

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140670.5 + phase: 0 /pseudo/partial
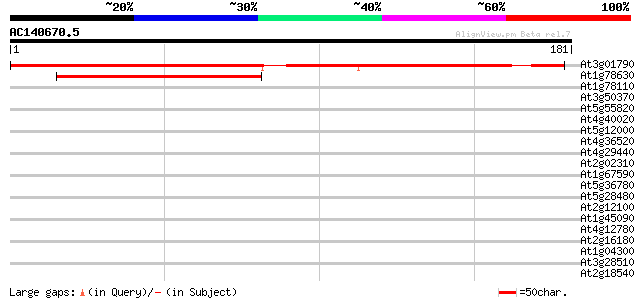
(181 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g01790 ribosomal protein L13 like protein 217 3e-57
At1g78630 unknown protein 65 2e-11
At1g78110 unknown protein 30 0.71
At3g50370 putative protein 29 1.2
At5g55820 unknown protein 28 2.1
At4g40020 putative protein 28 2.1
At5g12000 putative receptor - like kinase 28 2.7
At4g36520 trichohyalin like protein 28 2.7
At4g29440 putative protein 28 3.5
At2g02310 putative phloem-specific lectin 28 3.5
At1g67590 unknown protein 28 3.5
At5g36780 putative protein 27 4.6
At5g28480 putative protein 27 4.6
At2g12100 hypothetical protein 27 4.6
At1g45090 hypothetical protein 27 4.6
At4g12780 auxilin-like protein 27 6.0
At2g16180 hypothetical protein 27 6.0
At1g04300 unknown protein 27 6.0
At3g28510 hypothetical protein 27 7.8
At2g18540 putative vicilin storage protein (globulin-like) 27 7.8
>At3g01790 ribosomal protein L13 like protein
Length = 205
Score = 217 bits (552), Expect = 3e-57
Identities = 119/194 (61%), Positives = 135/194 (69%), Gaps = 28/194 (14%)
Query: 1 RAAAGIKRIKLDGLRWRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNA 60
+A AGIKRI LDGLRWRVFDA+GQ+LGRLASQI+TV+ KDKPTY PNRDDGD+CIVLNA
Sbjct: 17 KAVAGIKRINLDGLRWRVFDARGQVLGRLASQISTVLQAKDKPTYCPNRDDGDICIVLNA 76
Query: 61 KDIAVTGRKLTDKVYYWHTG-----KEL*RIRWPKTLQMLFAKLFCA*FQKTTYVM---- 111
K+I TGRKLTDK Y WHTG KE ++L+ AK +K + M
Sbjct: 77 KEIGFTGRKLTDKFYRWHTGYIGHLKE-------RSLKDQMAKDPTEVIRKAVWRMLPSN 129
Query: 112 ------DRKLRIFPGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENA 165
DRKLRIF G EHPFGD+PLEP+VMPPR VREMRPRARRAMIRAQKKAEQ +
Sbjct: 130 NLRDDRDRKLRIFEGGEHPFGDKPLEPFVMPPRRVREMRPRARRAMIRAQKKAEQAE--- 186
Query: 166 EQRQNAEEMKNSKK 179
E+K KK
Sbjct: 187 ---NEGTEVKKGKK 197
>At1g78630 unknown protein
Length = 241
Score = 65.1 bits (157), Expect = 2e-11
Identities = 32/66 (48%), Positives = 45/66 (67%)
Query: 16 WRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTGRKLTDKVY 75
W V DA +ILGRLAS IA + GK+ +YTP+ D G IV+NA+ +AV+G+K K+Y
Sbjct: 105 WFVVDATDKILGRLASTIANHIRGKNLASYTPSVDMGAFVIVVNAEKVAVSGKKRNQKLY 164
Query: 76 YWHTGK 81
H+G+
Sbjct: 165 RRHSGR 170
>At1g78110 unknown protein
Length = 342
Score = 30.0 bits (66), Expect = 0.71
Identities = 20/62 (32%), Positives = 26/62 (41%), Gaps = 9/62 (14%)
Query: 127 DRPLEPYVMPPRTVREMRPRA---------RRAMIRAQKKAEQQQENAEQRQNAEEMKNS 177
D EP V PP + MR R+ R + Q+K E+Q+E E MK
Sbjct: 230 DTETEPAVPPPNALLLMRCRSAPAKSWLEERMKVKTEQEKREEQKEEKETEDQETSMKTK 289
Query: 178 KK 179
KK
Sbjct: 290 KK 291
>At3g50370 putative protein
Length = 2152
Score = 29.3 bits (64), Expect = 1.2
Identities = 17/45 (37%), Positives = 24/45 (52%), Gaps = 1/45 (2%)
Query: 138 RTVREMRPRARRAMIRAQKKA-EQQQENAEQRQNAEEMKNSKKSE 181
R RE R RR A++ A +QE E + AEE++ SK+ E
Sbjct: 507 RLAREQDERQRRLEEEAREAAFRNEQERLEATRRAEELRKSKEEE 551
>At5g55820 unknown protein
Length = 1826
Score = 28.5 bits (62), Expect = 2.1
Identities = 12/31 (38%), Positives = 22/31 (70%)
Query: 141 REMRPRARRAMIRAQKKAEQQQENAEQRQNA 171
+E++ +A A I+AQK+ ++ Q NAE+ + A
Sbjct: 1642 KELKRQAMDARIKAQKELKEDQNNAEKTRQA 1672
Score = 26.9 bits (58), Expect = 6.0
Identities = 14/61 (22%), Positives = 28/61 (44%)
Query: 121 SEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKS 180
+E DR L+ M ++ + ++ I +KK E +++ + +EM+ KK
Sbjct: 1532 AEQKENDRKLKKEAMKLERAKQEQENLKKQEIEKKKKEEDRKKKEAEMAWKQEMEKKKKE 1591
Query: 181 E 181
E
Sbjct: 1592 E 1592
>At4g40020 putative protein
Length = 615
Score = 28.5 bits (62), Expect = 2.1
Identities = 11/44 (25%), Positives = 25/44 (56%)
Query: 138 RTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
R++ + + +KK E++++ E+++N +E K SKK +
Sbjct: 378 RSLNRQESMPKEVVEVVEKKIEEKEKKEEKKENKKEKKESKKEK 421
>At5g12000 putative receptor - like kinase
Length = 703
Score = 28.1 bits (61), Expect = 2.7
Identities = 15/42 (35%), Positives = 23/42 (54%)
Query: 140 VREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
V EM RA + A +KA++ E QR+ EMK ++S+
Sbjct: 350 VAEMEKAKCRAALEAAEKAQRMAELEGQRRKQAEMKARRESQ 391
>At4g36520 trichohyalin like protein
Length = 1432
Score = 28.1 bits (61), Expect = 2.7
Identities = 14/46 (30%), Positives = 29/46 (62%), Gaps = 2/46 (4%)
Query: 138 RTVREMRPRAR--RAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
R ++E R +A R + A++KAEQ+++ EQ++ ++K + + E
Sbjct: 675 RKIKEAREKAENERRAVEAREKAEQERKMKEQQELELQLKEAFEKE 720
>At4g29440 putative protein
Length = 1071
Score = 27.7 bits (60), Expect = 3.5
Identities = 12/27 (44%), Positives = 18/27 (66%)
Query: 26 LGRLASQIATVVMGKDKPTYTPNRDDG 52
LG+L+S + V+ K+KP+ P DDG
Sbjct: 544 LGKLSSASTSQVLEKEKPSSPPTFDDG 570
>At2g02310 putative phloem-specific lectin
Length = 307
Score = 27.7 bits (60), Expect = 3.5
Identities = 16/52 (30%), Positives = 25/52 (47%), Gaps = 2/52 (3%)
Query: 16 WRVFDAKGQILGRLASQIATVVMGKDKPTYTPNRDDGDMCIVLNAKDIAVTG 67
WRVF +K + L V GK ++ G C++L AK++ +TG
Sbjct: 100 WRVFSSKKDLYFTLCYDPVLVEDGKK--SFWLETASGKKCVLLAAKELWITG 149
>At1g67590 unknown protein
Length = 347
Score = 27.7 bits (60), Expect = 3.5
Identities = 15/53 (28%), Positives = 30/53 (56%), Gaps = 11/53 (20%)
Query: 140 VREMRPRARRAMIRAQKK-----------AEQQQENAEQRQNAEEMKNSKKSE 181
+++M +A R RA++K AE+++ NAE + N + +K S+K++
Sbjct: 270 MKKMEVKAERMKARAEEKLANKLAATKRIAEERRANAEAKLNEKAVKTSEKAD 322
>At5g36780 putative protein
Length = 565
Score = 27.3 bits (59), Expect = 4.6
Identities = 18/61 (29%), Positives = 30/61 (48%), Gaps = 5/61 (8%)
Query: 122 EHPFGDRPLEPYVMPPRTVREMRPR-----ARRAMIRAQKKAEQQQENAEQRQNAEEMKN 176
E +R E + R V E + + A +A IRAQK+AE++ + E++ + N
Sbjct: 419 EEALKERKREEQLEKARLVMERKRKLQEKAAAKAAIRAQKEAEKKLKECEKKAKKKAAAN 478
Query: 177 S 177
S
Sbjct: 479 S 479
>At5g28480 putative protein
Length = 1248
Score = 27.3 bits (59), Expect = 4.6
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query: 119 PGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQR 168
P S H D P E +TV ++ R +A R +K AEQ Q + ++
Sbjct: 535 PRSSHK-SDSPSEKVERVTKTVEKVNKRVEKANKRVEKAAEQVQRKSVKK 583
>At2g12100 hypothetical protein
Length = 728
Score = 27.3 bits (59), Expect = 4.6
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query: 119 PGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQR 168
P S H D P E +TV ++ R +A R +K AEQ Q + ++
Sbjct: 493 PRSSHK-SDSPSEKVERVTKTVEKVNKRVEKANKRVEKAAEQVQRKSVKK 541
>At1g45090 hypothetical protein
Length = 1210
Score = 27.3 bits (59), Expect = 4.6
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 1/50 (2%)
Query: 119 PGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQR 168
P S H D P E +TV ++ R +A R +K AEQ Q + ++
Sbjct: 529 PRSSHK-SDSPSEKVERVTKTVEKVNKRVEKANKRVEKAAEQVQRKSVKK 577
>At4g12780 auxilin-like protein
Length = 904
Score = 26.9 bits (58), Expect = 6.0
Identities = 13/35 (37%), Positives = 20/35 (57%)
Query: 138 RTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAE 172
R R +A +A A++KAE+ A++R NAE
Sbjct: 602 RAAAGARDKAAKAAAEAREKAEKAAAEAKERANAE 636
>At2g16180 hypothetical protein
Length = 1218
Score = 26.9 bits (58), Expect = 6.0
Identities = 15/55 (27%), Positives = 26/55 (47%), Gaps = 1/55 (1%)
Query: 119 PGSEHPFGDRPLEPYVMPPRTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEE 173
P + H D P E +TV ++ R +A R +K AEQ Q + ++ ++
Sbjct: 514 PRASHK-SDSPSEKVERVTKTVEKVNKRVEKANKRVEKAAEQVQRKSVKKSTKQK 567
>At1g04300 unknown protein
Length = 1082
Score = 26.9 bits (58), Expect = 6.0
Identities = 13/40 (32%), Positives = 25/40 (62%), Gaps = 2/40 (5%)
Query: 142 EMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
E RA+R +KK++++Q A+Q++N + K+ +K E
Sbjct: 439 ETEQRAKRGAAEREKKSKKKQ--AKQKRNKNKGKDKRKEE 476
>At3g28510 hypothetical protein
Length = 530
Score = 26.6 bits (57), Expect = 7.8
Identities = 16/45 (35%), Positives = 29/45 (63%), Gaps = 3/45 (6%)
Query: 138 RTVREMRPRARR-AMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
+T+ E + +AR+ A +KKAE++ + ++ + AEE K KK+E
Sbjct: 455 KTLEEEKEKARKLAEEEEKKKAEKEAKKMKKAEEAEEKK--KKTE 497
>At2g18540 putative vicilin storage protein (globulin-like)
Length = 699
Score = 26.6 bits (57), Expect = 7.8
Identities = 13/44 (29%), Positives = 23/44 (51%)
Query: 138 RTVREMRPRARRAMIRAQKKAEQQQENAEQRQNAEEMKNSKKSE 181
R RE R RR +++ E+ ++ E+R+ EEM ++ E
Sbjct: 531 RKEREEVERKRREEQERKRREEEARKREEERKREEEMAKRREQE 574
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.140 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,769,939
Number of Sequences: 26719
Number of extensions: 145995
Number of successful extensions: 706
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 683
Number of HSP's gapped (non-prelim): 35
length of query: 181
length of database: 11,318,596
effective HSP length: 93
effective length of query: 88
effective length of database: 8,833,729
effective search space: 777368152
effective search space used: 777368152
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 57 (26.6 bits)
Medicago: description of AC140670.5


