

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140721.1 - phase: 0
(313 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
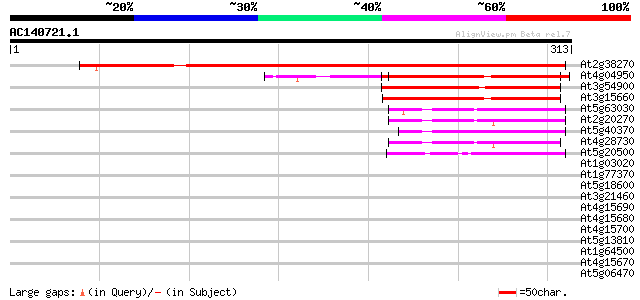
At2g38270 unknown protein 364 e-101
At4g04950 putative thioredoxin 101 6e-22
At3g54900 unknown protein 100 1e-21
At3g15660 unknown protein 86 3e-17
At5g63030 glutaredoxin-like protein 45 4e-05
At2g20270 putative glutaredoxin 44 9e-05
At5g40370 glutaredoxin -like protein 44 1e-04
At4g28730 unknown protein 42 4e-04
At5g20500 glutaredoxin 42 6e-04
At1g03020 putative glutaredoxin 39 0.003
At1g77370 unknown protein 39 0.004
At5g18600 glutaredoxin -like protein 38 0.006
At3g21460 glutaredoxin, putative 38 0.006
At4g15690 glutaredoxin 38 0.008
At4g15680 glutaredoxin 37 0.018
At4g15700 glutaredoxin homolog 36 0.024
At5g13810 unknown protein 36 0.031
At1g64500 unknown protein 35 0.040
At4g15670 glutaredoxin 35 0.053
At5g06470 putative protein 35 0.069
>At2g38270 unknown protein
Length = 293
Score = 364 bits (935), Expect = e-101
Identities = 176/273 (64%), Positives = 223/273 (81%), Gaps = 8/273 (2%)
Query: 40 QNTPTLSL--NFHPNSSTNLRPISLKPYHPRKPQSWLVAMAVKNLADTSPVTVSPENDGL 97
+NTP ++L F P+ S +L+ P + +S+ +A AVK+L +T + ++
Sbjct: 25 RNTPVITLYSRFTPSFSFPSLSFTLRDTAPSRRRSFFIASAVKSLTETELLPITE----- 79
Query: 98 TGEELPSGAGVYAVYDKNGELQFIGLSRNIAATVLAHRKSVPELCGSFKVGVVDEPDRES 157
+ +PS +GVYAVYDK+ ELQF+G+SRNIAA+V AH KSVPELCGS KVG+V+EPD+
Sbjct: 80 -ADSIPSASGVYAVYDKSDELQFVGISRNIAASVSAHLKSVPELCGSVKVGIVEEPDKAV 138
Query: 158 LTQAWKSWMEEHIKITGKVPPGNESGNATWVRPQPKKKADLRLTPGRHVQLTVPLEELVD 217
LTQAWK W+EEHIK+TGKVPPGN+SGN T+V+ P+KK+D+RLTPGRHV+LTVPLEEL+D
Sbjct: 139 LTQAWKLWIEEHIKVTGKVPPGNKSGNNTFVKQTPRKKSDIRLTPGRHVELTVPLEELID 198
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
+LVKE+KVVAFIKGSRSAP CGFSQ+V+GILE +GVDYE+VDVLD++YN+GLRETLK YS
Sbjct: 199 RLVKESKVVAFIKGSRSAPQCGFSQRVVGILESQGVDYETVDVLDDEYNHGLRETLKNYS 258
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
NWPTFPQIF+ GELVGGCDILTSM E GE+A +
Sbjct: 259 NWPTFPQIFVKGELVGGCDILTSMYENGELANI 291
>At4g04950 putative thioredoxin
Length = 488
Score = 101 bits (251), Expect = 6e-22
Identities = 48/101 (47%), Positives = 70/101 (68%), Gaps = 3/101 (2%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRE 271
L+ ++KL + V+ F+KG P CGFS+KV+ IL++ VD+ S D+L ++ +RE
Sbjct: 153 LKSRLEKLTNSHPVMLFMKGIPEEPRCGFSRKVVDILKEVNVDFGSFDILSDNE---VRE 209
Query: 272 TLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
LKK+SNWPTFPQ++ NGEL+GG DI +M+E GE+ FK
Sbjct: 210 GLKKFSNWPTFPQLYCNGELLGGADIAIAMHESGELKDAFK 250
Score = 96.7 bits (239), Expect = 1e-20
Identities = 54/167 (32%), Positives = 92/167 (54%), Gaps = 13/167 (7%)
Query: 143 GSFKVGVVDEPDRESLT--QAWKSWMEEHIKITGKVPPGNESGNATWVRPQPKKKADLRL 200
GSF + ++D+ R+ L W S+ + ++K G G + V K ++
Sbjct: 327 GSFDI-LLDDEVRQGLKVYSNWSSYPQLYVK-------GELMGGSDIVLEMQKSGELKKV 378
Query: 201 TPGRHVQLTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDV 260
+ + LE+ + L+ ++V+ F+KGS P CGFS KV+ L E V + S D+
Sbjct: 379 LTEKGITGEQSLEDRLKALINSSEVMLFMKGSPDEPKCGFSSKVVKALRGENVSFGSFDI 438
Query: 261 LDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
L ++ +R+ +K +SNWPTFPQ++ GEL+GGCDI+ ++E G++
Sbjct: 439 LTDEE---VRQGIKNFSNWPTFPQLYYKGELIGGCDIIMELSESGDL 482
Score = 84.0 bits (206), Expect = 1e-16
Identities = 41/100 (41%), Positives = 64/100 (64%), Gaps = 3/100 (3%)
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNY 267
L+ L ++ LV V+ F+KG P CGFS KV+ IL +E +++ S D+L +D
Sbjct: 279 LSETLRARLEGLVNSKPVMLFMKGRPEEPKCGFSGKVVEILNQEKIEFGSFDILLDD--- 335
Query: 268 GLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
+R+ LK YSNW ++PQ+++ GEL+GG DI+ M + GE+
Sbjct: 336 EVRQGLKVYSNWSSYPQLYVKGELMGGSDIVLEMQKSGEL 375
>At3g54900 unknown protein
Length = 173
Score = 100 bits (249), Expect = 1e-21
Identities = 48/100 (48%), Positives = 68/100 (68%), Gaps = 3/100 (3%)
Query: 208 LTVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNY 267
LT L++ ++KLV KVV F+KG+R P+CGFS V+ IL+ V +E V++L+ N
Sbjct: 67 LTPQLKDTLEKLVNSEKVVLFMKGTRDFPMCGFSNTVVQILKNLNVPFEDVNILE---NE 123
Query: 268 GLRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
LR+ LK+YSNWPTFPQ+++ GE GGCDI + GE+
Sbjct: 124 MLRQGLKEYSNWPTFPQLYIGGEFFGGCDITLEAFKTGEL 163
>At3g15660 unknown protein
Length = 169
Score = 85.5 bits (210), Expect = 3e-17
Identities = 38/99 (38%), Positives = 67/99 (67%), Gaps = 3/99 (3%)
Query: 209 TVPLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG 268
T L+++V+ VK+N V+ ++KG +P CGFS + +L++ V S ++L++
Sbjct: 62 TDSLKDIVENDVKDNPVMIYMKGVPESPQCGFSSLAVRVLQQYNVPISSRNILEDQE--- 118
Query: 269 LRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
L+ +K +S+WPTFPQIF+ GE +GG DI+ +M+++GE+
Sbjct: 119 LKNAVKSFSHWPTFPQIFIKGEFIGGSDIILNMHKEGEL 157
>At5g63030 glutaredoxin-like protein
Length = 125
Score = 45.4 bits (106), Expect = 4e-05
Identities = 30/103 (29%), Positives = 56/103 (54%), Gaps = 10/103 (9%)
Query: 212 LEELVDK---LVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG 268
+E +V+K +V VV F K CG+ Q+V +L + G ++ ++ LDE + G
Sbjct: 15 MEVVVNKAKEIVSAYPVVVFSK-----TYCGYCQRVKQLLTQLGATFKVLE-LDEMSDGG 68
Query: 269 -LRETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
++ L +++ T P +F+ G +GGCD + N++G++ L
Sbjct: 69 EIQSALSEWTGQTTVPNVFIKGNHIGGCDRVMETNKQGKLVPL 111
>At2g20270 putative glutaredoxin
Length = 179
Score = 44.3 bits (103), Expect = 9e-05
Identities = 31/101 (30%), Positives = 51/101 (49%), Gaps = 8/101 (7%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG--L 269
LEE V V EN VV + K C +S +V + + V+ V+ LD+ + G L
Sbjct: 74 LEETVKTTVAENPVVVYSK-----TWCSYSSQVKSLFKSLQVEPLVVE-LDQLGSEGSQL 127
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
+ L+K + T P +F+ G+ +GGC ++ KGE+ +
Sbjct: 128 QNVLEKITGQYTVPNVFIGGKHIGGCSDTLQLHNKGELEAI 168
>At5g40370 glutaredoxin -like protein
Length = 111
Score = 43.9 bits (102), Expect = 1e-04
Identities = 22/93 (23%), Positives = 49/93 (52%), Gaps = 5/93 (5%)
Query: 218 KLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKKYS 277
++V VV F K C + +V +L++ G +++V++ E ++ L +++
Sbjct: 8 EIVNSESVVVFSK-----TYCPYCVRVKELLQQLGAKFKAVELDTESDGSQIQSGLAEWT 62
Query: 278 NWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
T P +F+ G +GGCD +++++ G++ L
Sbjct: 63 GQRTVPNVFIGGNHIGGCDATSNLHKDGKLVPL 95
>At4g28730 unknown protein
Length = 174
Score = 42.0 bits (97), Expect = 4e-04
Identities = 29/98 (29%), Positives = 51/98 (51%), Gaps = 8/98 (8%)
Query: 212 LEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG--L 269
+EE + K V EN VV + K C + +V + ++ GV V+ LD+ G L
Sbjct: 69 MEESIRKTVTENTVVIYSK-----TWCSYCTEVKTLFKRLGVQPLVVE-LDQLGPQGPQL 122
Query: 270 RETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEV 307
++ L++ + T P +F+ G+ +GGC +N KG++
Sbjct: 123 QKVLERLTGQHTVPNVFVCGKHIGGCTDTVKLNRKGDL 160
>At5g20500 glutaredoxin
Length = 135
Score = 41.6 bits (96), Expect = 6e-04
Identities = 29/100 (29%), Positives = 50/100 (50%), Gaps = 5/100 (5%)
Query: 211 PLEELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLR 270
P + V K + +K+V F K P C ++ V L++ V Y V++ + + + ++
Sbjct: 30 PEADFVKKTISSHKIVIFSKSY--CPYCKKAKSVFRELDQ--VPYV-VELDEREDGWSIQ 84
Query: 271 ETLKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
L + T PQ+F+NG+ +GG D E GE+A L
Sbjct: 85 TALGEIVGRRTVPQVFINGKHLGGSDDTVDAYESGELAKL 124
>At1g03020 putative glutaredoxin
Length = 102
Score = 39.3 bits (90), Expect = 0.003
Identities = 29/101 (28%), Positives = 49/101 (47%), Gaps = 7/101 (6%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYG-LRET 272
E + L+++ VV F K S C S + ++ G + +V LDE N +
Sbjct: 2 EKISNLLEDKPVVIFSKTS-----CCMSHSIKSLISGYGAN-STVYELDEMSNGPEIERA 55
Query: 273 LKKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
L + PT P +F+ ELVGG + L S+ + ++A L ++
Sbjct: 56 LVELGCKPTVPAVFIGQELVGGANQLMSLQVRNQLASLLRR 96
>At1g77370 unknown protein
Length = 130
Score = 38.9 bits (89), Expect = 0.004
Identities = 28/95 (29%), Positives = 46/95 (47%), Gaps = 5/95 (5%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
V + NK+V F K P C S+++ L++E E D D ++ L +
Sbjct: 37 VQNAILSNKIVIFSKSY--CPYCLRSKRIFSQLKEEPFVVELDQREDGDQ---IQYELLE 91
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGL 310
+ T PQ+F+NG+ +GG D L + E G++ L
Sbjct: 92 FVGRRTVPQVFVNGKHIGGSDDLGAALESGQLQKL 126
>At5g18600 glutaredoxin -like protein
Length = 102
Score = 38.1 bits (87), Expect = 0.006
Identities = 24/100 (24%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
+++ K+V E VV + K S C S + +L G + ++ + + + L
Sbjct: 2 DMITKMVMERPVVIYSKSS-----CCMSHTIKTLLCDFGANPAVYELDEISRGREIEQAL 56
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ P P +F+ GELVGG + + S++ G + + K+
Sbjct: 57 LRLGCSPAVPGVFIGGELVGGANEVMSLHLNGSLIPMLKR 96
>At3g21460 glutaredoxin, putative
Length = 102
Score = 38.1 bits (87), Expect = 0.006
Identities = 23/100 (23%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
++V +L + VV F K + C S + + ++GV V++ + Y + L
Sbjct: 2 DVVARLASQRAVVIFSKST-----CCMSHAIKRLFYEQGVSPAIVEIDQDMYGKDIEWAL 56
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ PT P +F+ G+ VG + + +++ G + L K+
Sbjct: 57 ARLGCSPTVPAVFVGGKFVGTANTVMTLHLNGSLKILLKE 96
>At4g15690 glutaredoxin
Length = 102
Score = 37.7 bits (86), Expect = 0.008
Identities = 24/100 (24%), Positives = 47/100 (47%), Gaps = 5/100 (5%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E + K++ E VV F K S C S + + GV+ ++ + + + + L
Sbjct: 2 ENLQKMISEKSVVIFSKNS-----CCMSHTIKTLFLDFGVNPTIYELDEINIGREIEQAL 56
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ PT P +F+ G+LVGG + + S++ + + K+
Sbjct: 57 AQLGCSPTVPVVFIGGQLVGGANQVMSLHLNRSLVPMLKR 96
>At4g15680 glutaredoxin
Length = 102
Score = 36.6 bits (83), Expect = 0.018
Identities = 23/98 (23%), Positives = 46/98 (46%), Gaps = 5/98 (5%)
Query: 216 VDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETLKK 275
+ K++ E VV F K S C S + + GV+ ++ + + + + L +
Sbjct: 4 LQKMISEKSVVIFSKNS-----CCMSHTIKTLFIDFGVNPTIYELDEINRGKEIEQALAQ 58
Query: 276 YSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
PT P +F+ G+LVGG + + S++ + + K+
Sbjct: 59 LGCSPTVPVVFIGGQLVGGANQVMSLHLNRSLVPMLKR 96
>At4g15700 glutaredoxin homolog
Length = 102
Score = 36.2 bits (82), Expect = 0.024
Identities = 24/100 (24%), Positives = 45/100 (45%), Gaps = 5/100 (5%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E + K++ E VV F K S C S + + GV+ ++ + + L
Sbjct: 2 ENLQKMISEKSVVIFSKNS-----CCMSHTIKTLFLDLGVNPTIYELDEISRGKEIEHAL 56
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ PT P +F+ G+LVGG + + S++ + + K+
Sbjct: 57 AQLGCSPTVPVVFIGGQLVGGANQVMSLHLNRSLVPMLKR 96
>At5g13810 unknown protein
Length = 274
Score = 35.8 bits (81), Expect = 0.031
Identities = 14/33 (42%), Positives = 24/33 (72%)
Query: 281 TFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ PQ+F+ G+ VGG D++ S+ E GE+A + K+
Sbjct: 185 SLPQVFIMGKYVGGADVIKSLFEIGELAKILKE 217
>At1g64500 unknown protein
Length = 368
Score = 35.4 bits (80), Expect = 0.040
Identities = 22/71 (30%), Positives = 41/71 (56%), Gaps = 2/71 (2%)
Query: 243 KVIGILEKEGVDYESVDV-LDEDYNYGLRETLKKYSNWPTFPQIFLNGELVGGCDILTSM 301
+V I+E++GV + DV LD L+E L+ ++ P++F+ G +GG +T+M
Sbjct: 233 RVRAIMEQQGVVVDERDVSLDAGVLSELKELLQDEASVAP-PRVFVKGRYLGGAAEVTAM 291
Query: 302 NEKGEVAGLFK 312
NE G++ + +
Sbjct: 292 NENGKLGRVLR 302
>At4g15670 glutaredoxin
Length = 102
Score = 35.0 bits (79), Expect = 0.053
Identities = 23/100 (23%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query: 214 ELVDKLVKENKVVAFIKGSRSAPLCGFSQKVIGILEKEGVDYESVDVLDEDYNYGLRETL 273
E + K+ E +V F K S C S + + GV+ ++ + + + + L
Sbjct: 2 EKLQKMTSEKSLVIFSKNS-----CCMSHTIKTLFLDLGVNPTIYELDEINRGKEIEQAL 56
Query: 274 KKYSNWPTFPQIFLNGELVGGCDILTSMNEKGEVAGLFKK 313
+ PT P +F+ G+LVGG + + S++ + + K+
Sbjct: 57 AQLGCSPTVPVVFIGGQLVGGANQVMSLHLNRSLIPMLKR 96
>At5g06470 putative protein
Length = 239
Score = 34.7 bits (78), Expect = 0.069
Identities = 25/93 (26%), Positives = 45/93 (47%), Gaps = 5/93 (5%)
Query: 222 ENKVVAFIKGSRSA-PLCGFSQKVIGILEKEGVDYESVDV-LDEDYNYGLRETLKKYSNW 279
E+ VV + G RS ++V +LE V Y DV +D ++ E +
Sbjct: 88 EDSVVFYTTGLRSVRKTFEACRRVRFLLENHQVMYRERDVSMDSEFR---EEMWRLLGGK 144
Query: 280 PTFPQIFLNGELVGGCDILTSMNEKGEVAGLFK 312
T P++F+ G +GG + + ++NE G++ L +
Sbjct: 145 VTSPRLFIRGRYIGGAEEVVALNENGKLKKLLQ 177
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.134 0.397
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,834,564
Number of Sequences: 26719
Number of extensions: 364643
Number of successful extensions: 747
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 17
Number of HSP's that attempted gapping in prelim test: 710
Number of HSP's gapped (non-prelim): 47
length of query: 313
length of database: 11,318,596
effective HSP length: 99
effective length of query: 214
effective length of database: 8,673,415
effective search space: 1856110810
effective search space used: 1856110810
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140721.1


