+IMD+ D GG FF+YG+GGTGKT
Sbjct: 447 NHFILDERKYDRAYLIEKHEEWLTMVTSEQKKIYDEIMDAVLHDRGGVFFVYGFGGTGKT 506
Query: 74 FIWKTSLAVVRSKGLIVLNAAS 95
F+WK A +RSKG I LN AS
Sbjct: 507 FLWKLLSAAIRSKGDISLNVAS 528
>At1g64410 unknown protein
Length = 753
Score = 85.5 bits (210), Expect = 4e-18
Identities = 42/82 (51%), Positives = 56/82 (68%)
Query: 14 NKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTGKT 73
N I+DE Y++ LT+ + L M+T EQ+ ++D+IMD D GG FF+YG+GGTGKT
Sbjct: 497 NHFILDERKYDRAYLTEKHEEWLTMVTLEQKKIYDEIMDVVLHDRGGVFFVYGFGGTGKT 556
Query: 74 FIWKTSLAVVRSKGLIVLNAAS 95
F+WK A +RSKG I LN AS
Sbjct: 557 FLWKLLSAAIRSKGDISLNVAS 578
>At2g14470 pseudogene
Length = 1265
Score = 84.7 bits (208), Expect = 7e-18
Identities = 47/95 (49%), Positives = 65/95 (67%), Gaps = 2/95 (2%)
Query: 2 PRPNTIDMPTFTNKLIVDELNYNKDE-LTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGG 60
P+P TID +N+LIV+EL YN++ L + + MLT EQR V+++I ++ + GG
Sbjct: 765 PKP-TIDGIDNSNRLIVEELRYNRESNLKEKHEEWKQMLTPEQRGVYNEITEAVFNNLGG 823
Query: 61 FFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
FF+YG+GGTGKTFIWKT A +R + IVLN AS
Sbjct: 824 VFFVYGFGGTGKTFIWKTLSATIRYRDQIVLNVAS 858
>At3g31980 hypothetical protein
Length = 1099
Score = 83.6 bits (205), Expect = 2e-17
Identities = 41/95 (43%), Positives = 62/95 (65%), Gaps = 1/95 (1%)
Query: 1 MPRPNTIDMPTFTNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGG 60
MP+P T + +N+ I +E NYN ++L + D +T EQ+ ++D+I+ + + GG
Sbjct: 581 MPKP-TKEGTDHSNRFITEEKNYNIEKLREDHDDWFNKMTSEQKGIYDEIIKAVLENSGG 639
Query: 61 FFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
FF+YG+GGT KTF+WKT A VR +GLI +N AS
Sbjct: 640 IFFVYGFGGTSKTFMWKTLSAAVRMRGLISVNVAS 674
>At3g30420 hypothetical protein
Length = 837
Score = 71.6 bits (174), Expect = 6e-14
Identities = 35/95 (36%), Positives = 55/95 (57%)
Query: 1 MPRPNTIDMPTFTNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGG 60
MP+P + N L+ E N D+ +T ++ L ++QR ++D ++ S +G
Sbjct: 311 MPQPEKSILEEVNNSLLRQEFQINIDKERETHANLFSKLNEQQRIIYDDVLKSVTNKEGK 370
Query: 61 FFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
FFLYG GGTGKTF++KT ++ +RS G V+ AS
Sbjct: 371 LFFLYGDGGTGKTFLYKTIISALRSNGKNVMPVAS 405
>At1g35940 hypothetical protein
Length = 1678
Score = 71.6 bits (174), Expect = 6e-14
Identities = 38/71 (53%), Positives = 49/71 (68%), Gaps = 1/71 (1%)
Query: 26 DELTK-TQTDMLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTGKTFIWKTSLAVVR 84
DE K T+ + MLT EQR V++ I ++ + GG FF+YG+GGTGKTFIWKT A +R
Sbjct: 1217 DEFPKPTRDEWKQMLTPEQRGVYNAITEAVFNNLGGVFFVYGFGGTGKTFIWKTLSAAIR 1276
Query: 85 SKGLIVLNAAS 95
+G IVLN AS
Sbjct: 1277 CRGQIVLNVAS 1287
>At4g04300 hypothetical protein
Length = 286
Score = 66.2 bits (160), Expect = 2e-12
Identities = 36/59 (61%), Positives = 42/59 (71%), Gaps = 1/59 (1%)
Query: 38 MLTDEQRCVHDKIMDSKGFDDGG-FFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
MLT EQR ++D+I ++ D GG FFF+YG GG GKTFIWKT AV RSKG LN AS
Sbjct: 1 MLTPEQRGIYDQITNAVFNDMGGVFFFVYGSGGIGKTFIWKTLAAVGRSKGQTCLNIAS 59
>At3g43350 putative protein
Length = 830
Score = 65.5 bits (158), Expect = 4e-12
Identities = 31/50 (62%), Positives = 37/50 (74%)
Query: 46 VHDKIMDSKGFDDGGFFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
++D+IMD D GG FF+YG+GGTGKTF+WK A VRSKG I LN AS
Sbjct: 183 IYDEIMDVVLHDRGGVFFVYGFGGTGKTFLWKLLSAAVRSKGDISLNVAS 232
>At5g35130 putative protein
Length = 414
Score = 57.4 bits (137), Expect = 1e-09
Identities = 32/94 (34%), Positives = 51/94 (54%)
Query: 1 MPRPNTIDMPTFTNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGG 60
MP+P+ + N L+ E YN + K + L +Q+ V+D +M+S G
Sbjct: 68 MPKPDLDVINELGNSLLTQESQYNVHKEKKDHIKLFSTLNVQQKEVYDAVMESVENGLGK 127
Query: 61 FFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAA 94
FFLYG G GKT+++ T ++++RS+ IVL A
Sbjct: 128 LFFLYGPRGIGKTYLYSTIISMLRSEKKIVLPVA 161
>At1g54430 hypothetical protein
Length = 1639
Score = 56.6 bits (135), Expect = 2e-09
Identities = 26/57 (45%), Positives = 39/57 (67%)
Query: 39 LTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
L ++QR ++D ++ S +G FFLYG GGTGKTF++KT ++ +RS G V+ AS
Sbjct: 1151 LNEQQRIIYDDVLKSVINKEGKLFFLYGAGGTGKTFLYKTIISALRSNGKNVMPVAS 1207
>At5g32630 putative protein
Length = 856
Score = 54.7 bits (130), Expect = 8e-09
Identities = 36/84 (42%), Positives = 47/84 (55%), Gaps = 10/84 (11%)
Query: 13 TNKLIVDELNYNKDELTKTQTD-MLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGTG 71
+N+L+VDEL YN D K + D ML E R ++D+I + F+D GGT
Sbjct: 597 SNRLVVDELRYNIDSNLKEKHDEWFQMLNTEHRGIYDEITGAI-FND--------LGGTE 647
Query: 72 KTFIWKTSLAVVRSKGLIVLNAAS 95
KT +WKT A R +G IVLN AS
Sbjct: 648 KTSMWKTLAAAFRCRGQIVLNVAS 671
>At5g37110 putative helicase
Length = 1307
Score = 52.0 bits (123), Expect = 5e-08
Identities = 24/58 (41%), Positives = 37/58 (63%)
Query: 13 TNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGGFFFLYGYGGT 70
TN LI+DE Y++D LT+ + MLT EQ+ ++D I+ + + G F+YG+GGT
Sbjct: 891 TNVLILDEKGYDRDNLTEKHAKWIKMLTPEQKSIYDDIIGAVNENVGVVVFVYGFGGT 948
>At3g30560 hypothetical protein
Length = 1473
Score = 50.4 bits (119), Expect = 1e-07
Identities = 28/60 (46%), Positives = 38/60 (62%), Gaps = 1/60 (1%)
Query: 1 MPRPNTIDMPTFTNKLIVDELNYNKDELTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGG 60
+P+ D P F N+LI+DE NYN++ L D L MLT EQ+ V+DKIMD+ + GG
Sbjct: 987 LPQLKPGDEPAF-NQLILDERNYNRETLKTIHDDWLKMLTTEQKKVYDKIMDAVLNNKGG 1045
>At3g31440 hypothetical protein
Length = 536
Score = 46.6 bits (109), Expect = 2e-06
Identities = 35/95 (36%), Positives = 45/95 (46%), Gaps = 26/95 (27%)
Query: 2 PRPNTIDMPTFTNKLIVDELNYNKDE-LTKTQTDMLLMLTDEQRCVHDKIMDSKGFDDGG 60
P+P T D +N+LIV+EL YN++ L + + MLT EQR
Sbjct: 67 PKP-TRDGIDNSNRLIVEELRYNRESNLKEKHEEWKQMLTSEQR---------------- 109
Query: 61 FFFLYGYGGTGKTFIWKTSLAVVRSKGLIVLNAAS 95
GT KT IWKT A +R + IVLN AS
Sbjct: 110 --------GTWKTIIWKTLFAAIRRRDQIVLNMAS 136
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.140 0.429
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 2,609,278
Number of Sequences: 26719
Number of extensions: 103719
Number of successful extensions: 333
Number of sequences better than 10.0: 42
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 296
Number of HSP's gapped (non-prelim): 42
length of query: 109
length of database: 11,318,596
effective HSP length: 85
effective length of query: 24
effective length of database: 9,047,481
effective search space: 217139544
effective search space used: 217139544
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC140721.12


