

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140549.6 - phase: 0 /pseudo
(305 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
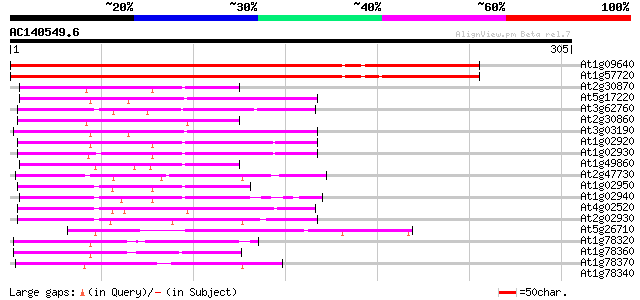
At1g09640 unknown protein 360 e-100
At1g57720 unknown protein 359 e-100
At2g30870 glutathione S-transferase (erd13) 69 4e-12
At5g17220 glutathione S-transferase-like protein 68 7e-12
At3g62760 Glutathione transferase III-like protein 64 8e-11
At2g30860 glutathione S-transferase 62 3e-10
At3g03190 glutathione S-transferase 59 3e-09
At1g02920 glutathione S-transferase 58 6e-09
At1g02930 glutathione S-transferase 58 6e-09
At1g49860 glutathione S-transferase, putative 56 2e-08
At2g47730 glutathione S-transferase (GST6) 56 3e-08
At1g02950 glutathione S-transferase (GST31) 52 5e-07
At1g02940 50 2e-06
At4g02520 Atpm24.1 glutathione S transferase 48 8e-06
At2g02930 putative glutathione S-transferase 47 1e-05
At5g26710 glutamyl-tRNA synthetase 47 1e-05
At1g78320 2,4-D inducible glutathione S-transferase like protein 44 8e-05
At1g78360 hypothetical protein 42 3e-04
At1g78370 unknown protein 42 5e-04
At1g78340 GST7 like protein 41 0.001
>At1g09640 unknown protein
Length = 414
Score = 360 bits (923), Expect = e-100
Identities = 183/256 (71%), Positives = 210/256 (81%), Gaps = 4/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGV+NKTP F+ MNPIGKVPVLETP+G VFESNAIARYV+RL G N+L GSSLI AQI+
Sbjct: 36 MGVTNKTPAFLKMNPIGKVPVLETPEGSVFESNAIARYVSRLNGDNSLNGSSLIEYAQIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEI A+I++ + PR+GF Y P EE AIS+LKRAL+ALN+HL NTYLVGHS
Sbjct: 96 QWIDFSSLEIYASILRWFGPRMGFMPYSAPAEEGAISTLKRALDALNTHLTSNTYLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
+TLADIIT NL +GFA ++ K FTSEFPHVERYFWT+VNQPNF K+LG VKQTEA+PPI
Sbjct: 156 ITLADIITVCNLNLGFATVMTKKFTSEFPHVERYFWTVVNQPNFTKVLGDVKQTEAVPPI 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
S K+ + +KP K+EPKK E PK+ EEEAPKPK KNPLDLLPPS M+LDDWK
Sbjct: 216 AS-KKAAQPAKP--KEEPKKKEAPVAEAPKLAEEEEAPKPKAKNPLDLLPPSPMVLDDWK 272
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 273 RLYSNTKSNFREVAIK 288
>At1g57720 unknown protein
Length = 413
Score = 359 bits (922), Expect = e-100
Identities = 179/256 (69%), Positives = 212/256 (81%), Gaps = 5/256 (1%)
Query: 1 MGVSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL-GHNNLFGSSLIHQAQID 59
MGV+NK+P+F+ MNPIGKVPVLETP+GP+FESNAIARYV+R G N+L GSSLI A I+
Sbjct: 36 MGVTNKSPEFLKMNPIGKVPVLETPEGPIFESNAIARYVSRKNGDNSLNGSSLIEYAHIE 95
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
QWIDFSSLEIDAN++K + PR+G+A + P EE AIS+LKR LEALN+HLA NT+LVGHS
Sbjct: 96 QWIDFSSLEIDANMLKWFAPRMGYAPFSAPAEEAAISALKRGLEALNTHLASNTFLVGHS 155
Query: 120 VTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQVKQTEAMPPI 179
VTLADI+T NL +GFA ++ K FTS FPHVERYFWT+VNQP F+K+LG KQTEA+PP+
Sbjct: 156 VTLADIVTICNLNLGFATVMTKKFTSAFPHVERYFWTMVNQPEFKKVLGDAKQTEAVPPV 215
Query: 180 PSAKQQPKESKPKTKDEPKKVAKSEPEKPKVEVEEEAPKPKPKNPLDLLPPSKMILDDWK 239
P+ K+ P+ +KP K+EPKK A E PK EEEAPKPK KNPLDLLPPS M+LDDWK
Sbjct: 216 PT-KKAPQPAKP--KEEPKKAA-PVAEAPKPAEEEEAPKPKAKNPLDLLPPSPMVLDDWK 271
Query: 240 RLYSNTKRNFREVAIK 255
RLYSNTK NFREVAIK
Sbjct: 272 RLYSNTKSNFREVAIK 287
>At2g30870 glutathione S-transferase (erd13)
Length = 215
Score = 68.6 bits (166), Expect = 4e-12
Identities = 39/128 (30%), Positives = 67/128 (51%), Gaps = 9/128 (7%)
Query: 6 KTPQFINMNPIGKVPVLETPDGPVFESNAIARYVA---RLGHNNLFGSSLIHQAQIDQWI 62
+ P+++ + P GK+PVL D +FES AI RY+A R +L G ++ + Q++QW+
Sbjct: 40 RQPEYLAIQPFGKIPVLVDGDYKIFESRAIMRYIAEKYRSQGPDLLGKTIEERGQVEQWL 99
Query: 63 DFSSLEIDANIMKL-----YLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVG 117
D + ++ L + P +GF ++E+ L L+ + L+ N YL G
Sbjct: 100 DVEATSYHPPLLALTLNIVFAPLMGFPADEKVIKESE-EKLAEVLDVYEAQLSKNEYLAG 158
Query: 118 HSVTLADI 125
V+LAD+
Sbjct: 159 DFVSLADL 166
>At5g17220 glutathione S-transferase-like protein
Length = 214
Score = 67.8 bits (164), Expect = 7e-12
Identities = 50/170 (29%), Positives = 80/170 (46%), Gaps = 9/170 (5%)
Query: 6 KTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL---GHNNLFGSSLIHQAQIDQWI 62
K P+ + P G+VP +E D +FES AIARY A NL G SL H+A +DQW
Sbjct: 41 KKPEHLLRQPFGQVPAIEDGDFKLFESRAIARYYATKFADQGTNLLGKSLEHRAIVDQWA 100
Query: 63 D-----FSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVG 117
D F+ L I + PRLG VE+ + L L+ N+ L+ N +L G
Sbjct: 101 DVETYYFNVLAQPLVINLIIKPRLGEKCDVVLVEDLKV-KLGVVLDIYNNRLSSNRFLAG 159
Query: 118 HSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKIL 167
T+AD+ + + + R++ + ++P+++K++
Sbjct: 160 EEFTMADLTHMPAMGYLMSITDINQMVKARGSFNRWWEEISDRPSWKKLM 209
>At3g62760 Glutathione transferase III-like protein
Length = 219
Score = 64.3 bits (155), Expect = 8e-11
Identities = 49/173 (28%), Positives = 80/173 (45%), Gaps = 15/173 (8%)
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQ------AQI 58
+K P F++MNP GKVP L+ D +FES AI Y+A + G+ L A +
Sbjct: 40 HKLPSFLSMNPFGKVPALQDDDLTLFESRAITAYIAEKHRDK--GTDLTRHEDPKEAAIV 97
Query: 59 DQWIDFSSLEIDANI-----MKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNT 113
W + + + I + +P G + VEEN + +L + L+ L
Sbjct: 98 KLWSEVEAHHFNPAISAVIHQLIVVPLQGESPNAAIVEEN-LENLGKILDVYEERLGKTK 156
Query: 114 YLVGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKI 166
YL G + TLAD+ Y F K + ++ P+V+ ++ L ++P F K+
Sbjct: 157 YLAGDTYTLADLHHVPYTYY-FMKTIHAGLINDRPNVKAWWEDLCSRPAFLKV 208
>At2g30860 glutathione S-transferase
Length = 215
Score = 62.4 bits (150), Expect = 3e-10
Identities = 38/128 (29%), Positives = 64/128 (49%), Gaps = 7/128 (5%)
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVA---RLGHNNLFGSSLIHQAQIDQW 61
+K P ++ + P G VP + D +FES A+ RYVA R +L G ++ + Q++QW
Sbjct: 39 HKQPAYLALQPFGTVPAVVDGDYKIFESRAVMRYVAEKYRSQGPDLLGKTVEDRGQVEQW 98
Query: 62 IDFSSLEIDANIMKLYLPRLGFATYFPPVEENAI----SSLKRALEALNSHLAHNTYLVG 117
+D + ++ L L + + P +E I L L+ +HL+ + YL G
Sbjct: 99 LDVEATTYHPPLLNLTLHIMFASVMGFPSDEKLIKESEEKLAGVLDVYEAHLSKSKYLAG 158
Query: 118 HSVTLADI 125
V+LAD+
Sbjct: 159 DFVSLADL 166
>At3g03190 glutathione S-transferase
Length = 214
Score = 58.9 bits (141), Expect = 3e-09
Identities = 44/173 (25%), Positives = 81/173 (46%), Gaps = 9/173 (5%)
Query: 3 VSNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL---GHNNLFGSSLIHQAQID 59
+ K PQ + P G+VP +E +FES AIARY A +L G +L +A +D
Sbjct: 38 LEQKKPQHLLRQPFGQVPAIEDGYLKLFESRAIARYYATKYADQGTDLLGKTLEGRAIVD 97
Query: 60 QWID-----FSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTY 114
QW++ F ++ + + ++ P+ G VEE + + L+ + LA N Y
Sbjct: 98 QWVEVENNYFYAVALPLVMNVVFKPKSGKPCDVALVEELKV-KFDKVLDVYENRLATNRY 156
Query: 115 LVGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKIL 167
L G TLAD+ + + + + ++ R++ + +P ++K++
Sbjct: 157 LGGDEFTLADLSHMPGMRYIMNETSLSGLVTSRENLNRWWNEISARPAWKKLM 209
>At1g02920 glutathione S-transferase
Length = 209
Score = 58.2 bits (139), Expect = 6e-09
Identities = 52/171 (30%), Positives = 70/171 (40%), Gaps = 10/171 (5%)
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARL---GHNNLFGSSLIHQAQIDQW 61
+K FI NP GKVP E D +FES AI +Y+A N L A I
Sbjct: 41 HKKEPFIFRNPFGKVPAFEDGDFKLFESRAITQYIAHFYSDKGNQLVSLGSKDIAGIAMG 100
Query: 62 IDFSSLEIDANIMKL-----YLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLV 116
I+ S E D KL P G T VEE + L + L+ L + YL
Sbjct: 101 IEIESHEFDPVGSKLVWEQVLKPLYGMTTDKTVVEEEE-AKLAKVLDVYEHRLGESKYLA 159
Query: 117 GHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKIL 167
TL D+ T + K F E PHV + + ++P+ +K+L
Sbjct: 160 SDKFTLVDLHTIPVIQYLLGTPTKKLF-DERPHVSAWVADITSRPSAKKVL 209
>At1g02930 glutathione S-transferase
Length = 208
Score = 58.2 bits (139), Expect = 6e-09
Identities = 54/172 (31%), Positives = 74/172 (42%), Gaps = 13/172 (7%)
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVAR----LGHNNLFGSSLIHQAQIDQ 60
+K FI NP GKVP E D +FES AI +Y+A G+N L S+ A I
Sbjct: 41 HKKEPFILRNPFGKVPAFEDGDFKIFESRAITQYIAHEFSDKGNNLL--STGKDMAIIAM 98
Query: 61 WIDFSSLEIDANIMKL-----YLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYL 115
I+ S E D KL P G T VEE + L + L+ L + YL
Sbjct: 99 GIEIESHEFDPVGSKLVWEQVLKPLYGMTTDKTVVEEEE-AKLAKVLDVYEHRLGESKYL 157
Query: 116 VGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKIL 167
TL D+ T + K F E PHV + + ++P+ +K+L
Sbjct: 158 ASDHFTLVDLHTIPVIQYLLGTPTKKLF-DERPHVSAWVADITSRPSAQKVL 208
>At1g49860 glutathione S-transferase, putative
Length = 254
Score = 56.2 bits (134), Expect = 2e-08
Identities = 44/129 (34%), Positives = 64/129 (49%), Gaps = 10/129 (7%)
Query: 6 KTPQFIN-MNPIGKVPVLETPDGPVFESNAIARYVARLGHN---NLFGSSLIHQAQIDQW 61
KT F++ +NP G+VPVLE D +FE AI RY+A + NL +A + W
Sbjct: 43 KTKTFLSTLNPFGEVPVLEDGDLKLFEPKAITRYLAEQYKDVGTNLLPDDPKKRAIMSMW 102
Query: 62 IDFSS---LEIDANIMK--LYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLV 116
++ S L I + ++K + P G AT V+EN L L + L + YL
Sbjct: 103 MEVDSNQFLPIASTLIKELIINPYQGLATDDTAVQENK-EKLSEVLNIYETRLGESPYLA 161
Query: 117 GHSVTLADI 125
G S +LAD+
Sbjct: 162 GESFSLADL 170
>At2g47730 glutathione S-transferase (GST6)
Length = 263
Score = 55.8 bits (133), Expect = 3e-08
Identities = 50/183 (27%), Positives = 83/183 (45%), Gaps = 21/183 (11%)
Query: 4 SNKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQ------AQ 57
++K + +NP G++P LE D +FES AI +Y+A + G LI Q A
Sbjct: 88 AHKQEAHLALNPFGQIPALEDGDLTLFESRAITQYLAE--EYSEKGEKLISQDCKKVKAT 145
Query: 58 IDQWIDFSSLEIDANIMKLYLPRL-----GFATYFPPVEENAISSLKRALEALNSHLAHN 112
+ W+ + D N KL R+ G T P + L++ L+ + LA +
Sbjct: 146 TNVWLQVEGQQFDPNASKLAFERVFKGMFGMTT-DPAAVQELEGKLQKVLDVYEARLAKS 204
Query: 113 TYLVGHSVTLADI--ITTSNLYIGF-AKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQ 169
+L G S TLAD+ + + +G +K+L S P V + + +P + K++
Sbjct: 205 EFLAGDSFTLADLHHLPAIHYLLGTDSKVLFDS----RPKVSEWIKKISARPAWAKVIDL 260
Query: 170 VKQ 172
KQ
Sbjct: 261 QKQ 263
>At1g02950 glutathione S-transferase (GST31)
Length = 243
Score = 51.6 bits (122), Expect = 5e-07
Identities = 38/138 (27%), Positives = 66/138 (47%), Gaps = 14/138 (10%)
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIH------QAQI 58
+KT F+++NP G+VPV E ++ES AI +Y+A + + G+ L++ A +
Sbjct: 62 HKTEPFLSLNPFGQVPVFEDGSVKLYESRAITQYIAYVHSSR--GTQLLNLRSHETMATL 119
Query: 59 DQWIDFSSLEIDANIMKL-----YLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNT 113
W++ + + D KL P G T V+EN + L++ L L +
Sbjct: 120 TMWMEIEAHQFDPPASKLTWEQVIKPIYGLETDQTIVKENE-AILEKVLNIYEKRLEESR 178
Query: 114 YLVGHSVTLADIITTSNL 131
+L +S TL D+ N+
Sbjct: 179 FLACNSFTLVDLHHLPNI 196
>At1g02940
Length = 256
Score = 49.7 bits (117), Expect = 2e-06
Identities = 44/176 (25%), Positives = 79/176 (44%), Gaps = 28/176 (15%)
Query: 6 KTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIHQAQID------ 59
K P F+ +NP G+VPV + ES AI+ Y+A + + G+ L++
Sbjct: 77 KKPSFLAINPFGQVPVFLDGGLKLTESRAISEYIATVHKSR--GTQLLNYKSYKTMGTQR 134
Query: 60 QWIDFSSLEIDANIMKL-----YLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTY 114
W+ S E D L P G T + V E + L++ L+ L ++++
Sbjct: 135 MWMAIESFEFDPLTSTLTWEQSIKPMYGLKTDYKVVNETE-AKLEKVLDIYEERLKNSSF 193
Query: 115 LVGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKILGQV 170
L +S T+AD+ N+ + L+ + H +R F VN+P+ R+ + ++
Sbjct: 194 LASNSFTMADLYHLPNI-----QYLMDT------HTKRMF---VNRPSVRRWVAEI 235
>At4g02520 Atpm24.1 glutathione S transferase
Length = 212
Score = 47.8 bits (112), Expect = 8e-06
Identities = 43/174 (24%), Positives = 72/174 (40%), Gaps = 15/174 (8%)
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLIH--QAQIDQW- 61
+K F++ NP G+VP E D +FES AI +Y+A N G++L+ I Q+
Sbjct: 41 HKKEPFLSRNPFGQVPAFEDGDLKLFESRAITQYIAHRYENQ--GTNLLQTDSKNISQYA 98
Query: 62 -----IDFSSLEIDANIMKLYLPRLGFATYFPPVEENAI----SSLKRALEALNSHLAHN 112
+ + D KL ++ + Y +E + + L + L+ + L
Sbjct: 99 IMAIGMQVEDHQFDPVASKLAFEQIFKSIYGLTTDEAVVAEEEAKLAKVLDVYEARLKEF 158
Query: 113 TYLVGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKI 166
YL G + TL D+ + K FT E P V + + +P K+
Sbjct: 159 KYLAGETFTLTDLHHIPAIQYLLGTPTKKLFT-ERPRVNEWVAEITKRPASEKV 211
>At2g02930 putative glutathione S-transferase
Length = 212
Score = 47.4 bits (111), Expect = 1e-05
Identities = 44/177 (24%), Positives = 74/177 (40%), Gaps = 19/177 (10%)
Query: 5 NKTPQFINMNPIGKVPVLETPDGPVFESNAIARYVARLGHNNLFGSSLI--HQAQIDQWI 62
+K F++ NP G+VP E D +FES AI +Y+A N G++L+ I Q+
Sbjct: 41 HKKEPFLSRNPFGQVPAFEDGDLKLFESRAITQYIAHRYENQ--GTNLLPADSKNIAQYA 98
Query: 63 DFS-SLEIDANIMKLYLPRLGFATYF---------PPVEENAISSLKRALEALNSHLAHN 112
S ++++A+ +L + F V + L + L+ + L
Sbjct: 99 IMSIGIQVEAHQFDPVASKLAWEQVFKFNYGLNTDQAVVAEEEAKLAKVLDVYEARLKEF 158
Query: 113 TYLVGHSVTLADI--ITTSNLYIGFAKLLVKSFTSEFPHVERYFWTLVNQPNFRKIL 167
YL G + TL D+ I +G K +E P V + + +P K+L
Sbjct: 159 KYLAGETFTLTDLHHIPVIQYLLGTP---TKKLFTERPRVNEWVAEITKRPASEKVL 212
>At5g26710 glutamyl-tRNA synthetase
Length = 719
Score = 47.0 bits (110), Expect = 1e-05
Identities = 41/196 (20%), Positives = 88/196 (43%), Gaps = 34/196 (17%)
Query: 32 SNAIARYVARLGHN--NLFGSSLIHQAQIDQWIDFSSLEIDANIMKLYLPRLGFATYFPP 89
+ + RYV R + +G++ +QID+W+D++S+
Sbjct: 53 ATVLLRYVGRSAKKLPDFYGNNAFDSSQIDEWVDYASVFSSG------------------ 94
Query: 90 VEENAISSLKRALEALNSHLAHNTYLVGHSVTLADIITTSNLYIGFAKLLVKSFTSEFPH 149
S + A ++ +L +T+LVGHS+++AD+ S L + + ++
Sbjct: 95 ------SEFENACGRVDKYLESSTFLVGHSLSIADVAIWSALAGTGQRWESLRKSKKYQS 148
Query: 150 VERYFWTLVNQPNFRKILGQVKQTEAMPPI--PSAKQQPKESKPKTKDEPKKVAKSEPEK 207
+ R+F +++++ + ++L +V T P A + K+S+ K + + K E +
Sbjct: 149 LVRWFNSILDE--YSEVLNKVLATYVKKGSGKPVAAPKSKDSQQAVKGDGQDKGKPEVDL 206
Query: 208 PKVEVEEE----APKP 219
P+ E+ + AP+P
Sbjct: 207 PEAEIGKVKLRFAPEP 222
>At1g78320 2,4-D inducible glutathione S-transferase like protein
Length = 220
Score = 44.3 bits (103), Expect = 8e-05
Identities = 42/136 (30%), Positives = 59/136 (42%), Gaps = 15/136 (11%)
Query: 3 VSNKTPQFINMNPI-GKVPVLETPDGPVFESNAIARYVARL--GHNNLFGSSLIHQAQID 59
+SNK+P + MNPI K+PVL P+ ES +Y+ L N + S +AQ
Sbjct: 37 LSNKSPLLLQMNPIHKKIPVLIHEGKPICESIIQVQYIDELWPDTNPILPSDPYQRAQAR 96
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
W D+ ID K Y+P + +E A L+ L+S L Y G+
Sbjct: 97 FWADY----ID---KKTYVPCKALWSESGEKQEAAKIEFIEVLKTLDSELGDKYYFGGNE 149
Query: 120 VTLADIITTSNLYIGF 135
L DI +IGF
Sbjct: 150 FGLVDI-----AFIGF 160
>At1g78360 hypothetical protein
Length = 222
Score = 42.4 bits (98), Expect = 3e-04
Identities = 38/127 (29%), Positives = 53/127 (40%), Gaps = 8/127 (6%)
Query: 3 VSNKTPQFINMNPIGK-VPVLETPDGPVFESNAIARYVARL--GHNNLFGSSLIHQAQID 59
++NK+P + MNPI K +PVL PV ES +Y+ + +N+ S H+AQ
Sbjct: 38 INNKSPLLLEMNPIHKTIPVLIHNGKPVLESLIQIQYIDEVWSDNNSFLPSDPYHRAQAL 97
Query: 60 QWIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHS 119
W DF ID R +AT +E A L+ L L Y G
Sbjct: 98 FWADF----IDKKEQLYVCGRKTWATKGEELEA-ANKEFIEILKTLQCELGEKPYFGGDK 152
Query: 120 VTLADII 126
DI+
Sbjct: 153 FGFVDIV 159
>At1g78370 unknown protein
Length = 217
Score = 41.6 bits (96), Expect = 5e-04
Identities = 45/150 (30%), Positives = 61/150 (40%), Gaps = 12/150 (8%)
Query: 4 SNKTPQFINMNPI-GKVPVLETPDGPVFESNAIARYV--ARLGHNNLFGSSLIHQAQIDQ 60
SNK+P + NPI K+PVL PV ES + +YV A N F S +AQ
Sbjct: 38 SNKSPLLLQSNPIHKKIPVLVHNGKPVCESLNVVQYVDEAWPEKNPFFPSDPYGRAQARF 97
Query: 61 WIDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSV 120
W DF + K++ + +E A++ L S L Y G S
Sbjct: 98 WADFVDKKFTDAQFKVWGKK-------GEEQEAGKKEFIEAVKILESELGDKPYFGGDSF 150
Query: 121 TLADI--ITTSNLYIGFAKLLVKSFTSEFP 148
DI IT S+ + + K S SE P
Sbjct: 151 GYVDISLITFSSWFQAYEKFGNFSIESESP 180
>At1g78340 GST7 like protein
Length = 218
Score = 40.8 bits (94), Expect = 0.001
Identities = 36/127 (28%), Positives = 54/127 (42%), Gaps = 10/127 (7%)
Query: 5 NKTPQFINMNPIGK-VPVLETPDGPVFESNAIARYVARL--GHNNLFGSSLIHQAQIDQW 61
+K+P + MNP+ K +PVL PV ES + +Y+ + N + S +AQ W
Sbjct: 39 DKSPLLLQMNPVHKKIPVLIHNGKPVCESMNVVQYIDEVWSDKNPILPSDPYQRAQARFW 98
Query: 62 IDFSSLEIDANIMKLYLPRLGFATYFPPVEENAISSLKRALEALNSHLAHNTYLVGHSVT 121
+DF +D KL+ P +E A AL+ L + L Y G +
Sbjct: 99 VDF----VDT---KLFEPADKIWQTKGEEQETAKKEYIEALKILETELGDKPYFGGDTFG 151
Query: 122 LADIITT 128
DI T
Sbjct: 152 FVDIAMT 158
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,076,384
Number of Sequences: 26719
Number of extensions: 317781
Number of successful extensions: 2383
Number of sequences better than 10.0: 144
Number of HSP's better than 10.0 without gapping: 67
Number of HSP's successfully gapped in prelim test: 81
Number of HSP's that attempted gapping in prelim test: 1894
Number of HSP's gapped (non-prelim): 397
length of query: 305
length of database: 11,318,596
effective HSP length: 99
effective length of query: 206
effective length of database: 8,673,415
effective search space: 1786723490
effective search space used: 1786723490
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140549.6


