

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.7 + phase: 0
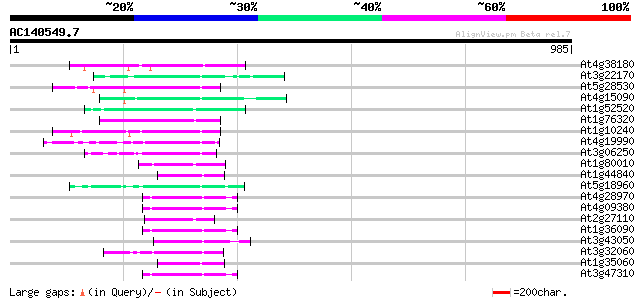
(985 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g38180 unknown protein (At4g38180) 83 9e-16
At3g22170 far-red impaired response protein, putative 70 6e-12
At5g28530 far-red impaired response protein (FAR1) - like 65 1e-10
At4g15090 unknown protein 65 2e-10
At1g52520 F6D8.26 65 2e-10
At1g76320 putative phytochrome A signaling protein 64 5e-10
At1g10240 unknown protein 64 5e-10
At4g19990 putative protein 60 8e-09
At3g06250 unknown protein 60 8e-09
At1g80010 hypothetical protein 59 1e-08
At1g44840 hypothetical protein 59 1e-08
At5g18960 FAR1 - like protein 59 2e-08
At4g28970 putative protein 58 2e-08
At4g09380 putative protein 57 5e-08
At2g27110 Mutator-like transposase 57 5e-08
At1g36090 hypothetical protein 57 5e-08
At3g43050 putative protein 57 7e-08
At3g32060 hypothetical protein 56 9e-08
At1g35060 hypothetical protein 55 2e-07
At3g47310 putative protein 55 3e-07
>At4g38180 unknown protein (At4g38180)
Length = 788
Score = 82.8 bits (203), Expect = 9e-16
Identities = 77/324 (23%), Positives = 141/324 (42%), Gaps = 23/324 (7%)
Query: 106 RLVCERSGSHIVPKKKPKHANTGS----RKCGCLFMISGYQSKQTKEWGLNILNGVHNHP 161
+ VC + G + +K+ K + GC +S + + + +W ++ HNH
Sbjct: 118 QFVCAKEGFRNMNEKRTKDREIKRPRTITRVGCKASLS-VKMQDSGKWLVSGFVKDHNHE 176
Query: 162 MEPALEGHILAG--RLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHC----MTNVKQVY 215
+ P + H L ++ K ++ L + M PR I+ L + T V
Sbjct: 177 LVPPDQVHCLRSHRQISGPAKTLIDTLQAAGMGPRRIMSALIKEYGGISKVGFTEVDCRN 236
Query: 216 NERQQIWKANRGDKKPLQFLISKLEEHNYT----YYSRTQLESNTIEDIFWAHPTSIKLF 271
R K+ G+ +Q L+ L + N +YS E ++ ++FWA P +I F
Sbjct: 237 YMRNNRQKSIEGE---IQLLLDYLRQMNADNPNFFYSVQGSEDQSVGNVFWADPKAIMDF 293
Query: 272 NNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKL 331
+F + D+TY++N YR+P GV G F+ +E E +FVW+
Sbjct: 294 THFGDTVTFDTTYRSNRYRLPFAPFTGVNHHGQPILFGCAFIINETEASFVWLFNTWLAA 353
Query: 332 LSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCV-LNCKYPLGFKK 390
+S+ + I TD D + A+ HVFP + C +H+ +++ + K+P F+
Sbjct: 354 MSAHPPVS--ITTDHDAVIRAAIMHVFPGARHRFCKWHILKKCQEKLSHVFLKHP-SFES 410
Query: 391 D-GKEVSNRDVVKKIMNAWKAMVE 413
D K V+ + V+ W ++++
Sbjct: 411 DFHKCVNLTESVEDFERCWFSLLD 434
>At3g22170 far-red impaired response protein, putative
Length = 814
Score = 70.1 bits (170), Expect = 6e-12
Identities = 70/336 (20%), Positives = 136/336 (39%), Gaps = 37/336 (11%)
Query: 148 EWGLNILNGVHNHPMEPALEGHILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHC 207
+W ++ HNH + PA + E +KI + K + +I LK+
Sbjct: 170 KWVIHSFVREHNHELLPAQA-------VSEQTRKIYAAMAK-QFAEYKTVISLKSDSKSS 221
Query: 208 MTNVKQVYNERQQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQL-ESNTIEDIFWAHPT 266
E+ + GD K L +S+++ N ++ L + ++++FW
Sbjct: 222 F--------EKGRTLSVETGDFKILLDFLSRMQSLNSNFFYAVDLGDDQRVKNVFWVDAK 273
Query: 267 SIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLT 326
S + +F V+ +D+TY N Y+MP+ VGV +G ++ E + W++
Sbjct: 274 SRHNYGSFCDVVSLDTTYVRNKYKMPLAIFVGVNQHYQYMVLGCALISDESAATYSWLME 333
Query: 327 MLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPL 386
+ + + KV++T+ D+ + V +FP + +HV V +
Sbjct: 334 TWLRAIGGQ--APKVLITELDVVMNSIVPEIFPNTRHCLFLWHVLMKVSENL-------- 383
Query: 387 GFKKDGKEVSNRDVVKKIMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLD 446
G+ V D + K + +S + +A ++ + + F + D M +L
Sbjct: 384 -----GQVVKQHD--NFMPKFEKCIYKSGKDEDFAR---KWYKNLARFGLKDDQWMISLY 433
Query: 447 EVKDKIVRAWTDHVLHLGCRTTNRVESAHALLKKYL 482
E + K + VL G T+ R +S +A KY+
Sbjct: 434 EDRKKWAPTYMTDVLLAGMSTSQRADSINAFFDKYM 469
>At5g28530 far-red impaired response protein (FAR1) - like
Length = 700
Score = 65.5 bits (158), Expect = 1e-10
Identities = 74/325 (22%), Positives = 137/325 (41%), Gaps = 36/325 (11%)
Query: 76 DELLEWVRRQANKAGFTIVTQRSSLINPM--FR--LVCERSGSHIVPKKKPKHANTGSRK 131
DE E+ A K+GF+I RS+ + +R VC RSG + P+KK + RK
Sbjct: 80 DEAFEYYSTFARKSGFSIRKARSTESQNLGVYRRDFVCYRSGFN-QPRKKANVEHPRERK 138
Query: 132 ---CGCLFMISGYQSKQ----TKEWGLNILNGVHNHPMEPALEGHILAG--RLKEDDKKI 182
CGC + Y +K+ W ++ + VHNH + + +L ++++ D++
Sbjct: 139 SVRCGCDGKL--YLTKEVVDGVSHWYVSQFSNVHNHELLEDDQVRLLPAYRKIQQSDQER 196
Query: 183 VRDLTKSKMLPRNILIHL----------------KNQRPHCMTNVKQVYNERQQIWKANR 226
+ L+K+ P N ++ L K+ R K V + +
Sbjct: 197 ILLLSKAG-FPVNRIVKLLELEKGVVSGQLPFIEKDVRNFVRACKKSVQENDAFMTEKRE 255
Query: 227 GDKKPLQFLISKLEEHNYTY-YSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYK 285
D L L E + + Y T E+ +E+I WA+ S++ ++ F V+V D++Y+
Sbjct: 256 SDTLELLECCKGLAERDMDFVYDCTSDENQKVENIAWAYGDSVRGYSLFGDVVVFDTSYR 315
Query: 286 TNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTD 345
+ Y + + G+ + +G + E +F W L + + + + I+TD
Sbjct: 316 SVPYGLLLGVFFGIDNNGKAMLLGCVLLQDESCRSFTWALQTFVRFMRGR--HPQTILTD 373
Query: 346 RDMSLMKAVAHVFPESYALNCFFHV 370
D L A+ P + + H+
Sbjct: 374 IDTGLKDAIGREMPNTNHVVFMSHI 398
>At4g15090 unknown protein
Length = 768
Score = 65.1 bits (157), Expect = 2e-10
Identities = 74/339 (21%), Positives = 131/339 (37%), Gaps = 32/339 (9%)
Query: 158 HNHPMEPALEGHILAGR-LKEDDKKIVRDLTKSKMLPRNILIHL-------KNQRPHCMT 209
HNH + PAL H R +K +K + L + + + + KN T
Sbjct: 89 HNHELLPALAYHFRIQRNVKLAEKNNIDILHAVSERTKKMYVEMSRQSGGYKNIGSLLQT 148
Query: 210 NVKQVYNERQQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQL-ESNTIEDIFWAHPTSI 268
+V ++ + + GD + L ++++ N ++ L E + ++FWA S
Sbjct: 149 DVSSQVDKGRYL-ALEEGDSQVLLEYFKRIKKENPKFFYAIDLNEDQRLRNLFWADAKSR 207
Query: 269 KLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTML 328
+ +F V+ D+TY ++P+ +GV +G + E E FVW++
Sbjct: 208 DDYLSFNDVVSFDTTYVKFNDKLPLALFIGVNHHSQPMLLGCALVADESMETFVWLIKTW 267
Query: 329 RKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGF 388
+ + + KVI+TD+D LM AV+ + P + +HV + + K F
Sbjct: 268 LRAMGGR--APKVILTDQDKFLMSAVSELLPNTRHCFALWHVLEKIPEYFSHVMKRHENF 325
Query: 389 -KKDGKEVSNRDVVKKIMNAWKAMVESPNQQLYANALVEFKDSCSDFPIFVDYAMTTLDE 447
K K + + W MV S F + D + L E
Sbjct: 326 LLKFNKCIFRSWTDDEFDMRWWKMV-------------------SQFGLENDEWLLWLHE 366
Query: 448 VKDKIVRAWTDHVLHLGCRTTNRVESAHALLKKYLDNSV 486
+ K V + V G T+ R ES ++ KY+ +
Sbjct: 367 HRQKWVPTFMSDVFLAGMSTSQRSESVNSFFDKYIHKKI 405
>At1g52520 F6D8.26
Length = 703
Score = 65.1 bits (157), Expect = 2e-10
Identities = 69/289 (23%), Positives = 116/289 (39%), Gaps = 14/289 (4%)
Query: 131 KCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGHILAGRLKEDDKKI---VRDLT 187
+ GC MI Q +K W + + HNH L G L +K K + V D
Sbjct: 152 RTGCPAMIRMRQV-DSKRWRVVEVTLDHNH-----LLGCKLYKSVKRKRKCVSSPVSDAK 205
Query: 188 KSKMLPRNILIHLKNQRPHCMTNVK-QVYNERQQIWKANRGDKKPLQFLISKLEEHNYTY 246
K+ ++ + N P+ N K Q + RGD + +++ N +
Sbjct: 206 TIKLYRACVVDNGSNVNPNSTLNKKFQNSTGSPDLLNLKRGDSAAIYNYFCRMQLTNPNF 265
Query: 247 YSRTQL-ESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLT 305
+ + + + ++FWA S + F V+ +DS+Y + + +P+ GV T
Sbjct: 266 FYLMDVNDEGQLRNVFWADAFSKVSCSYFGDVIFIDSSYISGKFEIPLVTFTGVNHHGKT 325
Query: 306 YSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALN 365
+ GF+ E E++ W +L+ LS + IVTDR L A++ VFP S+
Sbjct: 326 TLLSCGFLAGETMESYHW---LLKVWLSVMKRSPQTIVTDRCKPLEAAISQVFPRSHQRF 382
Query: 366 CFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRDVVKKIMNAWKAMVES 414
H+ + ++ Y K K V V + AW MV +
Sbjct: 383 SLTHIMRKIPEKLGGLHNYDAVRKAFTKAVYETLKVVEFEAAWGFMVHN 431
>At1g76320 putative phytochrome A signaling protein
Length = 670
Score = 63.5 bits (153), Expect = 5e-10
Identities = 47/215 (21%), Positives = 96/215 (43%), Gaps = 4/215 (1%)
Query: 158 HNHPMEPALEGHILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRP-HCMTNVKQVYN 216
HNH + P + + R E K L + K P HL + + +
Sbjct: 91 HNHDLLPEQAHYFRSHRNTELVKSNDSRLRRKKNTPLTDCKHLSAYHDLDFIDGYMRNQH 150
Query: 217 ERQQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQL-ESNTIEDIFWAHPTSIKLFNNFP 275
++ + + GD + L + +++E N ++ E + + ++FW I+ + +F
Sbjct: 151 DKGRRLVLDTGDAEILLEFLMRMQEENPKFFFAVDFSEDHLLRNVFWVDAKGIEDYKSFS 210
Query: 276 TVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSK 335
V+ +++Y + Y++P+ VGV +G G + + +VW+ M L++
Sbjct: 211 DVVSFETSYFVSKYKVPLVLFVGVNHHVQPVLLGCGLLADDTVYTYVWL--MQSWLVAMG 268
Query: 336 MNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHV 370
KV++TD++ ++ A+A V PE+ C +HV
Sbjct: 269 GQKPKVMLTDQNNAIKAAIAAVLPETRHCYCLWHV 303
>At1g10240 unknown protein
Length = 680
Score = 63.5 bits (153), Expect = 5e-10
Identities = 74/317 (23%), Positives = 140/317 (43%), Gaps = 28/317 (8%)
Query: 76 DELLEWVRRQANKAGFTIVTQRSSLINPMFR------LVCERSGS---HIVPKKKPKHAN 126
D E+ A + GF+I R+ + + + VC R+G+ + + KP+ N
Sbjct: 58 DTAYEFYSTFAKRCGFSIRRHRTEGKDGVGKGLTRRYFVCHRAGNTPIKTLSEGKPQR-N 116
Query: 127 TGSRKCGC--LFMISGYQSKQTKEWGLNILNGVHNHPM-EPALEGHILAGR-LKEDDKKI 182
S +CGC IS + EW + HNH + EP + A R + + DK
Sbjct: 117 RRSSRCGCQAYLRISKLTELGSTEWRVTGFANHHNHELLEPNQVRFLPAYRSISDADKSR 176
Query: 183 VRDLTKSKMLPRNILIHLKNQRPHCMT------NVKQVYNERQQIWKANRGDKKPLQFL- 235
+ +K+ + + ++ L+ ++ C+ K V N Q K + D+ + FL
Sbjct: 177 ILMFSKTGISVQQMMRLLELEK--CVEPGFLPFTEKDVRNLLQSFKKLDPEDEN-IDFLR 233
Query: 236 -ISKLEEHNYTYYSRTQLESNT-IEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPM 293
++E + + L++N +E+I W++ +SI+ + F +V D+T++ + MP+
Sbjct: 234 MCQSIKEKDPNFKFEFTLDANDKLENIAWSYASSIQSYELFGDAVVFDTTHRLSAVEMPL 293
Query: 294 FEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKA 353
VGV + + G + E ++ W L ++ K + I+TD +M L +A
Sbjct: 294 GIWVGVNNYGVPCFFGCVLLRDENLRSWSWALQAFTGFMNGK--APQTILTDHNMCLKEA 351
Query: 354 VAHVFPESYALNCFFHV 370
+A P + C + V
Sbjct: 352 IAGEMPATKHALCIWMV 368
>At4g19990 putative protein
Length = 672
Score = 59.7 bits (143), Expect = 8e-09
Identities = 72/316 (22%), Positives = 133/316 (41%), Gaps = 47/316 (14%)
Query: 59 EVDTAKFFSDDVKSKNRDELLEWVRRQANKAGFTIVTQ--RSSLINPMF---RLVCERSG 113
E+D + F ++++E E+ + AN GFT + + R S + F + VC R G
Sbjct: 20 EIDEGREF------ESKEEAFEFYKEYANSVGFTTIIKASRRSRMTGKFIDAKFVCTRYG 73
Query: 114 SHIVPKKKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGHILAG 173
S KK+ G+ G+ Q ++ G ++ + + +
Sbjct: 74 S----KKEDIDTGLGT---------DGFNIPQARKRGR-----INRSSSKTDCKAFLHVK 115
Query: 174 RLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQVYNERQQIWKANRGDKKPLQ 233
R ++D + +VR L K H +++++ + R+++ K N K ++
Sbjct: 116 R-RQDGRWVVRSLVKE---------HNHEIFTGQADSLREL-SGRRKLEKLNGAIVKEVK 164
Query: 234 FLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPM 293
KLE+ + + ++ +IFW + F V+ +D+T+ N Y++P+
Sbjct: 165 S--RKLEDGDVERLLNFFTDMQSLRNIFWVDAKGRFDYTCFSDVVSIDTTFIKNEYKLPL 222
Query: 294 FEVVGVTSTDLTYSVGFG-FMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMK 352
GV +GFG +T E + FVW+ K + +VI+T D L +
Sbjct: 223 VAFTGVNHHGQFLLLGFGLLLTDESKSGFVWLFRAWLKAMHG--CRPRVILTKHDQMLKE 280
Query: 353 AVAHVFPESYALNCFF 368
AV VFP S +CF+
Sbjct: 281 AVLEVFPSS--RHCFY 294
>At3g06250 unknown protein
Length = 764
Score = 59.7 bits (143), Expect = 8e-09
Identities = 58/232 (25%), Positives = 98/232 (42%), Gaps = 25/232 (10%)
Query: 132 CGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPALEGHILAGRLKEDDKKIVRDLTKSKM 191
CG I + + + W ++ LN HNH +EP G+ KKI D+T
Sbjct: 251 CGAYMRI---KRQDSGGWIVDRLNKDHNHDLEP--------GKKNAGMKKITDDVTGG-- 297
Query: 192 LPRNILIHLKNQRPHCMTNVKQVYNERQQIWKANRGDKKPLQFLISKLEEHNYTYYSRTQ 251
L LI L + H + + + W L + SK E +Y+ +
Sbjct: 298 LDSVDLIELNDLSNHISSTRENTIGKE---WYPVL-----LDYFQSKQAEDMGFFYA-IE 348
Query: 252 LESN-TIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGF 310
L+SN + IFWA S + F +V D++Y+ Y +P +G +G
Sbjct: 349 LDSNGSCMSIFWADSRSRFACSQFGDAVVFDTSYRKGDYSVPFATFIGFNHHRQPVLLGG 408
Query: 311 GFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESY 362
+ E +E F W+ + +S + + +V D+D+ + +AVA VFP ++
Sbjct: 409 ALVADESKEAFSWLFQTWLRAMSGR--RPRSMVADQDLPIQQAVAQVFPGTH 458
>At1g80010 hypothetical protein
Length = 696
Score = 59.3 bits (142), Expect = 1e-08
Identities = 39/157 (24%), Positives = 75/157 (46%), Gaps = 7/157 (4%)
Query: 226 RGDKKPLQ--FLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDST 283
RG + LQ F +L N+ Y + ++ ++FW + +++F VL+ D+T
Sbjct: 240 RGGFRALQDFFFQIQLSSPNFLYLMDLA-DDGSLRNVFWIDARARAAYSHFGDVLLFDTT 298
Query: 284 YKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNM-HKVI 342
+N Y +P+ VG+ T +G G + + E +VW + R L+ + ++
Sbjct: 299 CLSNAYELPLVAFVGINHHGDTILLGCGLLADQSFETYVW---LFRAWLTCMLGRPPQIF 355
Query: 343 VTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCV 379
+T++ ++ AV+ VFP ++ HV N+ Q V
Sbjct: 356 ITEQCKAMRTAVSEVFPRAHHRLSLTHVLHNICQSVV 392
>At1g44840 hypothetical protein
Length = 926
Score = 58.9 bits (141), Expect = 1e-08
Identities = 31/118 (26%), Positives = 59/118 (49%), Gaps = 2/118 (1%)
Query: 260 IFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEE 319
+F A SI F + +LV+D T+ Y+ + G + Y +GF + E +E
Sbjct: 557 MFLAFGASIAGFRHLRRILVVDGTHLKGKYKGVLLTSSGQDANFQVYPLGFAVVDSENDE 616
Query: 320 NFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQR 377
++ W T L ++++ + I++DR S++ AV VFP++ C H+ N++ +
Sbjct: 617 SWTWFFTKLERIIADSKTL--TILSDRHSSILVAVKRVFPQANHGACIIHLCRNIQTK 672
>At5g18960 FAR1 - like protein
Length = 788
Score = 58.5 bits (140), Expect = 2e-08
Identities = 60/310 (19%), Positives = 120/310 (38%), Gaps = 38/310 (12%)
Query: 106 RLVCERSGSHIVPKKKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPMEPA 165
R VC R G + CG I + + + W ++ LN HNH +EP
Sbjct: 256 RFVCSREGFQ----------HPSRMGCGAYMRI---KRQDSGGWIVDRLNKDHNHDLEPG 302
Query: 166 LEGHILAGRLKEDDKKIVR--DLTKSKMLPRNILIHLKNQRPHCMTNVKQVYNERQQIWK 223
+ ++ +D + DL + N H+K R N + W
Sbjct: 303 KKNDAGMKKIPDDGTGGLDSVDLIELNDFGNN---HIKKTRE----------NRIGKEWY 349
Query: 224 ANRGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDST 283
D F + E+ + Y + + + IFWA + + F +V D++
Sbjct: 350 PLLLD----YFQSRQTEDMGFFYAVELDVNNGSCMSIFWADSRARFACSQFGDSVVFDTS 405
Query: 284 YKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIV 343
Y+ Y +P ++G +G + E +E F+W+ + +S + + IV
Sbjct: 406 YRKGSYSVPFATIIGFNHHRQPVLLGCAMVADESKEAFLWLFQTWLRAMSGR--RPRSIV 463
Query: 344 TDRDMSLMKAVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKD-GKEVSNRDVVK 402
D+D+ + +A+ VFP ++ + ++ ++ + +P FK + K + +
Sbjct: 464 ADQDLPIQQALVQVFPGAHHRYSAWQIREKERENLI---PFPSEFKYEYEKCIYQTQTIV 520
Query: 403 KIMNAWKAMV 412
+ + W A++
Sbjct: 521 EFDSVWSALI 530
Score = 31.2 bits (69), Expect = 3.0
Identities = 31/120 (25%), Positives = 45/120 (36%), Gaps = 30/120 (25%)
Query: 48 GGSGVLPLQPHEVDTAKFFSDDVKSKNRDELLEWVRRQANKAGFTIVT-----QRSSLIN 102
GGSGV P E DTA +E E+ A + GF + T R+
Sbjct: 37 GGSGVEPYVGLEFDTA------------EEAREFYNAYAARTGFKVRTGQLYRSRTDGTV 84
Query: 103 PMFRLVCERSGSHIVPKKKPKHANTGSRKCGCLFMISGYQSKQTKEWGLNILNGVHNHPM 162
R VC + G + + + GC I Q + T +W L+ + HNH +
Sbjct: 85 SSRRFVCSKEGFQL------------NSRTGCTAFIR-VQRRDTGKWVLDQIQKEHNHEL 131
>At4g28970 putative protein
Length = 914
Score = 58.2 bits (139), Expect = 2e-08
Identities = 42/167 (25%), Positives = 76/167 (45%), Gaps = 14/167 (8%)
Query: 234 FLISKLEEHNYTYYSRTQLESNT-IEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMP 292
+++ K+ TY +LE + +F A I+ F V+V+D+T+ +Y
Sbjct: 369 YMLEKVNPGTVTY---VELEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGM 425
Query: 293 MFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMK 352
+ Y + FG + EK+ +++W L L+ + S + V ++DR S+ K
Sbjct: 426 LVIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLEKLKTVYSDVPGL--VFISDRHQSIKK 483
Query: 353 AVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRD 399
AV V+P + C +H+ N++ R ++ KDG V RD
Sbjct: 484 AVKTVYPNALHAACIWHLCQNMRDRVTID--------KDGAAVKFRD 522
>At4g09380 putative protein
Length = 960
Score = 57.0 bits (136), Expect = 5e-08
Identities = 42/167 (25%), Positives = 76/167 (45%), Gaps = 14/167 (8%)
Query: 234 FLISKLEEHNYTYYSRTQLESNT-IEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMP 292
+++ K+ TY +LE + +F A I+ F V+V+D+T+ +Y
Sbjct: 369 YMLEKVNPGTVTY---VELEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGM 425
Query: 293 MFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMK 352
+ Y + FG + EK+ +++W L L+ + S + V ++DR S+ K
Sbjct: 426 LVIATAQDPNHHHYPLAFGIIDSEKDVSWIWFLENLKTVYSDVPGL--VFISDRHQSIKK 483
Query: 353 AVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRD 399
AV V+P + C +H+ N++ R ++ KDG V RD
Sbjct: 484 AVKTVYPNALHAACIWHLCQNMRDRVKID--------KDGAAVKFRD 522
>At2g27110 Mutator-like transposase
Length = 851
Score = 57.0 bits (136), Expect = 5e-08
Identities = 31/124 (25%), Positives = 60/124 (48%), Gaps = 5/124 (4%)
Query: 238 KLEEHNYTYYSRTQL-ESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEV 296
+++ N ++ QL E N + ++FWA S + +F + +D+ Y+ N +R+P
Sbjct: 206 RMQAENPGFFYAVQLDEDNQMSNVFWADSRSRVAYTHFGDTVTLDTRYRCNQFRVPFAPF 265
Query: 297 VGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKV-IVTDRDMSLMKAVA 355
GV G + E + +F+W + + L++ + V +VTD+D ++ A
Sbjct: 266 TGVNHHGQAILFGCALILDESDTSFIW---LFKTFLTAMRDQPPVSLVTDQDRAIQIAAG 322
Query: 356 HVFP 359
VFP
Sbjct: 323 QVFP 326
>At1g36090 hypothetical protein
Length = 645
Score = 57.0 bits (136), Expect = 5e-08
Identities = 42/167 (25%), Positives = 76/167 (45%), Gaps = 14/167 (8%)
Query: 234 FLISKLEEHNYTYYSRTQLESNT-IEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMP 292
+++ K+ TY +LE + +F A I+ F V+V+D+T+ +Y
Sbjct: 329 YMLEKVNPGTVTY---VELEGEKKFKYLFIALGACIEGFRAMRKVIVVDATHLKTVYGGM 385
Query: 293 MFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMK 352
+ Y + FG + EK+ +++W L L+ + S + V ++DR S+ K
Sbjct: 386 LVIATAHDPNHHHYPLAFGIIDSEKDVSWIWFLEKLKTVYSDVPRL--VFISDRHQSIKK 443
Query: 353 AVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRD 399
AV V+P + C +H+ N++ R ++ KDG V RD
Sbjct: 444 AVKTVYPNALHAACIWHLCQNMRDRVKID--------KDGAAVKFRD 482
>At3g43050 putative protein
Length = 608
Score = 56.6 bits (135), Expect = 7e-08
Identities = 42/170 (24%), Positives = 74/170 (42%), Gaps = 16/170 (9%)
Query: 253 ESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGF 312
E + +F+A I+ FN V+++D+T+ + + Y + FG
Sbjct: 310 EKKNFQYLFFALGACIEGFNAMRKVIIVDATHLKIAQGGVLVVALAQDLEHHHYPIDFGV 369
Query: 313 MTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQA 372
+ EK+ ++ W L+ ++ + V V+DR+ SLMK+ ++P S C +H+
Sbjct: 370 LDGEKDVSWSWFFEKLKSVIPDSSEL--VFVSDRNQSLMKSQRELYPLSQHGCCIYHLCQ 427
Query: 373 NVKQRCVLNCKYPLGFKKDGKEVSNRDVVKKIMNAWKAMVESPNQQLYAN 422
NVK C + K +G KK M KA E+ +LY +
Sbjct: 428 NVKGACNYSKKKVVG--------------KKFMECAKAYTEAEFLKLYGD 463
>At3g32060 hypothetical protein
Length = 487
Score = 56.2 bits (134), Expect = 9e-08
Identities = 46/210 (21%), Positives = 98/210 (45%), Gaps = 12/210 (5%)
Query: 166 LEGHILAGRLKEDDKKIVRDLTKSKMLPRNILIHLKNQRPHCMTNVKQVYNERQQIWKAN 225
L H L G+++ +I+ +L ++K+ + + + H + ++K R++ +K
Sbjct: 165 LHDHFL-GQVETPVPRIIMELVQTKLGVKVSYSTVLRGKYHAIYDLK---GSREESYK-- 218
Query: 226 RGDKKPLQFLISKLEEHNYTYYSRTQLESNTIEDIFWAHPTSIKLFNNFPTVLVMDSTYK 285
D +++ K+ + TY E++ + +F A SI+ F VL++D+T+
Sbjct: 219 --DINCYLYMLKKVNDGTVTYLKLD--ENDKFQYVFVALGASIEGFRVMRKVLIVDATHL 274
Query: 286 TNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTD 345
N Y + Y + F + E + ++ W L+ ++ + V +TD
Sbjct: 275 KNGYGGVLVFASAQDPNRHHYIIAFAVLDGENDASWEWFFEKLKTVVPDTSEL--VFMTD 332
Query: 346 RDMSLMKAVAHVFPESYALNCFFHVQANVK 375
R+ SL+KA+ +V+ ++ C +H+ NVK
Sbjct: 333 RNASLIKAIRNVYTAAHHGYCIWHLSQNVK 362
>At1g35060 hypothetical protein
Length = 873
Score = 55.1 bits (131), Expect = 2e-07
Identities = 31/118 (26%), Positives = 57/118 (48%), Gaps = 2/118 (1%)
Query: 260 IFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMPMFEVVGVTSTDLTYSVGFGFMTHEKEE 319
+F A SI+ F + VLV+D T+ Y+ + G + Y + F + E ++
Sbjct: 508 MFLAFGASIQGFKHLRRVLVVDGTHLKGKYKGVLLTASGQDANFQVYPLAFAVVDSENDD 567
Query: 320 NFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMKAVAHVFPESYALNCFFHVQANVKQR 377
+ W T L ++++ N I++DR S+ V VFP+++ C H+ N++ R
Sbjct: 568 AWTWFFTKLERIIAD--NNTLTILSDRHESIKVGVKKVFPQAHHGACIIHLCRNIQAR 623
>At3g47310 putative protein
Length = 735
Score = 54.7 bits (130), Expect = 3e-07
Identities = 40/167 (23%), Positives = 74/167 (43%), Gaps = 14/167 (8%)
Query: 234 FLISKLEEHNYTYYSRTQLESNT-IEDIFWAHPTSIKLFNNFPTVLVMDSTYKTNMYRMP 292
+++ K+ TY +LE + +F A I+ F V+V+D+T+ +Y
Sbjct: 318 YMLEKVNPGTVTY---VELEGEKKFKYLFIALGACIEGFRTMRKVIVVDATHLKTVYGGM 374
Query: 293 MFEVVGVTSTDLTYSVGFGFMTHEKEENFVWVLTMLRKLLSSKMNMHKVIVTDRDMSLMK 352
+ Y + FG + E + +++W L L+ + S + V ++DR S+ K
Sbjct: 375 LVIATAQDPNHHHYPLAFGIIDSENDVSWIWFLEKLKTVYSDVPGL--VFISDRHQSIKK 432
Query: 353 AVAHVFPESYALNCFFHVQANVKQRCVLNCKYPLGFKKDGKEVSNRD 399
V V+P + C +H+ N++ R ++ KDG V RD
Sbjct: 433 VVKTVYPNALHAACIWHLCQNMRDRVKID--------KDGAAVKFRD 471
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,957,720
Number of Sequences: 26719
Number of extensions: 1109587
Number of successful extensions: 3738
Number of sequences better than 10.0: 117
Number of HSP's better than 10.0 without gapping: 51
Number of HSP's successfully gapped in prelim test: 67
Number of HSP's that attempted gapping in prelim test: 3528
Number of HSP's gapped (non-prelim): 211
length of query: 985
length of database: 11,318,596
effective HSP length: 109
effective length of query: 876
effective length of database: 8,406,225
effective search space: 7363853100
effective search space used: 7363853100
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC140549.7


