

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.11 + phase: 0
(607 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
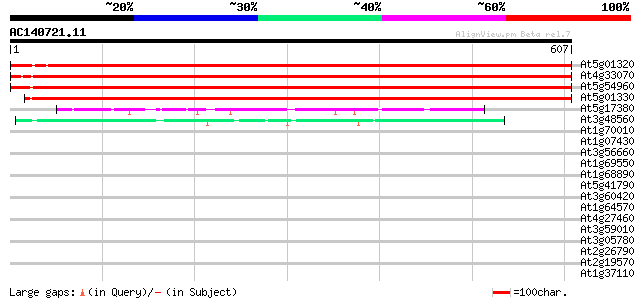
Score E
Sequences producing significant alignments: (bits) Value
At5g01320 pyruvate decarboxylase-like protein 1030 0.0
At4g33070 pyruvate decarboxylase-1 (Pdc1) 1029 0.0
At5g54960 pyruvate decarboxylase 1024 0.0
At5g01330 pyruvate decarboxylase-like protein 994 0.0
At5g17380 2-hydroxyphytanoyl-CoA lyase-like protein 84 2e-16
At3g48560 acetolactate synthase 82 9e-16
At1g70010 hypothetical protein 31 1.7
At1g07430 protein phosphatase 2C, putative 30 3.0
At3g56660 bZip transcription factor AtbZip49 30 3.9
At1g69550 putative disease resistance protein 30 3.9
At1g68890 putative menaquinone biosynthesis protein 30 3.9
At5g41790 myosin heavy chain-like protein 30 5.1
At3g60420 unknown protein 30 5.1
At1g64570 hypothetical protein 29 6.6
At4g27460 unknown protein 29 8.7
At3g59010 pectinesterase precursor-like protein 29 8.7
At3g05780 putative mitochondrial LON ATP-dependent protease 29 8.7
At2g26790 putative salt-inducible protein 29 8.7
At2g19570 cytidine deaminase 1 (cda1) 29 8.7
At1g37110 29 8.7
>At5g01320 pyruvate decarboxylase-like protein
Length = 603
Score = 1030 bits (2662), Expect = 0.0
Identities = 502/607 (82%), Positives = 546/607 (89%), Gaps = 4/607 (0%)
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGI 60
MDT +G+++ CK DI + P++ V+TIQ S + +SESTLG HL+RRLV+ G+
Sbjct: 1 MDTKIGAIDTCKPTTGDIGSPPSN--AVATIQDSAPITT--TSESTLGRHLSRRLVQAGV 56
Query: 61 TDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGL 120
TD+F+VPGDFNLTLLDHLIAEP+L NIGCCNELNAGYAADGYARSRGVGACVVTFTVGGL
Sbjct: 57 TDVFSVPGDFNLTLLDHLIAEPELNNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGL 116
Query: 121 SVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVV 180
SV+NAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVV
Sbjct: 117 SVLNAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVV 176
Query: 181 NNLEDAHEMIDTAISTALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMGLE 240
NNLEDAHE ID AI+TALKESKPVYISISCNLA+ PHPTF+R+PVPF L+P++SN MGLE
Sbjct: 177 NNLEDAHEQIDKAIATALKESKPVYISISCNLAATPHPTFARDPVPFDLTPRMSNTMGLE 236
Query: 241 AAVEAAAEVLNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDHKH 300
AAVEA E LNKAVKPV+V GPKLRVAKA EAF+ELAD S YP +VMPS KGL+PE+H H
Sbjct: 237 AAVEATLEFLNKAVKPVMVGGPKLRVAKASEAFLELADASGYPLAVMPSTKGLVPENHPH 296
Query: 301 FIGTFWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIGN 360
FIGT+WGAVST FC+EIVESADAY+FAGPIFNDYSSVGYSLLLKKEKAIIV PDRVV+ N
Sbjct: 297 FIGTYWGAVSTPFCSEIVESADAYIFAGPIFNDYSSVGYSLLLKKEKAIIVHPDRVVVAN 356
Query: 361 GPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHIQN 420
GPTFGCVLM DF LAKR+KRN TAYENY RIFVPEG P+K +P EPLRVN +FQHIQ
Sbjct: 357 GPTFGCVLMSDFFRELAKRVKRNETAYENYERIFVPEGKPLKCKPGEPLRVNAMFQHIQK 416
Query: 421 MLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIA 480
MLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQA PEKRV++
Sbjct: 417 MLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQATPEKRVLS 476
Query: 481 CIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAI 540
IGDGSFQVTAQD+STM+R GQK IIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAI
Sbjct: 477 FIGDGSFQVTAQDISTMIRNGQKAIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAI 536
Query: 541 HNGQGHCWTTKVFCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSSAN 600
HNG+G CWTTKV EEELVEAI+TAT KKD LCFIEVIVHKDDTSKELLEWGSRVS+AN
Sbjct: 537 HNGEGKCWTTKVRYEEELVEAIKTATTEKKDSLCFIEVIVHKDDTSKELLEWGSRVSAAN 596
Query: 601 SRPPNPQ 607
RPPNPQ
Sbjct: 597 GRPPNPQ 603
>At4g33070 pyruvate decarboxylase-1 (Pdc1)
Length = 607
Score = 1029 bits (2660), Expect = 0.0
Identities = 499/609 (81%), Positives = 548/609 (89%), Gaps = 4/609 (0%)
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASS--ESTLGSHLARRLVEV 58
MDT +GS++ CK P N + +PT NGTV+TI S + ++ + ++TLG HLARRLV+
Sbjct: 1 MDTKIGSIDDCK-PTNGDVCSPT-NGTVATIHNSVPSSAITINYCDATLGRHLARRLVQA 58
Query: 59 GITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVG 118
G+TD+F+VPGDFNLTLLDHL+AEP L IGCCNELNAGYAADGYARSRGVGACVVTFTVG
Sbjct: 59 GVTDVFSVPGDFNLTLLDHLMAEPDLNLIGCCNELNAGYAADGYARSRGVGACVVTFTVG 118
Query: 119 GLSVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQA 178
GLSV+NAIAGAYSENLP+ICIVGGPNSND+GTNRILHHTIGLPDFSQELRCFQTVTCYQA
Sbjct: 119 GLSVLNAIAGAYSENLPLICIVGGPNSNDYGTNRILHHTIGLPDFSQELRCFQTVTCYQA 178
Query: 179 VVNNLEDAHEMIDTAISTALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMG 238
VVNNL+DAHE ID AISTALKESKPVYIS+SCNLA+IPH TFSR+PVPFSL+P+LSN MG
Sbjct: 179 VVNNLDDAHEQIDKAISTALKESKPVYISVSCNLAAIPHHTFSRDPVPFSLAPRLSNKMG 238
Query: 239 LEAAVEAAAEVLNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDH 298
LEAAVEA E LNKAVKPV+V GPKLRVAKAC+AF+ELAD S Y ++MPSAKG +PE H
Sbjct: 239 LEAAVEATLEFLNKAVKPVMVGGPKLRVAKACDAFVELADASGYALAMMPSAKGFVPEHH 298
Query: 299 KHFIGTFWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVI 358
HFIGT+WGAVST FC+EIVESADAY+FAGPIFNDYSSVGYSLLLKKEKAI+VQPDR+ +
Sbjct: 299 PHFIGTYWGAVSTPFCSEIVESADAYIFAGPIFNDYSSVGYSLLLKKEKAIVVQPDRITV 358
Query: 359 GNGPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHI 418
NGPTFGC+LM DF L+KR+KRN TAYENY RIFVPEG P+K E REPLRVN +FQHI
Sbjct: 359 ANGPTFGCILMSDFFRELSKRVKRNETAYENYHRIFVPEGKPLKCESREPLRVNTMFQHI 418
Query: 419 QNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRV 478
Q MLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQA PEKRV
Sbjct: 419 QKMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQASPEKRV 478
Query: 479 IACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVD 538
+A IGDGSFQVT QD+STMLR GQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVD
Sbjct: 479 LAFIGDGSFQVTVQDISTMLRNGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVD 538
Query: 539 AIHNGQGHCWTTKVFCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSS 598
AIHNG+G+CWT KV EEELVEAI TAT KKDCLCFIEVI+HKDDTSKELLEWGSRVS+
Sbjct: 539 AIHNGEGNCWTAKVRYEEELVEAITTATTEKKDCLCFIEVILHKDDTSKELLEWGSRVSA 598
Query: 599 ANSRPPNPQ 607
ANSRPPNPQ
Sbjct: 599 ANSRPPNPQ 607
>At5g54960 pyruvate decarboxylase
Length = 607
Score = 1024 bits (2648), Expect = 0.0
Identities = 499/609 (81%), Positives = 547/609 (88%), Gaps = 4/609 (0%)
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQKSPSTQSLASS--ESTLGSHLARRLVEV 58
MDT +GS++AC +DI P NG VST+Q + S S ++TLG +LARRLVE+
Sbjct: 1 MDTKIGSIDACNPTNHDIGGPP--NGGVSTVQNTSPLHSTTVSPCDATLGRYLARRLVEI 58
Query: 59 GITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVG 118
G+TD+F+VPGDFNLTLLDHLIAEP LK IGCCNELNAGYAADGYARSRGVGACVVTFTVG
Sbjct: 59 GVTDVFSVPGDFNLTLLDHLIAEPNLKLIGCCNELNAGYAADGYARSRGVGACVVTFTVG 118
Query: 119 GLSVINAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQA 178
GLSV+NAIAGAYSENLP+ICIVGGPNSND+GTNRILHHTIGLPDF+QELRCFQ VTC+QA
Sbjct: 119 GLSVLNAIAGAYSENLPLICIVGGPNSNDYGTNRILHHTIGLPDFTQELRCFQAVTCFQA 178
Query: 179 VVNNLEDAHEMIDTAISTALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMG 238
V+NNLE+AHE+IDTAISTALKESKPVYISISCNL +IP PTFSR PVPF L K+SN +G
Sbjct: 179 VINNLEEAHELIDTAISTALKESKPVYISISCNLPAIPLPTFSRHPVPFMLPMKVSNQIG 238
Query: 239 LEAAVEAAAEVLNKAVKPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDH 298
L+AAVEAAAE LNKAVKPVLV GPK+RVAKA +AF+ELAD S Y +VMPSAKG +PE H
Sbjct: 239 LDAAVEAAAEFLNKAVKPVLVGGPKMRVAKAADAFVELADASGYGLAVMPSAKGQVPEHH 298
Query: 299 KHFIGTFWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVI 358
KHFIGT+WGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRV I
Sbjct: 299 KHFIGTYWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVTI 358
Query: 359 GNGPTFGCVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHI 418
GNGP FGCVLMKDFLS LAKR+K NNT+YENY RI+VPEG P++ P E LRVNVLFQHI
Sbjct: 359 GNGPAFGCVLMKDFLSELAKRIKHNNTSYENYHRIYVPEGKPLRDNPNESLRVNVLFQHI 418
Query: 419 QNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRV 478
QNMLSSE+AV+AETGDSWFNCQKLKLP+GCGYEFQMQYGSIGWSVGATLGYAQA+P +RV
Sbjct: 419 QNMLSSESAVLAETGDSWFNCQKLKLPEGCGYEFQMQYGSIGWSVGATLGYAQAMPNRRV 478
Query: 479 IACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVD 538
IACIGDGSFQVTAQDVSTM+RCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYT V+
Sbjct: 479 IACIGDGSFQVTAQDVSTMIRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTAFVE 538
Query: 539 AIHNGQGHCWTTKVFCEEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSS 598
AIHNG+G CWT KV CEEELV+AI TAT +K+ CFIEVIVHKDDTSKELLEWGSRVS+
Sbjct: 539 AIHNGEGKCWTAKVRCEEELVKAINTATNEEKESFCFIEVIVHKDDTSKELLEWGSRVSA 598
Query: 599 ANSRPPNPQ 607
ANSRPPNPQ
Sbjct: 599 ANSRPPNPQ 607
>At5g01330 pyruvate decarboxylase-like protein
Length = 592
Score = 994 bits (2570), Expect = 0.0
Identities = 491/593 (82%), Positives = 531/593 (88%), Gaps = 4/593 (0%)
Query: 17 DIITTPTSNGTVSTIQKSPSTQS--LASSESTLGSHLARRLVEVGITDIFTVPGDFNLTL 74
D+ + P SNG V+TIQ S T + L SS +TLG HL+RRLV+ G+TDIFTVPGDFNL+L
Sbjct: 2 DVRSLP-SNG-VATIQDSAPTAATILGSSAATLGRHLSRRLVQAGVTDIFTVPGDFNLSL 59
Query: 75 LDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGLSVINAIAGAYSENL 134
LD LIA P+L NIGCCNELNAGYAADGYARSRGVGACVVTFTVGGLSV+NAIAGAYSENL
Sbjct: 60 LDQLIANPELNNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGLSVLNAIAGAYSENL 119
Query: 135 PVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNNLEDAHEMIDTAI 194
PVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVN+LEDAHE ID AI
Sbjct: 120 PVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNHLEDAHEQIDKAI 179
Query: 195 STALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPMGLEAAVEAAAEVLNKAV 254
+TAL+ESKPVYISISCNLA+IPHPTF+ PVPF L+P+LSN LEAAVEA E LNKAV
Sbjct: 180 ATALRESKPVYISISCNLAAIPHPTFASYPVPFDLTPRLSNKDCLEAAVEATLEFLNKAV 239
Query: 255 KPVLVAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVSTAFC 314
KPV+V GPKLRVAKA +AF+ELAD S YP +VMPSAKG +PE+H HFIGT+WGAVST FC
Sbjct: 240 KPVMVGGPKLRVAKARDAFVELADASGYPVAVMPSAKGFVPENHPHFIGTYWGAVSTLFC 299
Query: 315 AEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIGNGPTFGCVLMKDFLS 374
+EIVESADAY+FAGPIFNDYSSVGYSLLLKKEKAIIV PD VV+ NGPTFGCV M +F
Sbjct: 300 SEIVESADAYIFAGPIFNDYSSVGYSLLLKKEKAIIVHPDSVVVANGPTFGCVRMSEFFR 359
Query: 375 ALAKRLKRNNTAYENYFRIFVPEGLPVKSEPREPLRVNVLFQHIQNMLSSETAVIAETGD 434
LAKR+K N TAYENY RIFVPEG P+K +PREPLR+N +FQHIQ MLS+ETAVIAETGD
Sbjct: 360 ELAKRVKPNKTAYENYHRIFVPEGKPLKCKPREPLRINAMFQHIQKMLSNETAVIAETGD 419
Query: 435 SWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDGSFQVTAQDV 494
SWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQA PEKRV++ IGDGSFQVTAQDV
Sbjct: 420 SWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQATPEKRVLSFIGDGSFQVTAQDV 479
Query: 495 STMLRCGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAIHNGQGHCWTTKVFC 554
STM+R GQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAIHNG+G CWTTKV
Sbjct: 480 STMIRNGQKTIIFLINNGGYTIEVEIHDGPYNVIKNWNYTGLVDAIHNGEGKCWTTKVRY 539
Query: 555 EEELVEAIETATGPKKDCLCFIEVIVHKDDTSKELLEWGSRVSSANSRPPNPQ 607
EEELVEAI TAT KKD LCFIEVIVHKDDTSKELLEWGSRVS+AN RPPNPQ
Sbjct: 540 EEELVEAINTATLEKKDSLCFIEVIVHKDDTSKELLEWGSRVSAANGRPPNPQ 592
>At5g17380 2-hydroxyphytanoyl-CoA lyase-like protein
Length = 572
Score = 84.3 bits (207), Expect = 2e-16
Identities = 115/508 (22%), Positives = 208/508 (40%), Gaps = 84/508 (16%)
Query: 51 LARRLVEVGITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGA 110
+A+ L +G+T +F V G +T L ++ I NE +AGYAA Y G
Sbjct: 18 VAKSLSHLGVTHMFGVVG-IPVTSLASRAMALGIRFIAFHNEQSAGYAASAYGYLTGKPG 76
Query: 111 CVVTFTVGGLSVINAIAG---AYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQEL 167
+ TV G ++ +AG A+ P++ I G + D +G DF QEL
Sbjct: 77 --ILLTVSGPGCVHGLAGLSNAWVNTWPMVMISGSCDQRD----------VGRGDF-QEL 123
Query: 168 RCFQTVTCYQAVVNNLEDAHEMIDTAISTALKES---KP--VYISISCNLASIPHPTFSR 222
+ V + + +D E+ D +S L + +P Y+ I ++ R
Sbjct: 124 DQIEAVKAFSKLSEKAKDVREIPD-CVSRVLDRAVSGRPGGCYLDIPTDVL--------R 174
Query: 223 EPVPFSLSPKLSNPM-----------GLEAAVEAAAEVLNKAVKPVLVAGPKLRVAKACE 271
+ + S + KL + + L + +E+A +L KA +P++V G ++A +
Sbjct: 175 QKISESEADKLVDEVERSRKEEPIRGSLRSEIESAVSLLRKAERPLIVFGKGAAYSRAED 234
Query: 272 AFIELADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVSTAFCAEIVESADAYLFAGPIF 331
+L + + P+ P KGL+P+ H+ +TA + + D L G
Sbjct: 235 ELKKLVEITGIPFLPTPMGKGLLPDTHEF--------SATAARSLAIGKCDVALVVGARL 286
Query: 332 NDYSSVGYSLLLKKEKAIIV-----------QPDRVVIGNGPTFGCVLMKD--------- 371
N G S K+ I+ +P ++G+ T +L ++
Sbjct: 287 NWLLHFGESPKWDKDVKFILVDVSEEEIELRKPHLGIVGDAKTVIGLLNREIKDDPFCLG 346
Query: 372 ----FLSALAKRLKRNNTAYE-NYFRIFVPEGLPVKSEPREPLRVNVL-FQHIQNMLSSE 425
++ +++K+ K N E + VP P +R +L + ++ SE
Sbjct: 347 KSNSWVESISKKAKENGEKMEIQLAKDVVPFNF---LTPMRIIRDAILAVEGPSPVVVSE 403
Query: 426 TAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSIGWSVGATLGYAQAVPEKRVIACIGDG 485
A + G S ++ + G +G++G +G + A A P++ V+A GD
Sbjct: 404 GANTMDVGRSVLVQKEPRTRLDAG-----TWGTMGVGLGYCIAAAVASPDRLVVAVEGDS 458
Query: 486 SFQVTAQDVSTMLRCGQKTIIFLINNGG 513
F +A +V T++R +I + NNGG
Sbjct: 459 GFGFSAMEVETLVRYNLAVVIIVFNNGG 486
>At3g48560 acetolactate synthase
Length = 670
Score = 82.0 bits (201), Expect = 9e-16
Identities = 119/554 (21%), Positives = 219/554 (39%), Gaps = 50/554 (9%)
Query: 7 SLEACKSPCNDIITTPTSNGTVSTIQKSPST--QSLASSESTLGSH-LARRLVEVGITDI 63
S+ A + ++ TTP+ T P T A + G+ L L G+ +
Sbjct: 61 SISAVLNTTTNVTTTPSP-----TKPTKPETFISRFAPDQPRKGADILVEALERQGVETV 115
Query: 64 FTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRG-VGACVVTFTVGGLSV 122
F PG ++ + L ++N+ +E +AA+GYARS G G C+ T G ++
Sbjct: 116 FAYPGGASMEIHQALTRSSSIRNVLPRHEQGGVFAAEGYARSSGKPGICIATSGPGATNL 175
Query: 123 INAIAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNN 182
++ +A A +++P++ I G GT+ I + +++T + +V +
Sbjct: 176 VSGLADALLDSVPLVAITGQVPRRMIGTDAFQETPI--------VEVTRSITKHNYLVMD 227
Query: 183 LEDAHEMIDTAISTALK-ESKPVYISISCNL---ASIPH-PTFSREPVPFSLSPKLSNPM 237
+ED +I+ A A PV + + ++ +IP+ R P S PK
Sbjct: 228 VEDIPRIIEEAFFLATSGRPGPVLVDVPKDIQQQLAIPNWEQAMRLPGYMSRMPKPPEDS 287
Query: 238 GLEAAVEAAAEVLNKAVKPVL-VAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPE 296
LE V +++++ KPVL V G L + F+EL + P + G P
Sbjct: 288 HLEQIV----RLISESKKPVLYVGGGCLNSSDELGRFVEL---TGIPVASTLMGLGSYPC 340
Query: 297 DHK---HFIGTFWGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQP 353
D + H +G T + VE +D L G F+D + + K + +
Sbjct: 341 DDELSLHMLGMH----GTVYANYAVEHSDLLLAFGVRFDDRVTGKLEAFASRAKIVHIDI 396
Query: 354 DRVVIGNGPTFGCVLMKDFLSAL----------AKRLKRNNTAYENYFRIFVPEGLPVKS 403
D IG T + D AL A+ LK + + N + + P+
Sbjct: 397 DSAEIGKNKTPHVSVCGDVKLALQGMNKVLENRAEELKLDFGVWRNELNV-QKQKFPLSF 455
Query: 404 EP-REPLRVNVLFQHIQNMLSSETAVIAETGD-SWFNCQKLKLPKGCGYEFQMQYGSIGW 461
+ E + + + + + + G + Q K + G++G+
Sbjct: 456 KTFGEAIPPQYAIKVLDELTDGKAIISTGVGQHQMWAAQFYNYKKPRQWLSSGGLGAMGF 515
Query: 462 SVGATLGYAQAVPEKRVIACIGDGSFQVTAQDVSTMLRCGQKTIIFLINNGGYTIEVEIH 521
+ A +G + A P+ V+ GDGSF + Q+++T+ + L+NN + ++
Sbjct: 516 GLPAAIGASVANPDAIVVDIDGDGSFIMNVQELATIRVENLPVKVLLLNNQHLGMVMQWE 575
Query: 522 DGPYNVIKNWNYTG 535
D Y + + G
Sbjct: 576 DRFYKANRAHTFLG 589
>At1g70010 hypothetical protein
Length = 1315
Score = 31.2 bits (69), Expect = 1.7
Identities = 32/123 (26%), Positives = 57/123 (46%), Gaps = 20/123 (16%)
Query: 306 WGAVSTAFCAEIVESADAYLFAGPIFNDYSSVGYSLLLKKEKAIIVQPDRVVIGNGPTFG 365
WG S V++ D Y + I +DYS + LL+ + ++ VI PTF
Sbjct: 471 WGPFS-------VQTHDGYRYFLTIVDDYSRATWVYLLRNKSDVL-----TVI---PTFV 515
Query: 366 CVLMKDFLSALAKRLKRNNTAYENYFRIFVPEGL-PVKSEPREPLRVNVL---FQHIQNM 421
++ F + + K ++ +N N+ + + +G+ P S P P + +V+ QHI N+
Sbjct: 516 TMVENQFETTI-KGVRSDNAPELNFTQFYHSKGIVPYHSCPETPQQNSVVERKHQHILNV 574
Query: 422 LSS 424
S
Sbjct: 575 ARS 577
>At1g07430 protein phosphatase 2C, putative
Length = 442
Score = 30.4 bits (67), Expect = 3.0
Identities = 24/75 (32%), Positives = 35/75 (46%), Gaps = 11/75 (14%)
Query: 531 WNYTGLVDAIHNGQGH-CWTTKVFCEEELVEAI--ETATGPKKDCLCFIEVIVHKDDTSK 587
W+Y G+ D GH C C+E L E + E + K++ +E + D K
Sbjct: 155 WHYFGVYD------GHGCSHVAARCKERLHELVQEEALSDKKEEWKKMMERSFTRMD--K 206
Query: 588 ELLEWGSRVSSANSR 602
E++ WG V SAN R
Sbjct: 207 EVVRWGETVMSANCR 221
>At3g56660 bZip transcription factor AtbZip49
Length = 620
Score = 30.0 bits (66), Expect = 3.9
Identities = 18/63 (28%), Positives = 28/63 (43%), Gaps = 5/63 (7%)
Query: 15 CNDIITTPTSNGTVSTIQKSPSTQSLASSESTLGSHLARRLVEVGITDIFTVPGDFNLTL 74
C ++ S+ + + I SP TQ ++ T RR++ G+ DFNLT
Sbjct: 488 CTEVFQFDVSSNSGAIIPASPHTQQCKNTSDTQKGKKNRRILSGGLP-----VSDFNLTK 542
Query: 75 LDH 77
DH
Sbjct: 543 EDH 545
>At1g69550 putative disease resistance protein
Length = 1398
Score = 30.0 bits (66), Expect = 3.9
Identities = 24/92 (26%), Positives = 43/92 (46%), Gaps = 8/92 (8%)
Query: 1 MDTNLGSLEACKSPCNDIITTPTSNGTVSTIQK--SPSTQSLASSESTLGSHLA-RRLVE 57
+ T + LE S C+ +I P+S G + I+ SL S++G+ + RL
Sbjct: 710 LSTAINLLEMVLSDCSSLIELPSSIGNATNIKSLDIQGCSSLLKLPSSIGNLITLPRLDL 769
Query: 58 VGITDIFTVPGDFNLTLLDHLIAEPKLKNIGC 89
+G + + +P + +LI P+L +GC
Sbjct: 770 MGCSSLVELPSS-----IGNLINLPRLDLMGC 796
>At1g68890 putative menaquinone biosynthesis protein
Length = 894
Score = 30.0 bits (66), Expect = 3.9
Identities = 24/118 (20%), Positives = 49/118 (41%), Gaps = 3/118 (2%)
Query: 41 ASSESTLGSHLARRLVEVGITDIFTVPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAAD 100
A+ + S + +G+T PG + L P + C +E + + A
Sbjct: 337 ANINAVWASAIIEECTRLGLTYFCVAPGSRSSHLAIAAANHPLTTCLACFDERSLAFHAI 396
Query: 101 GYARSRGVGACVVTFTVGGLS-VINAIAGAYSENLPVICIVGG--PNSNDFGTNRILH 155
GYA+ A ++T + +S ++ A+ A + LP++ + P G N+ ++
Sbjct: 397 GYAKGSLKPAVIITSSGTAVSNLLPAVVEASEDFLPLLLLTADRPPELQGVGANQAIN 454
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 29.6 bits (65), Expect = 5.1
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query: 557 ELVEAIETATGPKKDCLCFIEVI-VHKDDTSKELLEWGSRVSSANSR 602
EL E+ +T T + + F+EV HK D+S ++ E +RV SA +
Sbjct: 542 ELAESKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQ 588
>At3g60420 unknown protein
Length = 270
Score = 29.6 bits (65), Expect = 5.1
Identities = 38/175 (21%), Positives = 72/175 (40%), Gaps = 26/175 (14%)
Query: 148 FGTNRILHHTIGLPDF----SQELRCFQTVTCYQAVVNNLEDAHEMIDTAISTALKESKP 203
F T + + +G+P S LRC QT + A ++ ++ + + ++ +K
Sbjct: 51 FRTGQRIRSQLGVPIHRVFVSPFLRCIQTASEVVAALSAVDFDPIAMSSKDVLSIDNTK- 109
Query: 204 VYISISCNLASIPHPTFSREPV-----PFSLSPKLSNPMGLEAAVEAAAEVLNKAVKPVL 258
+ ++I L+ IPHP F + V F M E V++ +++ K V
Sbjct: 110 IKVAIEFGLSEIPHPIFIKSEVAPKDGKFDFKISDLEAMFPEGTVDSNVDMVYKEVP--- 166
Query: 259 VAGPKLRVAKACEAFIELADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVSTAF 313
++ +AF + K+ + ++ L+ + T WGAVS AF
Sbjct: 167 ------EWGESAQAFEDRYYKTVKILAEKYPSENLL-------LVTHWGAVSVAF 208
>At1g64570 hypothetical protein
Length = 1239
Score = 29.3 bits (64), Expect = 6.6
Identities = 22/75 (29%), Positives = 35/75 (46%), Gaps = 2/75 (2%)
Query: 212 LASIPHPTFSREPVPFSLSPKLSN--PMGLEAAVEAAAEVLNKAVKPVLVAGPKLRVAKA 269
L ++PH E V + LS S+ P G + A + A L ++ + VA +AK
Sbjct: 417 LFTLPHQEVGGEIVNYPLSSPSSSKSPSGQQQAKKTLAATLVESAQKQSVALVHKDIAKL 476
Query: 270 CEAFIELADKSAYPY 284
+ F+ L S YP+
Sbjct: 477 AKRFMPLFKVSLYPH 491
>At4g27460 unknown protein
Length = 391
Score = 28.9 bits (63), Expect = 8.7
Identities = 19/54 (35%), Positives = 22/54 (40%), Gaps = 2/54 (3%)
Query: 500 CGQKTIIFLINNGGYTIEVEIHDGPYNVIK--NWNYTGLVDAIHNGQGHCWTTK 551
C I LI G + V IH PY K N N + NGQ CW T+
Sbjct: 108 CSLIEAIDLIIKGAQNLIVPIHTKPYTKKKQHNDNVSVTTTTHSNGQRFCWITQ 161
>At3g59010 pectinesterase precursor-like protein
Length = 529
Score = 28.9 bits (63), Expect = 8.7
Identities = 31/110 (28%), Positives = 42/110 (38%), Gaps = 21/110 (19%)
Query: 28 VSTIQKSPSTQSLASSESTLGSHLARRLVEVGITDIFTVPGDFNLTLLDHLIAEPKLKNI 87
VS QKS S+ +L L H D T + LL+ + E +
Sbjct: 182 VSDKQKSSSSSNLTGGRKLLSDH-----------DFPTWVSSSDRKLLEASVEELRP--- 227
Query: 88 GCCNELNAGYAADGYARSRGVGACVVTFTVG-GLSVINAIAGAYSENLPV 136
+A AADG V + + G G SVI+ AG Y ENL +
Sbjct: 228 ------HAVVAADGSGTHMSVAEALASLEKGSGRSVIHLTAGTYKENLNI 271
>At3g05780 putative mitochondrial LON ATP-dependent protease
Length = 924
Score = 28.9 bits (63), Expect = 8.7
Identities = 14/47 (29%), Positives = 23/47 (48%)
Query: 340 SLLLKKEKAIIVQPDRVVIGNGPTFGCVLMKDFLSALAKRLKRNNTA 386
+L K + P+ +GP+ GC ++ FLS K+L R + A
Sbjct: 805 NLFFANSKLHLHVPEGATPKDGPSAGCTMITSFLSLAMKKLVRKDLA 851
>At2g26790 putative salt-inducible protein
Length = 799
Score = 28.9 bits (63), Expect = 8.7
Identities = 45/183 (24%), Positives = 73/183 (39%), Gaps = 17/183 (9%)
Query: 66 VPGDFNLTLLDHLIAEPKLKNIGCCNELNAGYAADGYARSRGVGACVVTFTVGGLSVINA 125
+P F T++ H NEL + + RG+ VVT+TV L
Sbjct: 623 IPDLFTYTIMIHTYCR--------LNELQKAESLFEDMKQRGIKPDVVTYTVL-LDRYLK 673
Query: 126 IAGAYSENLPVICIVGGPNSNDFGTNRILHHTIGLPDFSQELRCFQTVTCYQAVVNNLED 185
+ + E V VG +++ IGL ++ C+ + Q +NNLE
Sbjct: 674 LDPEHHETCSVQGEVGKRKASEV-LREFSAAGIGL-----DVVCYTVLIDRQCKMNNLEQ 727
Query: 186 AHEMIDTAISTALKESKPVYISISCNLASIPHPTFSREPVPFSLSPKLSNPM-GLEAAVE 244
A E+ D I + L+ Y ++ + + + V LS K + P EAAV+
Sbjct: 728 AAELFDRMIDSGLEPDMVAYTTLISSYFRKGYIDMAVTLVT-ELSKKYNIPSESFEAAVK 786
Query: 245 AAA 247
+AA
Sbjct: 787 SAA 789
>At2g19570 cytidine deaminase 1 (cda1)
Length = 301
Score = 28.9 bits (63), Expect = 8.7
Identities = 23/78 (29%), Positives = 39/78 (49%), Gaps = 4/78 (5%)
Query: 277 ADKSAYPYSVMPSAKGLIPEDHKHFIGTFWGAVSTAFCAEI--VESADAYLFAGPIFNDY 334
A++S PYS+ PS L+ D K + G W S A+ + V++A A Y
Sbjct: 201 ANRSYAPYSLCPSGVSLVDCDGKVYRG--WYMESAAYNPSMGPVQAALVDYVANGGGGGY 258
Query: 335 SSVGYSLLLKKEKAIIVQ 352
+ ++L++KE A++ Q
Sbjct: 259 ERIVGAVLVEKEDAVVRQ 276
>At1g37110
Length = 1356
Score = 28.9 bits (63), Expect = 8.7
Identities = 23/91 (25%), Positives = 38/91 (41%), Gaps = 2/91 (2%)
Query: 400 PVKSEPREPLRVNVLFQHIQNMLSSETAVIAETGDSWFNCQKLKLPKGCGYEFQMQYGSI 459
PVK E E + N++ HI +++ + A T D W K + Q
Sbjct: 73 PVKIEQSEQAK-NIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYTQLKLY 131
Query: 460 GWSVGATLGYAQAVPE-KRVIACIGDGSFQV 489
+ + +T+ Q V E R++A +G QV
Sbjct: 132 SFKMVSTMTIDQNVDEFLRIVAELGSLEIQV 162
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,815,620
Number of Sequences: 26719
Number of extensions: 594768
Number of successful extensions: 1459
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1442
Number of HSP's gapped (non-prelim): 25
length of query: 607
length of database: 11,318,596
effective HSP length: 105
effective length of query: 502
effective length of database: 8,513,101
effective search space: 4273576702
effective search space used: 4273576702
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140721.11


