

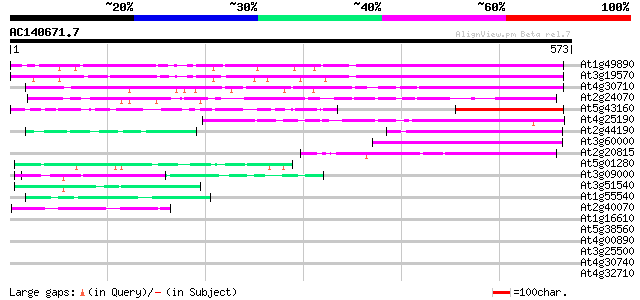
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140671.7 - phase: 0
(573 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g49890 unknown protein 370 e-102
At3g19570 unknown protein 344 9e-95
At4g30710 putative protein 249 3e-66
At2g24070 unknown protein 181 1e-45
At5g43160 putative protein 132 4e-31
At4g25190 putative protein 109 5e-24
At2g44190 unknown protein 105 9e-23
At3g60000 putative protein 95 9e-20
At2g20815 putative protein 82 6e-16
At5g01280 putative protein 47 4e-05
At3g09000 unknown protein 46 5e-05
At3g51540 unknown protein 43 4e-04
At1g55540 unknown protein 43 4e-04
At2g40070 En/Spm-like transposon protein 42 7e-04
At1g16610 arginine/serine-rich protein 42 0.001
At5g38560 putative protein 40 0.004
At4g00890 39 0.006
At3g25500 formin-like protein AHF1 39 0.010
At4g30740 putative protein 37 0.023
At4g32710 putative protein kinase 37 0.030
>At1g49890 unknown protein
Length = 659
Score = 370 bits (949), Expect = e-102
Identities = 268/632 (42%), Positives = 361/632 (56%), Gaps = 101/632 (15%)
Query: 1 MVTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMY----------- 49
MV A +TT +N P RP L ++N R+P+ ++V SRY+
Sbjct: 1 MVAAAISTTDPRN----PPRDRPQSL---TNNGGQRRPRGKQVPSRYLSPSPSHSVSSTT 53
Query: 50 ---------SSSSSSSFSSPPRRTN------SPLVNRTVNSTKQKSTPSVTPAAFVKRSQ 94
+SSSSSS SS RT+ SPL++R+ +T S TP+ KRSQ
Sbjct: 54 TTTTTTTTTTSSSSSSSSSAILRTSKRYPSPSPLLSRS--TTNSASNSIKTPSLLPKRSQ 111
Query: 95 STERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLR 154
S +R+R + E A KML TS RSLSVSFQGE+FS+P+SK K ++T S R
Sbjct: 112 SVDRRRPSAVSVTVGTEMSAATKMLITSTRSLSVSFQGEAFSLPISKKKETTSTPV-SHR 170
Query: 155 RSTPERRKVPATVTTPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQA-----NFM 209
+STPERR+ +TP DQ ENS + DQ RWP S++ + N +
Sbjct: 171 KSTPERRR-----STPV---------RDQRENS--KPVDQQRWPGASRRGNSESVVPNSL 214
Query: 210 NRSLDCGVLLRSSNGFGSNVVRSLRNSLLD--PRVSHDGATLRS--------------ES 253
+RSLDCG R G G L NS++D PRVS +G +
Sbjct: 215 SRSLDCGSD-RGKLGSGFVGRSMLHNSMIDESPRVSVNGRLSLDLGGRDEYLDIGDDIQR 273
Query: 254 NKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGG------KPQRSARAIVVPARFLQE 307
N G + + SD +SV+SGS++GV + G G K + R I+ ARF QE
Sbjct: 274 RPNNGLTSSVSCDFTASDTDSVSSGSTNGVQECGSGVNGEISKSKSLPRNIMASARFWQE 333
Query: 308 ATN----------PSSRNGGIGNRSTVPPKLLVPKKSVFDS-PASSPRGIVNNRLQGSPI 356
+ P S + G+ S++ K + K+ D+ P SSPRG+ SP+
Sbjct: 334 TNSRLRRLQDPGSPLSSSPGL-KTSSISSKFGLSKRFSSDAVPLSSPRGMA------SPV 386
Query: 357 R-SAVRPASPSKLGTPSPRSPSRGVS-PCRGRNGVASSLSS-RFVNEPSVLSFAVDVPRG 413
R SA+R ASPSKL + SP+R +S P R RNGV+ +++ N PS+LSF+ D+ RG
Sbjct: 387 RGSAIRSASPSKLWATTTSSPARALSSPSRARNGVSDQMNAYNRNNTPSILSFSADIRRG 446
Query: 414 KIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLRESVV 473
KIGE+RV+DAH LRLL+NR +QWRFVNARAD+++ VQ LN+EK+L+ AWV+ S+LR SV
Sbjct: 447 KIGEDRVMDAHLLRLLYNRDLQWRFVNARADSTVMVQRLNAEKNLWNAWVSISELRHSVT 506
Query: 474 AKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPVFGGA 533
KRI L LL+Q LKL SIL QM +LE+W++LDR +S SLSGATE+LKASTLRLP+ G
Sbjct: 507 LKRIKLLLLRQKLKLASILRGQMGFLEEWSLLDRDHSSSLSGATESLKASTLRLPIVGKT 566
Query: 534 KIDLLNLKEAICSAMDVMQAMASSICLLLPKV 565
+D+ +LK A+ SA+DVMQAM+SSI L KV
Sbjct: 567 VVDIQDLKHAVSSAVDVMQAMSSSIFSLTSKV 598
>At3g19570 unknown protein
Length = 644
Score = 344 bits (882), Expect = 9e-95
Identities = 255/625 (40%), Positives = 335/625 (52%), Gaps = 92/625 (14%)
Query: 1 MVTAISTTTTTKNTKRAPTPTRP----PLLPSESDNAIVRKPKAREVTSRYMY------- 49
MV AI NT P R P L + + R A+ V SRY+
Sbjct: 1 MVAAIPQGAAISNTDSKNPPPRDRQDKPQLTANNGGLQRRPRAAKNVPSRYLSPSPSHST 60
Query: 50 -----------SSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTER 98
+SSSSS +R SPL++RT NS S TP++ KRSQS +R
Sbjct: 61 TTTTTTATSTSTSSSSSVILRSSKRYPSPLLSRTTNSA---SNLVYTPSSLPKRSQSVDR 117
Query: 99 QRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTP 158
+R E A KML TS RSLSVSFQGE+FS P+SK K + V+ R+ TP
Sbjct: 118 RRP-SAVSDTRTEMSAATKMLITSTRSLSVSFQGEAFSFPISKKKETATPVSH--RKCTP 174
Query: 159 ERRKVPATVTTPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQA-----NFMNRSL 213
ERR+ TP DQ ENS + DQ WP S++ + N ++RS+
Sbjct: 175 ERRRA-----TPV---------RDQRENS--KPVDQQLWPGASRRGSSESVVPNSLSRSV 218
Query: 214 DCGVLLRSSNG--FGSNVV-RSLRNSLLDPRVSHDGAT------------LRSESNKNGG 258
D S +G GS V RS+ RVS DG +R E+
Sbjct: 219 DSD----SDDGRKLGSGFVGRSMLQHSQSSRVSGDGRLNLGFVGGDGMLEMRDENKARQS 274
Query: 259 SEP----VIEPELVPSDNESVTSGSSSGVLDYGGGKPQRSA---RAIVVPARFLQEATNP 311
+ P + + SD +SV+SGS++G + G G+ ++ R + +F QE +
Sbjct: 275 THPRLASSVSCDFTASDTDSVSSGSTNGAHECGSGEVSKTRSLPRNGMASTKFWQETNSR 334
Query: 312 SSRNGGIGNR---------STVPPKLLVPKKSVFDSP-ASSPRGIVNNRLQGSPIRSAVR 361
R G+ S++ K K+ DSP SSPRG+ SPIR A R
Sbjct: 335 LRRMQDPGSPQCSSPSSRISSISSKFSQSKRFSSDSPLTSSPRGMT------SPIRGATR 388
Query: 362 PASPSKLGTPSPRSPSR-GVSPCRGRNGVASSLSSRFVNEPSVLSFAVDVPRGKIGENRV 420
PASPSKL + +P+R SP R RNGV+ +++ PS+L F+ D+ RGKIGE+RV
Sbjct: 389 PASPSKLWATATSAPARTSSSPSRVRNGVSEQMNAYNRTLPSILCFSADIRRGKIGEDRV 448
Query: 421 IDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLRESVVAKRIMLQ 480
+DAH LRLL+NR +QWRF NARAD++L VQ L++EK L+ AWV+ S+LR SV KRI L
Sbjct: 449 MDAHLLRLLYNRDLQWRFANARADSTLMVQRLSAEKILWNAWVSISELRHSVTLKRIKLL 508
Query: 481 LLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPVFGGAKIDLLNL 540
L++Q LKL SIL EQM YLE+W++LDR +S SLSGATEALKASTLRLPV G A +D+ +L
Sbjct: 509 LMRQKLKLASILKEQMCYLEEWSLLDRNHSNSLSGATEALKASTLRLPVSGKAVVDIQDL 568
Query: 541 KEAICSAMDVMQAMASSICLLLPKV 565
K A+ SA+DVM AM SSI L KV
Sbjct: 569 KHAVSSAVDVMHAMVSSIFSLTSKV 593
>At4g30710 putative protein
Length = 644
Score = 249 bits (636), Expect = 3e-66
Identities = 215/597 (36%), Positives = 305/597 (51%), Gaps = 75/597 (12%)
Query: 17 APTPTRPPLLPSESDNAIV--RKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVN 74
A TR LLPS+ +NA+V R+P+ EV+SRY + + + R SP V R
Sbjct: 4 ATDTTRRRLLPSDKNNAVVATRRPRTMEVSSRYRSPTPTKNG------RCPSPSVTRPTV 57
Query: 75 STKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNG--ETPVAQKMLFT----------S 122
S+ +S + + ++ ST TP + + P + + L T +
Sbjct: 58 SSSSQSVAAKRAVSAERKRPSTPPSPTSPSTPIRDLSIDLPASSRRLSTGRLPESLWPST 117
Query: 123 ARSLSVSFQGESFSIPVSKAKPP--SATVTQSLRRSTPERRKVPATVTT----PTIGRGR 176
RSLSVSFQ +S S+PVSK + P S++ ++LR S+ +K A T+ PT R R
Sbjct: 118 MRSLSVSFQSDSVSVPVSKKERPVSSSSGDRTLRPSSNIAQKHKAETTSVSRKPTPERKR 177
Query: 177 V-----NGNSDQTENSI------SRSGDQHRWPAK-SQQNQANFMNRSLDCGVLLRSSNG 224
N SD +ENS SR +QHRWP++ + +N +NRSLD G ++S G
Sbjct: 178 SPLKGKNNVSDLSENSKPVDGPHSRLIEQHRWPSRIGGKITSNSLNRSLDLGD--KASRG 235
Query: 225 F---GSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGS-- 279
G + SLR L +S L S+ ++ P +N + TSG+
Sbjct: 236 IPTSGPGMGPSLRRMSLP--LSSSSRPLHKTSSNTSSYGGLVSPTKSEDNNIARTSGAQR 293
Query: 280 --SSGVLDYGGGKPQRSARAIVVPARFLQEAT--------NPSSRNGGIGNRSTVPPKLL 329
S+G LD AR +PA + A+ + S G +R P + L
Sbjct: 294 LLSAGSLDRAT-LATAVARLHPLPAPGSRPASPSRTSFLSSSSISRGMSTSRGVSPSRGL 352
Query: 330 VPKKSVFDSPASSP-RGIVNNRLQGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRNG 388
P + + + SP RG+ +R G+ RP++P PSRG+SP R R
Sbjct: 353 SPSRGLSPTRGLSPSRGLSPSR--GTNTSCFARPSTP----------PSRGISPSRIRQT 400
Query: 389 VASSLSSRFVNEPSVLSFAVDVPRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLS 448
S+ SS SVLSF DV +GK + + D H LRLLHNR +QWRF ARA++ +
Sbjct: 401 TTSTQSS---TTTSVLSFITDVKKGKKA-SYIEDVHQLRLLHNRYLQWRFAIARAESVMY 456
Query: 449 VQTLNSEKSLYAAWVATSKLRESVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRV 508
+Q L SE++L+ W A S+L++ V +RI LQ LK +KL S+LN+QM+ LEDWA L+R
Sbjct: 457 IQRLTSEETLFNVWHAISELQDHVTRQRIGLQQLKLEIKLNSLLNDQMVSLEDWATLERD 516
Query: 509 YSGSLSGATEALKASTLRLPVFGGAKIDLLNLKEAICSAMDVMQAMASSICLLLPKV 565
+ SL GA L+A+TLRLP GG K D +LK A+ SA+DVMQAM SSI LL KV
Sbjct: 517 HVSSLVGAISDLEANTLRLPATGGTKADTESLKAAMSSALDVMQAMGSSIWSLLSKV 573
>At2g24070 unknown protein
Length = 560
Score = 181 bits (459), Expect = 1e-45
Identities = 183/585 (31%), Positives = 254/585 (43%), Gaps = 160/585 (27%)
Query: 19 TPTRPPLLPSESDN--AIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNST 76
T RPPL PSE +N ++ R+ + EV+SRY + + + RR SP+V RT
Sbjct: 12 TSPRPPLAPSEKNNVGSVTRRARTMEVSSRYRSPTPTKT------RRCPSPIVTRTA--- 62
Query: 77 KQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGET----PVAQKMLFT----------S 122
PS +P +F+KR+ S ER R P + PV+ + L T +
Sbjct: 63 -----PSSSPESFLKRAVSAERNRGPSTPTTPVSDVLVDLPVSSRRLSTGRLPESLWPST 117
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSAT---------------------VTQSLRRSTPERR 161
RSLSVSFQ +S S+PVSK + P T T R+ TPER+
Sbjct: 118 MRSLSVSFQSDSVSVPVSKKEKPLVTSSTDRTLRPSSSNIAHKQQSETTSVTRKQTPERK 177
Query: 162 KVPATVTTPTIGRGRVNGNSDQTENSISRSGD-------QHRWPAKSQQNQANFMNRSLD 214
+ P +G+ N + Q+ENS G QHRW + + NRS D
Sbjct: 178 RSPL--------KGK-NVSPGQSENSKPMDGSHSMLIPPQHRWSGRIRG------NRSFD 222
Query: 215 CGVLLRSSNGFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNES 274
G D V L ++S+++ S I DN
Sbjct: 223 LG----------------------DKAVRRVSLPLSNKSSRHKKSSSDITRLFSCYDNGR 260
Query: 275 VTSGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKS 334
+ SS+ D + + +P L + P SR P +S
Sbjct: 261 LEVSSSTTSEDSSSTESLKHFSTSSLPR--LHPMSAPGSRTAS-------------PSRS 305
Query: 335 VFDSPASS-PRGIVNNRLQGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRNGVASSL 393
F S +SS RG+ +R SP+R + S + + +P PSRGVSP R R SS
Sbjct: 306 SFSSSSSSNSRGMSPSR-GVSPMRGLSPVGNRSLVRSSTP--PSRGVSPSRIRQTAQSSS 362
Query: 394 SSRFVNEPSVLSFAVDVPRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLN 453
++ SVLSF DV +GK + D H LRLL+NR QWRF NARA+ VQ
Sbjct: 363 TN-----TSVLSFIADVKKGK-KATYIEDVHQLRLLYNRYSQWRFANARAEGVSYVQ--- 413
Query: 454 SEKSLYAAWVATSKLRESVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSL 513
S++AK M+ LEDWA+++R + SL
Sbjct: 414 -----------------SLIAK--------------------MVCLEDWAMVEREHISSL 436
Query: 514 SGATEALKASTLRLPVFGGAKIDLLNLKEAICSAMDVMQAMASSI 558
+GA L+A+TLRLP+ GG K DL +LK A+ SA+DVMQ+M SSI
Sbjct: 437 AGAIGDLEANTLRLPLAGGTKADLGSLKLAMSSALDVMQSMGSSI 481
>At5g43160 putative protein
Length = 445
Score = 132 bits (333), Expect = 4e-31
Identities = 71/110 (64%), Positives = 86/110 (77%)
Query: 456 KSLYAAWVATSKLRESVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSG 515
+ LY AW + S L SV KRI +Q LKQ+LKLISILN QM +LE+W ++DR Y GSL G
Sbjct: 275 RRLYNAWRSISNLYNSVSMKRIEMQHLKQNLKLISILNMQMGHLEEWLVIDRNYMGSLVG 334
Query: 516 ATEALKASTLRLPVFGGAKIDLLNLKEAICSAMDVMQAMASSICLLLPKV 565
A EALK STL LPV GA +++ ++K+AICSA+DVMQAMASSICLLLPKV
Sbjct: 335 AAEALKGSTLCLPVDCGAMVNVQSVKDAICSAVDVMQAMASSICLLLPKV 384
Score = 89.0 bits (219), Expect = 7e-18
Identities = 102/334 (30%), Positives = 148/334 (43%), Gaps = 80/334 (23%)
Query: 1 MVTAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSP 60
M A + + N K+ +PP PSES N R+PK R+V SRY+ +SS S
Sbjct: 4 MTAATISPSFNANVKQ----NKPPSFPSESSN---RRPKTRDVASRYL-GGTSSFFHQSS 55
Query: 61 PRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLF 120
P+R SP+V R V TPS + R QST R+ E A++ML
Sbjct: 56 PKRCQSPIVTRPV-------TPS---SVATNRPQSTPRRESLD-----RREVSKAERMLL 100
Query: 121 TSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGN 180
TS RSL SFQ +SF+ T ERRK ++ T G G+
Sbjct: 101 TSGRSLFASFQADSFT------------------PGTLERRKTTSSATISKSGGGK---- 138
Query: 181 SDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDP 240
Q + +S +WP Q + + +RS+D + G G+ V R+L++S++
Sbjct: 139 --QEKLKLS-----DQWPRSLQPSCLS--SRSVDFTDTRKKLIGSGNGVARALQDSMV-- 187
Query: 241 RVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIVV 300
SN+ E + +L + ESV+SGSS+ G GK AR VV
Sbjct: 188 ------------SNRPVSRERITSVDL---ETESVSSGSSN-----GRGK-MLPARGNVV 226
Query: 301 PARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKS 334
AR Q+ PSS G+ + +V +L PK++
Sbjct: 227 KARVSQDRLEPSSH--GL-RKISVDSSVLSPKEA 257
>At4g25190 putative protein
Length = 443
Score = 109 bits (272), Expect = 5e-24
Identities = 99/379 (26%), Positives = 178/379 (46%), Gaps = 48/379 (12%)
Query: 198 PAKSQQNQANFMNRSLDCGVLLRSS--NGFGSNVVRSLRNSLLDPRVSHDGATLRSESNK 255
P N++ ++ S+ V L +S + S+ S NS ++ +T RS S
Sbjct: 10 PPSPNNNRSRTISSSISLPVSLNASLSSSTSSSSSSSPSNSSKRVMITRSQSTTRS-SRP 68
Query: 256 NGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIVVPARFLQEATNPSSRN 315
G S+ ++P+ N S+S + G+ + S AR+L++ T S R+
Sbjct: 69 IGSSDSKSGENIIPARN------SASRSQEINNGRSRESF------ARYLEQRTRGSPRS 116
Query: 316 GGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAVRPASPSKLGTPSPRS 375
+R P ASSP + + S +++ + ++P+ +P
Sbjct: 117 NA-SSRGVKPG-------------ASSPSAWALSPGRLSTMKTPLSSSAPTTSMCMTP-- 160
Query: 376 PSRGVSPCRGRNGVASSLSSRFVNEPSVLSFAVDVPRGKIGENRVIDAHSLRLLHNRLMQ 435
P VS + R+G +++ VL + + + K+ + D H R+ NRL+Q
Sbjct: 161 PESPVSKAKIRSGGGGAVAG-------VLKYFM--AQKKVSPVQEEDYHRFRIFQNRLLQ 211
Query: 436 WRFVNARADASLSVQTLNSEKSLYAAWVATSKLRESVVAKRIMLQLLKQHLKLISILNEQ 495
WRFVNAR +A+++ +N E L+ W+ K+R VV I +Q L+Q +K+ +L+ Q
Sbjct: 212 WRFVNARTEATMANLKINVEDQLFWVWLRIYKMRNYVVENLIEIQRLRQDIKVREVLSLQ 271
Query: 496 MIYLEDWAILDRVYSGSLSGATEALKASTLRLPVFGGA--------KIDLLNLKEAICSA 547
M L +W+ +D S +LS T L A ++RLP+ GA +ID++++ E + A
Sbjct: 272 MPLLNEWSKIDAKNSEALSKLTRKLHALSVRLPLVHGATKLIIWKLQIDMVSIHEEMVIA 331
Query: 548 MDVMQAMASSICLLLPKVY 566
++VM + I LP+++
Sbjct: 332 IEVMDEIEDVIIKFLPRIH 350
Score = 30.0 bits (66), Expect = 3.6
Identities = 29/113 (25%), Positives = 48/113 (41%), Gaps = 23/113 (20%)
Query: 22 RPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLV----------NR 71
RPP P+ + + + + V+ SSS+SSS SS P ++ ++ +R
Sbjct: 9 RPPS-PNNNRSRTISSSISLPVSLNASLSSSTSSSSSSSPSNSSKRVMITRSQSTTRSSR 67
Query: 72 TVNSTKQKSTPSVTPAA-FVKRSQSTERQRQRQ-----------GTPRPNGET 112
+ S+ KS ++ PA RSQ R R+ G+PR N +
Sbjct: 68 PIGSSDSKSGENIIPARNSASRSQEINNGRSRESFARYLEQRTRGSPRSNASS 120
>At2g44190 unknown protein
Length = 474
Score = 105 bits (261), Expect = 9e-23
Identities = 65/179 (36%), Positives = 94/179 (52%), Gaps = 4/179 (2%)
Query: 386 RNGVASSLSSRFVNEPSVLSFAVDVPRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADA 445
+NGV SL N D + K + D HSL+LLHNR +QWRF NA A+
Sbjct: 238 KNGVGLSLPPVAPNS----KIQADTKKQKKALGQQADVHSLKLLHNRYLQWRFANANAEV 293
Query: 446 SLSVQTLNSEKSLYAAWVATSKLRESVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAIL 505
Q +E+ Y+ + S+L +SV KRI LQ L++ + I+ Q LE WA+L
Sbjct: 294 KTQSQKAQAERMFYSLGLKMSELSDSVQRKRIELQHLQRVKAVTEIVESQTPSLEQWAVL 353
Query: 506 DRVYSGSLSGATEALKASTLRLPVFGGAKIDLLNLKEAICSAMDVMQAMASSICLLLPK 564
+ +S SL TEAL ++LRLP+ K++ L EA+ A M+ + +I L+PK
Sbjct: 354 EDEFSTSLLETTEALLNASLRLPLDSKIKVETKELAEALVVASKSMEGIVQNIGNLVPK 412
Score = 43.1 bits (100), Expect = 4e-04
Identities = 47/174 (27%), Positives = 66/174 (37%), Gaps = 27/174 (15%)
Query: 17 APTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNST 76
AP P PP R+P+ REV+SR+M SSSSS SS + L T NS
Sbjct: 19 APAPVPPPS---------TRRPRVREVSSRFMSPISSSSSSSS--SSSAGDLHQLTSNSP 67
Query: 77 KQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFS 136
+ + +ST QR R+ G+ + +ARSL F +
Sbjct: 68 RHHHQH--------QNQRSTSAQRMRRQLKMQEGD----ENRPSETARSLDSPFPLQQ-- 113
Query: 137 IPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSDQTENSISR 190
V K P + + R P T P R R+N T ++ +R
Sbjct: 114 --VDGGKNPKQHIRSKPLKENGHRLDTPTTAMLPPPSRSRLNQQRLLTASAATR 165
>At3g60000 putative protein
Length = 451
Score = 95.1 bits (235), Expect = 9e-20
Identities = 62/197 (31%), Positives = 96/197 (48%), Gaps = 3/197 (1%)
Query: 371 PSPRSPSRGVS-PCRGRNGVASSLSSRF--VNEPSVLSFAVDVPRGKIGENRVIDAHSLR 427
P P S +R VS P N SS SR P D + + ++ D HSL+
Sbjct: 197 PVPSSLNRSVSSPSSSCNVRESSSFSRLGLPLPPMAPKVPADTKKQRKVTEQLEDVHSLK 256
Query: 428 LLHNRLMQWRFVNARADASLSVQTLNSEKSLYAAWVATSKLRESVVAKRIMLQLLKQHLK 487
LLHNR +QWRF NA A +E +++ S+L +SV KRI LQ L +
Sbjct: 257 LLHNRYLQWRFANANAQVKTQTHKTQTETMIHSFGSKISELHDSVQRKRIELQRLLKTKA 316
Query: 488 LISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLPVFGGAKIDLLNLKEAICSA 547
L++I Q LE W+ ++ YS S+S +A ++LRLP+ G +D L + + +A
Sbjct: 317 LLAITESQTPCLEQWSAIEEEYSTSVSQTIQAFSNASLRLPLDGDIMVDSKQLGDGLVAA 376
Query: 548 MDVMQAMASSICLLLPK 564
++ + ++ +PK
Sbjct: 377 SKIVDGITQNVGNYMPK 393
Score = 32.3 bits (72), Expect = 0.73
Identities = 43/160 (26%), Positives = 64/160 (39%), Gaps = 32/160 (20%)
Query: 19 TPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNSTKQ 78
TP PP R+P+ REV+SR+M +SSSS + L + T NS KQ
Sbjct: 15 TPAPPPS---------TRRPRVREVSSRFMSPVTSSSS-------SAGDLHSLTCNSPKQ 58
Query: 79 KSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIP 138
++RS S +R R RQ E ++ T+ARSL F
Sbjct: 59 HHLQHHQ----IQRSVSAQRLR-RQLKMADGDENRSSE----TAARSLDSPF-------T 102
Query: 139 VSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVN 178
+S+++ S R + P + P R R++
Sbjct: 103 LSQSRKSSKPSHLKPLNENSHRLETPTPMVPPPPSRSRLS 142
>At2g20815 putative protein
Length = 438
Score = 82.4 bits (202), Expect = 6e-16
Identities = 82/270 (30%), Positives = 129/270 (47%), Gaps = 23/270 (8%)
Query: 298 IVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIR 357
I V +++L + T S+ GN T KL +S DS ++ + NRLQ +
Sbjct: 97 IKVSSKYLHDLTAKPSK----GNNKT---KL----RSQDDSQRTNSSKGIENRLQRNNSV 145
Query: 358 SAVRPA------SPSK-LGTPSPRSPSRGVSPCRGRN-GVASSLSSRFVNEPSVLSFAVD 409
S + SP + L T + PS + P RG+ G +L F + S
Sbjct: 146 SRYGSSMSQWALSPGRSLDTQAVTVPSSKLKPPRGKGVGKLINLGFDFFRSKNKSSPFTS 205
Query: 410 VPRGKIGENRVIDAHSLRLLHNRLMQWRFVNARADASLSVQTLNSEKS-LYAAWVATSKL 468
+ K + AH L+L++NRL+QWRFVNARA ++ + EK+ L AW KL
Sbjct: 206 PLKPKTCDTE--SAHQLKLMNNRLLQWRFVNARA-CDVNKNVASQEKNQLLCAWDTLIKL 262
Query: 469 RESVVAKRIMLQLLKQHLKLISILNEQMIYLEDWAILDRVYSGSLSGATEALKASTLRLP 528
V+ +RI LQ +KL + Q+ +LE W ++ + SLS ++L + RLP
Sbjct: 263 NNLVLQERIKLQKKNLEMKLNYVFLSQVKHLEAWEDMEIQHLSSLSIIRDSLHSVLSRLP 322
Query: 529 VFGGAKIDLLNLKEAICSAMDVMQAMASSI 558
+ GAK++L + I +A V A+ S++
Sbjct: 323 LKEGAKVNLESAVSIIKNAEAVTDAIISTV 352
>At5g01280 putative protein
Length = 460
Score = 46.6 bits (109), Expect = 4e-05
Identities = 77/322 (23%), Positives = 122/322 (36%), Gaps = 67/322 (20%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTN 65
S+++++++T R PTPTR P++ + +A T+R +SSS++S + R +
Sbjct: 86 SSSSSSRSTSRPPTPTRKSKTPAKRPS--TPTSRATSTTTRATLTSSSTTSSTRSWSRPS 143
Query: 66 S-------------PLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQR--QRQGTP---- 106
S R ST Q++T S T R S + R TP
Sbjct: 144 SSSGTGTSRVTLTAARATRPTTSTDQQTTGSATSTRSNNRPMSAPNSKPGSRSSTPTRRP 203
Query: 107 -RPNGET---------PVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRS 156
PNG + P+++ L A + S E + +P + PS +LR +
Sbjct: 204 STPNGSSTVLRSKPTKPLSKPALSLEASPIVRSRPWEPYEMPGFSVEAPS-----NLRTT 258
Query: 157 TPERRKVPATVTTPTIGRGRVNGNSDQTENSISRSGDQHRWPAKSQQNQANFMNRSLDCG 216
P+R + ++ T +S SRS R AK Q +RS
Sbjct: 259 LPDRPQTASSSRTRAF-----------DASSSSRSASTERDVAKRQSCSP---SRS---- 300
Query: 217 VLLRSSNGFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIE------PELVPS 270
R+ NG + V SLR + DG + + N E V+ P L S
Sbjct: 301 ---RAPNGNVNGAVPSLRGQRAKTN-NDDGRLISHAAKGNQKVEKVVNMRKLATPRLTES 356
Query: 271 DNESVTSG---SSSGVLDYGGG 289
+ + G SS+G G G
Sbjct: 357 GSRRLGGGGGDSSAGKSSSGSG 378
Score = 30.4 bits (67), Expect = 2.8
Identities = 29/134 (21%), Positives = 54/134 (39%), Gaps = 23/134 (17%)
Query: 48 MYSSSSSSSFSSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQ-------- 99
++SSSS S P ++S +R T++ TP+ P+ R+ ST +
Sbjct: 73 LHSSSSVSGIRRPSSSSSSRSTSRPPTPTRKSKTPAKRPSTPTSRATSTTTRATLTSSST 132
Query: 100 -RQRQGTPRPNGETPV-AQKMLFTSARSL-------------SVSFQGESFSIPVSKAKP 144
+ RP+ + ++ T+AR+ + S + + + +KP
Sbjct: 133 TSSTRSWSRPSSSSGTGTSRVTLTAARATRPTTSTDQQTTGSATSTRSNNRPMSAPNSKP 192
Query: 145 PSATVTQSLRRSTP 158
S + T + R STP
Sbjct: 193 GSRSSTPTRRPSTP 206
>At3g09000 unknown protein
Length = 541
Score = 46.2 bits (108), Expect = 5e-05
Identities = 72/314 (22%), Positives = 117/314 (36%), Gaps = 52/314 (16%)
Query: 13 NTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRT 72
N AP +RP +L S N ++ S +SSS + RR +S +R+
Sbjct: 103 NQHDAPN-SRPTVLKSRLGNC------REDIVSGNNNKPQTSSSSVAGLRRPSSSGSSRS 155
Query: 73 VNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQG 132
+ + S TP R +T R TP A+ T+A + + G
Sbjct: 156 TSRPATPTRRSTTPTTSTSRPVTTRASNSRSSTPTSRATLTAARATTSTAAPRTTTTSSG 215
Query: 133 ESFSIPVSKAKP-PSATVTQS--LRRSTPERRKVPATVTTPTI--GRGRVNGNS-DQTEN 186
+ S +++ P PS+ ++ R +TP RR P+T T P+I + G S T N
Sbjct: 216 SARSATPTRSNPRPSSASSKKPVSRPATPTRR--PSTPTGPSIVSSKAPSRGTSPSPTVN 273
Query: 187 SISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDPRVSHDG 246
S+S++ + P+ + + + + GF +LR +L D VS
Sbjct: 274 SLSKAPSRGTSPSPTLNSSRPWKPPEMP---------GFSLEAPPNLRTTLADRPVS--- 321
Query: 247 ATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIVVPARFLQ 306
S S P GS SG ++ GGG + +
Sbjct: 322 ---ASRGRPGVASAP----------------GSRSGSIERGGGPTSGGS------GNARR 356
Query: 307 EATNPSSRNGGIGN 320
++ +PS IGN
Sbjct: 357 QSCSPSRGRAPIGN 370
Score = 44.3 bits (103), Expect = 2e-04
Identities = 48/161 (29%), Positives = 68/161 (41%), Gaps = 16/161 (9%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSS---SSSFSSPPR 62
S++ ++++T R TPTR P+ S + R VT+R S SS S + + R
Sbjct: 148 SSSGSSRSTSRPATPTRRSTTPTTSTS--------RPVTTRASNSRSSTPTSRATLTAAR 199
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKR--SQSTERQRQRQGTPRPNGETPVAQKMLF 120
T S RT +T S S TP R S S+++ R TP TP ++
Sbjct: 200 ATTSTAAPRTT-TTSSGSARSATPTRSNPRPSSASSKKPVSRPATPTRRPSTPTGPSIVS 258
Query: 121 TSA--RSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPE 159
+ A R S S S S S+ PS T+ S PE
Sbjct: 259 SKAPSRGTSPSPTVNSLSKAPSRGTSPSPTLNSSRPWKPPE 299
Score = 40.8 bits (94), Expect = 0.002
Identities = 46/190 (24%), Positives = 81/190 (42%), Gaps = 15/190 (7%)
Query: 7 TTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSF--SSPPRRT 64
TTTT+ + R+ TPTR PS + + +KP +R T S+ + S S P R
Sbjct: 209 TTTTSSGSARSATPTRSNPRPSSASS---KKPVSRPATPTRRPSTPTGPSIVSSKAPSRG 265
Query: 65 NSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGT--PRPNGETPVAQKMLFTS 122
SP + TVNS + + +P+ + S+ + + PN T +A + + S
Sbjct: 266 TSP--SPTVNSLSKAPSRGTSPSPTLNSSRPWKPPEMPGFSLEAPPNLRTTLADRPVSAS 323
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRS--TPERRKVPATVTTPTI----GRGR 176
V+ S S + + P++ + + RR +P R + P T ++ GR +
Sbjct: 324 RGRPGVASAPGSRSGSIERGGGPTSGGSGNARRQSCSPSRGRAPIGNTNGSLTGVRGRAK 383
Query: 177 VNGNSDQTEN 186
+ +N
Sbjct: 384 ASNGGSGCDN 393
Score = 37.0 bits (84), Expect = 0.030
Identities = 38/175 (21%), Positives = 63/175 (35%), Gaps = 3/175 (1%)
Query: 240 PRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGGGKPQRS--ARA 297
P+ S + +G S P + + T+ +S V + +RA
Sbjct: 134 PQTSSSSVAGLRRPSSSGSSRSTSRPATPTRRSTTPTTSTSRPVTTRASNSRSSTPTSRA 193
Query: 298 IVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIR 357
+ AR P + G+ + P P+ S S R R +P
Sbjct: 194 TLTAARATTSTAAPRTTTTSSGSARSATPTRSNPRPSSASSKKPVSRPATPTRRPSTPTG 253
Query: 358 -SAVRPASPSKLGTPSPRSPSRGVSPCRGRNGVASSLSSRFVNEPSVLSFAVDVP 411
S V +PS+ +PSP S +P RG + + SSR P + F+++ P
Sbjct: 254 PSIVSSKAPSRGTSPSPTVNSLSKAPSRGTSPSPTLNSSRPWKPPEMPGFSLEAP 308
>At3g51540 unknown protein
Length = 438
Score = 43.1 bits (100), Expect = 4e-04
Identities = 48/195 (24%), Positives = 78/195 (39%), Gaps = 14/195 (7%)
Query: 6 STTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSS---SSSFSSPPR 62
S + + R+ + RP +P+ +++ +K ++ + SS S S + R
Sbjct: 105 SESDPSSRPTRSGSTIRPSNIPTIRSSSVPKKTTTTQIQASASVSSPKRTVSRSLTPSSR 164
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
+T SP + ST +TPS A + QR R TPR + A T+
Sbjct: 165 KTPSPTSTPSRISTTTSTTPSFKTAG--------DAQRSRSLTPRAKPQI-AANASSRTN 215
Query: 123 ARSLSVSFQGE-SFSIPVSKAKPPSATVTQSLRRSTPERR-KVPATVTTPTIGRGRVNGN 180
RS SV+ + + S ++ + PP T +L P P T+P + R R G
Sbjct: 216 VRSSSVTSRPDRSTTVSATPRTPPRLKETVTLAFGRPVGHVNSPKRNTSPDVTRTRPKGR 275
Query: 181 SDQTENSISRSGDQH 195
S + SG H
Sbjct: 276 SASPSLIPTFSGTSH 290
Score = 37.0 bits (84), Expect = 0.030
Identities = 63/240 (26%), Positives = 96/240 (39%), Gaps = 48/240 (20%)
Query: 6 STTTTTKNTKRAPTPTR-----PPLLPSESDNAIVR-KPKAREVTSRYMYSSSSSSSFSS 59
+TT+TT + K A R P P + NA R ++ VTSR S++ S++ +
Sbjct: 178 TTTSTTPSFKTAGDAQRSRSLTPRAKPQIAANASSRTNVRSSSVTSRPDRSTTVSATPRT 237
Query: 60 PPR--RTNSPLVNRTV---NSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPV 114
PPR T + R V NS K+ ++P VT RP G +
Sbjct: 238 PPRLKETVTLAFGRPVGHVNSPKRNTSPDVTRT-------------------RPKGRS-- 276
Query: 115 AQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTP-ERRKVPATVTTPTIG 173
++ SL +F G S + K+ PSATV S R R V + +
Sbjct: 277 -------ASPSLIPTFSGTSHTTTTPKSIKPSATVADSTRPGRKLSRASVQMAINHLDLA 329
Query: 174 R-GRVNGNSDQT----ENSISRSGDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSN 228
R G+V+ ++ + +SI S R P S + + N + G RS G+N
Sbjct: 330 RNGKVSTHTFSSPMLYPHSIRSSSSGLRKPCGSSEGSCSSSNHEEEDG---RSLTKEGNN 386
Score = 31.2 bits (69), Expect = 1.6
Identities = 29/114 (25%), Positives = 53/114 (46%), Gaps = 18/114 (15%)
Query: 300 VPARFLQEATNPSSRNGGIGNRSTVPP--KLLVPKKSVF-----DSPASSPRGIVNNRLQ 352
VP R + ++ +R+G S +P VPKK+ + SSP+ V+ L
Sbjct: 101 VPLRSESDPSSRPTRSGSTIRPSNIPTIRSSSVPKKTTTTQIQASASVSSPKRTVSRSLT 160
Query: 353 GSPIRSAVRPASPSKLGTPSPRSP----------SRGVSPCRGRNGVASSLSSR 396
S ++ ++PS++ T + +P SR ++P R + +A++ SSR
Sbjct: 161 PSSRKTPSPTSTPSRISTTTSTTPSFKTAGDAQRSRSLTP-RAKPQIAANASSR 213
>At1g55540 unknown protein
Length = 1821
Score = 43.1 bits (100), Expect = 4e-04
Identities = 50/191 (26%), Positives = 72/191 (37%), Gaps = 26/191 (13%)
Query: 17 APTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNST 76
A +P+ P ++ S + + + P A S SS S+ T+S +R +ST
Sbjct: 1344 ALSPSSPEMVSSSTGQSSLFPPSAPT-------SQVSSDQASATSSLTDS---SRLFSST 1393
Query: 77 KQKSTPSVTPA-AFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESF 135
STP +TP AF ST P + P AQ S +S S
Sbjct: 1394 SLSSTPPITPPDAFQSPQVSTPSSAVPITEPVSEPKKPEAQSSSILSTQSTVDSVA---- 1449
Query: 136 SIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSDQTENSI-SRSGDQ 194
+AT TQ+ P T TP G ++G S T++S+ S +
Sbjct: 1450 ----------NATKTQNEPLPVKSEISNPGTTVTPVSSSGFLSGFSSGTQSSLASMAAPS 1499
Query: 195 HRWPAKSQQNQ 205
WP SQ Q
Sbjct: 1500 FSWPGSSQPQQ 1510
>At2g40070 En/Spm-like transposon protein
Length = 510
Score = 42.4 bits (98), Expect = 7e-04
Identities = 36/162 (22%), Positives = 68/162 (41%), Gaps = 27/162 (16%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPR 62
T +S +T+ +++ PT ++P + S ++ R S+ S+++ S+ P
Sbjct: 209 TPMSRSTSLSSSRLTPTASKPTTSTARSAGSVTR-------------STPSTTTKSAGPS 255
Query: 63 RTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTS 122
R+ +PL T S+ S P++ P+ + RS + R+ P+A T+
Sbjct: 256 RSTTPLSRSTARSSTPTSRPTLPPSKTISRSSTPTRR-------------PIASASAATT 302
Query: 123 ARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVP 164
+ ++S S P PS S R ++P R P
Sbjct: 303 TANPTISQIKPSSPAPAKPMPTPSKNPALS-RAASPTVRSRP 343
Score = 37.0 bits (84), Expect = 0.030
Identities = 40/171 (23%), Positives = 65/171 (37%), Gaps = 15/171 (8%)
Query: 11 TKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVN 70
T ++ + +RP L S N+ +TSR SS SS S RR +S
Sbjct: 96 TMMSQTGDSKSRPATLTSRLANSSTESAARNHLTSRQQTSSPGLSSSSGASRRPSS---- 151
Query: 71 RTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARS----- 125
+ + +TP+ + S+S+ R TP A + T++RS
Sbjct: 152 -SGGPGSRPATPTGRSSTLTANSKSS-----RPSTPTSRATVSSATRPSLTNSRSTVSAT 205
Query: 126 LSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGR 176
+ S S+ S+ P ++ T S RS + + TT + G R
Sbjct: 206 TKPTPMSRSTSLSSSRLTPTASKPTTSTARSAGSVTRSTPSTTTKSAGPSR 256
Score = 35.0 bits (79), Expect = 0.11
Identities = 84/383 (21%), Positives = 127/383 (32%), Gaps = 46/383 (12%)
Query: 38 PKAREVTSRYMYSSSSSSSFS---SPPRRTNSPLVNRTVNSTKQKST--PSVTPAAFVKR 92
P + ++ S + + +PP P + + T T PA R
Sbjct: 55 PSRKAAPDDFLNSEGDKNDYEWLLTPPGTPLFPSLEMESHRTMMSQTGDSKSRPATLTSR 114
Query: 93 --SQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVT 150
+ STE + T R +P S R S G + P ++ +A +
Sbjct: 115 LANSSTESAARNHLTSRQQTSSPGLSSSSGASRRPSSSGGPGSRPATPTGRSSTLTAN-S 173
Query: 151 QSLRRSTPERRKVPATVTTPTI--GRGRVNGNSDQTENSISRSGDQHRW------PAKSQ 202
+S R STP R ++ T P++ R V+ + T S S S R P S
Sbjct: 174 KSSRPSTPTSRATVSSATRPSLTNSRSTVSATTKPTPMSRSTSLSSSRLTPTASKPTTST 233
Query: 203 QNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPV 262
A + RS + + S L S TL + S P
Sbjct: 234 ARSAGSVTRSTPSTT---TKSAGPSRSTTPLSRSTARSSTPTSRPTLPPSKTISRSSTPT 290
Query: 263 IEPELVPSDNESVTSGSSSGVLDYGGGKPQRSARAIVVPARF----LQEATNPSSRNGGI 318
P S + + + S + KP A A +P L A +P+ R+
Sbjct: 291 RRPIASASAATTTANPTISQI------KPSSPAPAKPMPTPSKNPALSRAASPTVRSRPW 344
Query: 319 ------GNRSTVPPKLLVPKKSVFDSPASSPRGIVNNRLQGSPIRSAVRPASPSKLGTPS 372
G PP L + ++ + P S+ RG P + R S G P
Sbjct: 345 KPSDMPGFSLETPPNL---RTTLPERPLSATRG--------RPGAPSSRSGSVEPGGPPG 393
Query: 373 PRSPSRGVSPCRGRNGVASSLSS 395
R + SP RGR + SS SS
Sbjct: 394 GRPRRQSCSPSRGRAPMYSSGSS 416
>At1g16610 arginine/serine-rich protein
Length = 414
Score = 41.6 bits (96), Expect = 0.001
Identities = 72/344 (20%), Positives = 129/344 (36%), Gaps = 68/344 (19%)
Query: 58 SSPPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQK 117
+ P R SP V+ + + + +S +P+ + RS+S R +P +
Sbjct: 2 AKPSRGRRSPSVSGSSSRSSSRSRSGSSPSRSISRSRSRSRSLSSSSSPSRS-------- 53
Query: 118 MLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRV 177
S+ S S +G+S + P + + P ++ S+P ++ V ++
Sbjct: 54 ---VSSGSRSPPRRGKSPAGPARRGRSPPPPPSKG--ASSPSKKAVQESLVLHV------ 102
Query: 178 NGNSDQTENSISRS-GDQHRWPAKSQQNQANFMNRSLDCGVLLRSSNGFGSNVVRS---- 232
+S+SR+ + H + + ++D V L +G+ R+
Sbjct: 103 --------DSLSRNVNEAHLKEIFGNFGEVIHVEIAMDRAVNLPRGHGYVEFKARADAEK 154
Query: 233 ----LRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVPSDNESVTSGSSSGVLDYGG 288
+ + +D +V TL + +PV P + + +++ GG
Sbjct: 155 AQLYMDGAQIDGKVVKATFTLPPRQKVSSPPKPV---SAAPKRDAPKSDNAAADAEKDGG 211
Query: 289 GKPQRSARAIVVPARFLQEATNPSSRNG-GIGNRSTVPPKLLVPKKSVFDSPASSPRGIV 347
+ R T+P + G RS +P + L P++ DSP
Sbjct: 212 PRRPRE--------------TSPQRKTGLSPRRRSPLPRRGLSPRRRSPDSP-------- 249
Query: 348 NNRLQGSPIR------SAVRPASPSKLGTPSPRSPSRGVSPCRG 385
+ R GSPIR RPASPS+ +PS P R SP RG
Sbjct: 250 HRRRPGSPIRRRGDTPPRRRPASPSRGRSPSSPPPRRYRSPPRG 293
Score = 31.6 bits (70), Expect = 1.2
Identities = 35/115 (30%), Positives = 51/115 (43%), Gaps = 17/115 (14%)
Query: 290 KPQRSARAIVVPARFLQEATNPSSRNGGIGNRSTVPPKLLVPKKSVFDSPASSPRGIVNN 349
KP R R+ V + ++ SR+G +RS + +S S +SSP V++
Sbjct: 3 KPSRGRRSPSVSGSSSRSSSR--SRSGSSPSRSISRSR----SRSRSLSSSSSPSRSVSS 56
Query: 350 RLQGSPIRSAVRPASPSKLGTPSPRSPSRGVSPCRGRNGVASSLSSRFVNEPSVL 404
+ SP R PA P++ G P PS+G ASS S + V E VL
Sbjct: 57 GSR-SPPRRGKSPAGPARRGRSPPPPPSKG----------ASSPSKKAVQESLVL 100
>At5g38560 putative protein
Length = 681
Score = 40.0 bits (92), Expect = 0.004
Identities = 45/171 (26%), Positives = 58/171 (33%), Gaps = 12/171 (7%)
Query: 3 TAISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSS---FSS 59
TA T T AP P PP P +S +V V S SSS S +S
Sbjct: 20 TAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSSPPPPVVSSPPPSSSPPPSPPVITS 79
Query: 60 PPRRTNSPLVNRTVNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKML 119
PP S V ++ STP+ TP A + P P P +
Sbjct: 80 PPPTVASSPPPPVVIASPPPSTPATTPPAPPQTVSPPPPPDASPSPPAPTTTNPPPKP-- 137
Query: 120 FTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTP 170
S S GE+ S P PP + + +T PAT +P
Sbjct: 138 -------SPSPPGETPSPPGETPSPPKPSPSTPTPTTTTSPPPPPATSASP 181
>At4g00890
Length = 431
Score = 39.3 bits (90), Expect = 0.006
Identities = 53/234 (22%), Positives = 99/234 (41%), Gaps = 22/234 (9%)
Query: 14 TKRAPTPTRPPLLPSES-DNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRT 72
T+ + +PPLLPS+S D +R P + +S S S R+ SP ++
Sbjct: 194 TEEFESQPKPPLLPSKSIDETRLRSPLMSQASSP---PPLPSKSIDENETRSQSPPISPP 250
Query: 73 VNSTKQK----STPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARS-LS 127
+ + + S+PS P K S E + + P P+ ++ L + S +
Sbjct: 251 KSDKQARSQTHSSPSPPPLLSPKAS---ENHQSKSPMPPPSPTAQISLSSLKSPIPSPAT 307
Query: 128 VSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIGRGRVNGNSDQTENS 187
++ FS P+S+ P + SL + P + K P+ T T N + + +
Sbjct: 308 ITAPPPPFSSPLSQTTP---SPKPSLPQIEPNQIKSPSPKLTNTESHASPEQNLVKPDAN 364
Query: 188 ISRSGDQHRWPAKSQ--QNQANFMNRSLDCGVLLRSSNGFGSNVVRSLRNSLLD 239
+ ++ PAK++ +N+ ++ ++ +SN G N SL +S +D
Sbjct: 365 L-----MNKIPAKNEVPKNRGTLEKKTEPMIMMFINSNVQGFNTSLSLDSSSID 413
>At3g25500 formin-like protein AHF1
Length = 1051
Score = 38.5 bits (88), Expect = 0.010
Identities = 49/211 (23%), Positives = 85/211 (40%), Gaps = 24/211 (11%)
Query: 4 AISTTTTTKNTKRAPTPTRPPLLPSESDNAIVRKPK------------AREVTSRYMYSS 51
+IS + + K ++ P P +D VR P + EV ++ S
Sbjct: 301 SISPSMSPKRSEPKPPVISTPEPAELTDYRFVRSPSLSLASLSSGLKNSDEVGLNQIFRS 360
Query: 52 SSSSSFSSPP---RRTNSPLVNRTVNSTKQKSTPSVTPAAFVKR----SQSTERQRQRQG 104
+ +S ++ P ++ NSPL + ST + P+ TP A+++ S ST R Q
Sbjct: 361 PTVTSLTTSPENNKKENSPLSS---TSTSPERRPNDTPEAYLRSPSHSSASTSPYRCFQK 417
Query: 105 TPR--PNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKAKPPSATVTQSLRRSTPERRK 162
+P P + + Q + S S S G+ F + + S + + S S+PE+
Sbjct: 418 SPEVLPAFMSNLRQGLQSQLLSSPSNSHGGQGFLKQLDALRSRSPSSSSSSVCSSPEKAS 477
Query: 163 VPATVTTPTIGRGRVNGNSDQTENSISRSGD 193
+ VT+P + S + S S D
Sbjct: 478 HKSPVTSPKLSSRNSQSLSSSPDRDFSHSLD 508
Score = 34.3 bits (77), Expect = 0.19
Identities = 58/248 (23%), Positives = 94/248 (37%), Gaps = 23/248 (9%)
Query: 49 YSSSSSSSFSSPPRRTNSPLVN-RTVN----STKQKSTPSVTPAAFVKRSQSTERQRQRQ 103
YS S S P R P N R+VN S S+ S + F+ S S +R
Sbjct: 254 YSPRGSQSGREPLNRVGLPGQNPRSVNNDTISCSSSSSGSPGRSTFISISPSMSPKRSEP 313
Query: 104 GTPRPNGETPVAQKMLFTSARSLSVSFQGESFSIPVSKA--------KPPSATVTQSLRR 155
P + P A+ + RS S+S S + S P ++T S
Sbjct: 314 KPPVISTPEP-AELTDYRFVRSPSLSLASLSSGLKNSDEVGLNQIFRSPTVTSLTTSPEN 372
Query: 156 STPERRKVPATVTTPTIGRGRVNGNSD---QTENSISRSGDQHRWPAKSQQNQANFMN-- 210
+ E + +T T+P R N + ++ + S S +R KS + FM+
Sbjct: 373 NKKENSPLSSTSTSP---ERRPNDTPEAYLRSPSHSSASTSPYRCFQKSPEVLPAFMSNL 429
Query: 211 -RSLDCGVLLRSSNGFGSNVVRSLRNSLLDPRVSHDGATLRSESNKNGGSEPVIEPELVP 269
+ L +L SN G ++L S +++ S K PV P+L
Sbjct: 430 RQGLQSQLLSSPSNSHGGQGFLKQLDALRSRSPSSSSSSVCSSPEKASHKSPVTSPKLSS 489
Query: 270 SDNESVTS 277
+++S++S
Sbjct: 490 RNSQSLSS 497
>At4g30740 putative protein
Length = 91
Score = 37.4 bits (85), Expect = 0.023
Identities = 29/90 (32%), Positives = 41/90 (45%), Gaps = 17/90 (18%)
Query: 124 RSLSVSFQGESFSIPVSKAK------------PPSATVTQSLRRSTP--ERRKVPATVTT 169
R LSVSFQ +S S+PVSK + PS+ + Q + T R+ P +
Sbjct: 2 RILSVSFQSDSVSVPVSKKERLVSSSSGDRTLRPSSNIAQKHKAETTSVSRKPTPERKRS 61
Query: 170 PTIGRGRVNG---NSDQTENSISRSGDQHR 196
P G+ V+ NS + SR +QHR
Sbjct: 62 PLKGKNNVSDLSENSKPVDGPHSRLIEQHR 91
>At4g32710 putative protein kinase
Length = 731
Score = 37.0 bits (84), Expect = 0.030
Identities = 42/156 (26%), Positives = 58/156 (36%), Gaps = 17/156 (10%)
Query: 18 PTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVNRTVNSTK 77
P P P PS +A P + ++ SS S + PP T SP S
Sbjct: 89 PPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISP----PPRSLP 144
Query: 78 QKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQGESFSI 137
+STP V A S +R+ P+P+ P+ S + S+
Sbjct: 145 SESTPPVNTA-----SPPPPSPPRRRSGPKPSFPPPINSSPPNPSPNTPSLPETSPPPKP 199
Query: 138 PVSKAKPPSATVTQSLRRSTPERRKVPATVTTPTIG 173
P+S PS+ STP +K PA VT P G
Sbjct: 200 PLSTTPFPSS--------STPPPKKSPAAVTLPFFG 227
Score = 35.4 bits (80), Expect = 0.086
Identities = 38/154 (24%), Positives = 65/154 (41%), Gaps = 13/154 (8%)
Query: 15 KRAPTPTRPPLLPSESDNAIVRKPKAREVTSRYMYSSSSSSSFSSPPRRTNSPLVN--RT 72
K +P P+ PP N I P++ S +++S SPPRR + P +
Sbjct: 117 KPSPPPSSPPSETVPPGNTISPPPRSLPSESTPPVNTASPPP-PSPPRRRSGPKPSFPPP 175
Query: 73 VNSTKQKSTPSVTPAAFVKRSQSTERQRQRQGTPRPNGETPVAQKMLFTSARSLSVSFQG 132
+NS+ +P+ TP+ S + TP P+ TP +K S ++++ F G
Sbjct: 176 INSSPPNPSPN-TPSL---PETSPPPKPPLSTTPFPSSSTPPPKK----SPAAVTLPFFG 227
Query: 133 ESFSIPVSKAKPPSATVTQSLRRSTPERRKVPAT 166
+ +P PP V + +++P P T
Sbjct: 228 PAGQLPDGTVAPPIGPVIEP--KTSPAESISPGT 259
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.127 0.353
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,692,635
Number of Sequences: 26719
Number of extensions: 582963
Number of successful extensions: 2352
Number of sequences better than 10.0: 101
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 78
Number of HSP's that attempted gapping in prelim test: 2157
Number of HSP's gapped (non-prelim): 195
length of query: 573
length of database: 11,318,596
effective HSP length: 105
effective length of query: 468
effective length of database: 8,513,101
effective search space: 3984131268
effective search space used: 3984131268
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140671.7


