

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140670.16 + phase: 0
(235 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
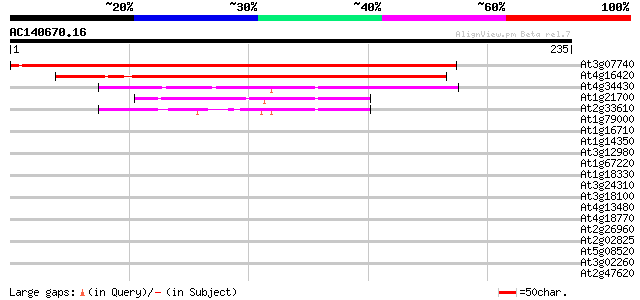
At3g07740 unknown protein 236 1e-62
At4g16420 transcriptional adaptor like protein 235 2e-62
At4g34430 putative protein 80 7e-16
At1g21700 putative transcriptional regulatory protein 60 1e-09
At2g33610 putative SWI/SNF complex subunit SW13 43 1e-04
At1g79000 hypothetical protein 40 0.001
At1g16710 hypothetical protein 39 0.002
At1g14350 putative transcription factor MYB124 (MYB124) 37 0.007
At3g12980 putative CREB-binding protein 37 0.009
At1g67220 hypothetical protein 37 0.009
At1g18330 hypothetical protein 36 0.015
At3g24310 putative transcription factor (MYB71) 35 0.026
At3g18100 putative transcription factor (MYB4R1) 35 0.034
At4g13480 myb-like protein 35 0.045
At4g18770 myb - like protein 34 0.059
At2g26960 putative transcription factor (MYB81) 34 0.059
At2g02825 MYB family transcription factor (MYB88) 34 0.059
At5g08520 unknown protein 34 0.077
At3g02260 unknown protein 34 0.077
At2g47620 SWI/SNF family like transcription activator 34 0.077
>At3g07740 unknown protein
Length = 548
Score = 236 bits (601), Expect = 1e-62
Identities = 107/187 (57%), Positives = 143/187 (76%), Gaps = 1/187 (0%)
Query: 1 MGRCCRAASRSSPIAADDNRSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKD 60
MGR + ASR + + +SKRKK++L +N S + G+ + K LY CNYC+KD
Sbjct: 1 MGRS-KLASRPAEEDLNPGKSKRKKISLGPENAAASISTGIEAGNERKPGLYCCNYCDKD 59
Query: 61 ISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLL 120
+SG +R KCAVC DFDLC+ECFSVGVEL HK++HPYRVMDNLSF L+ DW+A+EE+LL
Sbjct: 60 LSGLVRFKCAVCMDFDLCVECFSVGVELNRHKNSHPYRVMDNLSFSLVTSDWNADEEILL 119
Query: 121 LEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAM 180
LEA+ YGFGNW +VAD++G+K+ ++CI H+N+ Y+ SPCFP+PDLSH GK+K+ELLAM
Sbjct: 120 LEAIATYGFGNWKEVADHVGSKTTTECIKHFNSAYMQSPCFPLPDLSHTIGKSKDELLAM 179
Query: 181 AKGNQVK 187
+K + VK
Sbjct: 180 SKDSAVK 186
>At4g16420 transcriptional adaptor like protein
Length = 487
Score = 235 bits (599), Expect = 2e-62
Identities = 105/164 (64%), Positives = 134/164 (81%), Gaps = 4/164 (2%)
Query: 20 RSKRKKVALNADNLETSSAAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCI 79
R+++KK A N +N E++S G GK Y+C+YC KDI+GKIRIKCAVC DFDLCI
Sbjct: 17 RTRKKKNAANVENFESTSLVP-GAEGGGK---YNCDYCQKDITGKIRIKCAVCPDFDLCI 72
Query: 80 ECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNI 139
EC SVG E+TPHK +HPYRVM NL+FPLICPDWSA++EMLLLE L++YG GNW +VA+++
Sbjct: 73 ECMSVGAEITPHKCDHPYRVMGNLTFPLICPDWSADDEMLLLEGLEIYGLGNWAEVAEHV 132
Query: 140 GTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSGKNKEELLAMAKG 183
GTKSK QC++HY +Y+NSP FP+PD+SH +GKN++EL AMAKG
Sbjct: 133 GTKSKEQCLEHYRNIYLNSPFFPLPDMSHVAGKNRKELQAMAKG 176
>At4g34430 putative protein
Length = 827
Score = 80.5 bits (197), Expect = 7e-16
Identities = 49/152 (32%), Positives = 79/152 (51%), Gaps = 4/152 (2%)
Query: 38 AAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPY 97
AA + +G YHCN C+ D S K R C DFDLC ECF+ G + + S+ +
Sbjct: 104 AADELLKQEGPAVEYHCNSCSADCSRK-RYHCPKQADFDLCTECFNSG-KFSSDMSSSDF 161
Query: 98 RVMDNLSFPLI-CPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYV 156
+M+ P + W+ +E +LLLEAL+++ NWN++A+++ TK+K+QC+ H+ + +
Sbjct: 162 ILMEPAEAPGVGSGKWTDQETLLLLEALEIFK-ENWNEIAEHVATKTKAQCMLHFLQMPI 220
Query: 157 NSPCFPVPDLSHFSGKNKEELLAMAKGNQVKK 188
D K+ +L N V K
Sbjct: 221 EDAFLDQIDYKDPISKDTTDLAVSKDDNSVLK 252
>At1g21700 putative transcriptional regulatory protein
Length = 807
Score = 59.7 bits (143), Expect = 1e-09
Identities = 35/101 (34%), Positives = 57/101 (55%), Gaps = 5/101 (4%)
Query: 53 HCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLSF--PLICP 110
HCN+C++ + + + D LC +CF G + H RV D + F
Sbjct: 344 HCNHCSRPLP-TVYFQSQKKGDILLCCDCFHHGRFVVGHSCLDFVRV-DPMKFYGDQDGD 401
Query: 111 DWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHY 151
+W+ +E +LLLEA+++Y NW +AD++G+KSK+QCI H+
Sbjct: 402 NWTDQETLLLLEAVELYN-ENWVQIADHVGSKSKAQCILHF 441
>At2g33610 putative SWI/SNF complex subunit SW13
Length = 469
Score = 43.1 bits (100), Expect = 1e-04
Identities = 38/123 (30%), Positives = 57/123 (45%), Gaps = 24/123 (19%)
Query: 38 AAGMGITTDGKVSLYHCNYCNKDISGKIRIKCAVCQDFDL--CIECFSVGVELTPHKSNH 95
AA TT + + +CN C S I C C +DL C C+ +SN
Sbjct: 159 AASEPATTVKETAKRNCNGCKAICS----IACFACDKYDLTLCARCYV--------RSN- 205
Query: 96 PYRVMDNLS-FPLI------CPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCI 148
YRV N S F + P+WS +E +LLLEA+ YG +W VA ++ +++ C+
Sbjct: 206 -YRVGINSSEFKRVEISEESKPEWSDKEILLLLEAVMHYG-DDWKKVASHVIGRTEKDCV 263
Query: 149 DHY 151
+
Sbjct: 264 SQF 266
>At1g79000 hypothetical protein
Length = 1691
Score = 40.0 bits (92), Expect = 0.001
Identities = 21/45 (46%), Positives = 26/45 (57%), Gaps = 2/45 (4%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFS-VGVELTPHK-SNHP 96
CN C+ DI +C VC D+D+C CFS G PHK +NHP
Sbjct: 1518 CNACHLDIETGQGWRCEVCPDYDVCNACFSRDGGVNHPHKLTNHP 1562
>At1g16710 hypothetical protein
Length = 1706
Score = 38.9 bits (89), Expect = 0.002
Identities = 18/45 (40%), Positives = 25/45 (55%), Gaps = 2/45 (4%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECF-SVGVELTPHK-SNHP 96
CN C+ DI + +C VC D+D+C C+ G PHK + HP
Sbjct: 1533 CNVCHLDIESGLGWRCEVCPDYDVCNACYKKEGCINHPHKLTTHP 1577
Score = 28.5 bits (62), Expect = 3.2
Identities = 24/81 (29%), Positives = 35/81 (42%), Gaps = 9/81 (11%)
Query: 50 SLYHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLS-FPL- 107
S HC C ++G R C+ C+DF LC C+ + + HP D + FP+
Sbjct: 1412 SCTHC--CTLMVTGN-RWVCSQCKDFQLCDGCYE-AEQKREDRERHPVNQKDKHNIFPVE 1467
Query: 108 ---ICPDWSAEEEMLLLEALD 125
I D +E+L E D
Sbjct: 1468 IADIPTDTKDRDEILESEFFD 1488
>At1g14350 putative transcription factor MYB124 (MYB124)
Length = 436
Score = 37.4 bits (85), Expect = 0.007
Identities = 18/47 (38%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNS 158
WS EE+++L E + ++G NW +A KS QC + T Y+NS
Sbjct: 28 WSQEEDVILREQITLHGTENWAIIASKFKDKSTRQCRRRWYT-YLNS 73
Score = 30.0 bits (66), Expect = 1.1
Identities = 13/43 (30%), Positives = 25/43 (57%), Gaps = 1/43 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTV 154
WS EE+MLL EA ++G W ++A + ++ + + + T+
Sbjct: 80 WSPEEDMLLCEAQRVFG-NRWTEIAKVVSGRTDNAVKNRFTTL 121
>At3g12980 putative CREB-binding protein
Length = 1670
Score = 37.0 bits (84), Expect = 0.009
Identities = 15/51 (29%), Positives = 26/51 (50%), Gaps = 7/51 (13%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMDNLS 104
C C +++ +C VC D+D+C C+S G+ NHP+ ++ S
Sbjct: 1504 CAICQQELETAQGWRCEVCPDYDVCNACYSKGI-------NHPHSIISRPS 1547
Score = 28.5 bits (62), Expect = 3.2
Identities = 18/50 (36%), Positives = 24/50 (48%), Gaps = 4/50 (8%)
Query: 49 VSLYHC--NYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHP 96
V L HC + C +SG R C C++F +C +C V E K HP
Sbjct: 1378 VHLQHCCKHCCTLMVSGN-RWVCNQCKNFQICDKCHEV-EENRVEKEKHP 1425
>At1g67220 hypothetical protein
Length = 1357
Score = 37.0 bits (84), Expect = 0.009
Identities = 17/50 (34%), Positives = 24/50 (48%), Gaps = 3/50 (6%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSVGVELTPHKSNHPYRVMD 101
Y C C+K + +R C C++ LC C+ G EL H Y+ MD
Sbjct: 1097 YSCTRCSKAVLSGLRWFCEKCKNLHLCESCYDAGQEL---PGEHIYKRMD 1143
Score = 33.1 bits (74), Expect = 0.13
Identities = 11/30 (36%), Positives = 16/30 (52%)
Query: 54 CNYCNKDISGKIRIKCAVCQDFDLCIECFS 83
C C KD+S I C +C D+ C C++
Sbjct: 1225 CTACKKDVSTTIYFPCLLCPDYRACTGCYT 1254
>At1g18330 hypothetical protein
Length = 346
Score = 36.2 bits (82), Expect = 0.015
Identities = 17/39 (43%), Positives = 23/39 (58%), Gaps = 1/39 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDH 150
WS EE LEA+ +YG G W + ++IGTK+ Q H
Sbjct: 53 WSEEEHDRFLEAIKLYGRG-WRQIQEHIGTKTAVQIRSH 90
>At3g24310 putative transcription factor (MYB71)
Length = 269
Score = 35.4 bits (80), Expect = 0.026
Identities = 14/31 (45%), Positives = 21/31 (67%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTK 142
W+AEE+ LL++ + ++G G WN VA G K
Sbjct: 23 WTAEEDRLLIDYVQLHGEGRWNSVARLAGLK 53
>At3g18100 putative transcription factor (MYB4R1)
Length = 847
Score = 35.0 bits (79), Expect = 0.034
Identities = 19/69 (27%), Positives = 39/69 (55%), Gaps = 2/69 (2%)
Query: 83 SVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTK 142
S+G TP + Y+ N S ++ +W+AEE+ L A++++G +W VA+ + +
Sbjct: 471 SLGTNRTPFQCLARYQRSLNPS--ILKKEWTAEEDDQLRTAVELFGEKDWQSVANVLKGR 528
Query: 143 SKSQCIDHY 151
+ +QC + +
Sbjct: 529 TGTQCSNRW 537
Score = 35.0 bits (79), Expect = 0.034
Identities = 14/44 (31%), Positives = 28/44 (62%), Gaps = 1/44 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVY 155
W+ EE+ L EA+ +G+ +W+ VA N+ ++ +QC+ + +Y
Sbjct: 601 WTEEEDEKLREAIAEHGY-SWSKVATNLSCRTDNQCLRRWKRLY 643
Score = 30.8 bits (68), Expect = 0.65
Identities = 10/40 (25%), Positives = 25/40 (62%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHY 151
WS EE+ + A+ ++G NW+ ++ + ++++QC + +
Sbjct: 549 WSLEEDKRVKVAVTLFGSQNWHKISQFVPGRTQTQCRERW 588
Score = 30.8 bits (68), Expect = 0.65
Identities = 13/41 (31%), Positives = 24/41 (57%), Gaps = 1/41 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGT-KSKSQCIDHY 151
W+A E+ LL ++ +W D+A ++GT ++ QC+ Y
Sbjct: 445 WTAAEDKNLLRTIEQTSLTDWVDIAVSLGTNRTPFQCLARY 485
>At4g13480 myb-like protein
Length = 261
Score = 34.7 bits (78), Expect = 0.045
Identities = 14/31 (45%), Positives = 21/31 (67%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTK 142
W+AEE+ LL+E + ++G G WN V+ G K
Sbjct: 11 WTAEEDRLLIEYVRVHGEGRWNSVSKLAGLK 41
>At4g18770 myb - like protein
Length = 427
Score = 34.3 bits (77), Expect = 0.059
Identities = 12/46 (26%), Positives = 27/46 (58%)
Query: 107 LICPDWSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYN 152
L+ W+AEE+ +L++ ++ YG W+ +A + + QC + ++
Sbjct: 215 LVKGQWTAEEDRVLIQLVEKYGLRKWSHIAQVLPGRIGKQCRERWH 260
>At2g26960 putative transcription factor (MYB81)
Length = 427
Score = 34.3 bits (77), Expect = 0.059
Identities = 23/65 (35%), Positives = 32/65 (48%), Gaps = 6/65 (9%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNSPCFPVPDLSHFSG 171
W+ E+ LL+ +D +G GNWN V +N G S+C +VN PDL +
Sbjct: 25 WTQAEDNLLIAYVDKHGDGNWNAVQNNSGL---SRCGKSCRLRWVN---HLRPDLKKGAF 78
Query: 172 KNKEE 176
KEE
Sbjct: 79 TEKEE 83
>At2g02825 MYB family transcription factor (MYB88)
Length = 176
Score = 34.3 bits (77), Expect = 0.059
Identities = 17/47 (36%), Positives = 25/47 (53%), Gaps = 1/47 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTVYVNS 158
WS EE+ +L + + + G NW +A KS QC + T Y+NS
Sbjct: 33 WSPEEDDILRKQISLQGTENWAIIASKFNDKSTRQCRRRWYT-YLNS 78
Score = 28.1 bits (61), Expect = 4.2
Identities = 12/43 (27%), Positives = 24/43 (54%), Gaps = 1/43 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADNIGTKSKSQCIDHYNTV 154
WS EE+ LL EA ++G W ++A + ++ + + + T+
Sbjct: 85 WSPEEDTLLCEAQRLFG-NRWTEIAKVVSGRTDNAVKNRFTTL 126
>At5g08520 unknown protein
Length = 298
Score = 33.9 bits (76), Expect = 0.077
Identities = 14/46 (30%), Positives = 25/46 (53%), Gaps = 1/46 (2%)
Query: 112 WSAEEEMLLLEALDMYGFGNWNDVADN-IGTKSKSQCIDHYNTVYV 156
W+ +E L L LD YG G+W ++ N + T++ +Q H ++
Sbjct: 120 WTEDEHRLFLLGLDKYGKGDWRSISRNFVVTRTPTQVASHAQKYFI 165
>At3g02260 unknown protein
Length = 5079
Score = 33.9 bits (76), Expect = 0.077
Identities = 18/48 (37%), Positives = 23/48 (47%), Gaps = 3/48 (6%)
Query: 52 YHCNYCNKDISGKIRIKCAVCQDFDLCIECFSV---GVELTPHKSNHP 96
Y C+ C+ + R C VC DFDLC C+ V PH +HP
Sbjct: 2616 YCCDGCSTVPILRRRWHCTVCPDFDLCEACYEVLDADRLPPPHTRDHP 2663
>At2g47620 SWI/SNF family like transcription activator
Length = 512
Score = 33.9 bits (76), Expect = 0.077
Identities = 19/73 (26%), Positives = 41/73 (56%), Gaps = 6/73 (8%)
Query: 76 DLCIECFSVGVELTPHKSNHPYRVMDNLSFPLICPDWSAEEEMLLLEALDMYGFGNWNDV 135
++C +CF G + + ++++ N + + W+ EE +LLLE++ +G +W +
Sbjct: 197 NICEKCFKNG-NYGENNTADDFKLIGNSAAAV----WTEEEILLLLESVLKHG-DDWELI 250
Query: 136 ADNIGTKSKSQCI 148
+ ++ TKS+ CI
Sbjct: 251 SQSVSTKSRLDCI 263
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.135 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,655,774
Number of Sequences: 26719
Number of extensions: 242935
Number of successful extensions: 793
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 101
Number of HSP's successfully gapped in prelim test: 42
Number of HSP's that attempted gapping in prelim test: 657
Number of HSP's gapped (non-prelim): 172
length of query: 235
length of database: 11,318,596
effective HSP length: 96
effective length of query: 139
effective length of database: 8,753,572
effective search space: 1216746508
effective search space used: 1216746508
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC140670.16


