

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140547.5 - phase: 0
(603 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
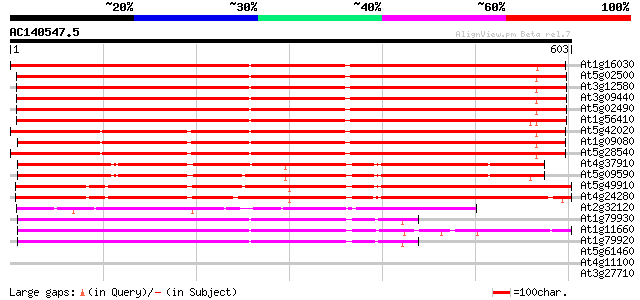
Score E
Sequences producing significant alignments: (bits) Value
At1g16030 unknown protein 815 0.0
At5g02500 dnaK-type molecular chaperone hsc70.1 793 0.0
At3g12580 putative protein 793 0.0
At3g09440 heat-shock protein (At-hsc70-3) 786 0.0
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 780 0.0
At1g56410 hypothetical protein 766 0.0
At5g42020 luminal binding protein (dbj|BAA13948.1) 660 0.0
At1g09080 putative luminal binding protein 658 0.0
At5g28540 luminal binding protein 658 0.0
At4g37910 heat shock protein 70 like protein 471 e-133
At5g09590 heat shock protein 70 (Hsc70-5) 462 e-130
At5g49910 heat shock protein 70 (Hsc70-7) 440 e-124
At4g24280 hsp 70-like protein 440 e-124
At2g32120 70kD heat shock protein 249 2e-66
At1g79930 putative heat-shock protein 249 2e-66
At1g11660 heat-shock protein like 249 4e-66
At1g79920 putative heat-shock protein (At1g79920) 248 9e-66
At5g61460 SMC-like protein (MIM) 40 0.004
At4g11100 unknown protein 39 0.008
At3g27710 putative protein 36 0.054
>At1g16030 unknown protein
Length = 646
Score = 815 bits (2105), Expect = 0.0
Identities = 416/611 (68%), Positives = 489/611 (79%), Gaps = 20/611 (3%)
Query: 1 MATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQ 60
MAT+ KAIGIDLGT+YSC+ VW N+RVEIIPNDQGNR TPSYVAFTDTERLIGDAAKNQ
Sbjct: 1 MATKSEKAIGIDLGTTYSCVGVWMNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQ 60
Query: 61 LAKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQ 120
+A P NTVFDAKRLIGR+FSD +VQ DI WPFKVV +KPMIVV+YK +EK FSP+
Sbjct: 61 VALNPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQFSPE 120
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EISSMVL K+KEVAE +LG V NAV+TVPAYFN+SQRQAT DAG I+G NV+RIINEPT
Sbjct: 121 EISSMVLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPT 180
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK + GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN
Sbjct: 181 AAAIAYGLDKKGTKAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGEDFDN 240
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
LVN V F RK+KKD I+ N +AL RLR+ACE+AKR LSS++QT IE+DSL GID
Sbjct: 241 RLVNHFVAEFRRKHKKD--IAGNARALRRLRTACERAKRTLSSTAQTTIEIDSLHEGIDF 298
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+ T++RA FEE+N DLF+KCM+ VEK L + K+ KS VH+ VLVGGST IPKIQQLL++
Sbjct: 299 YATISRARFEEMNMDLFRKCMDPVEKVLKDAKLDKSSVHDVVLVGGSTRIPKIQQLLQDF 358
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F KE CKSINPDEAVAYGAAVQAAIL EG +K++DLLLLDV P SLG+ET GGV
Sbjct: 359 F----NGKELCKSINPDEAVAYGAAVQAAILTGEGSEKVQDLLLLDVAPLSLGLETAGGV 414
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPR 480
M+VLIP+NT +P KKE VFS DNQ +LI+VYEGE A+T DN LLG FEL G PR
Sbjct: 415 MTVLIPRNTTVPCKKEQVFSTYADNQPGVLIQVYEGERARTRDNNLLGTFELKGIPPAPR 474
Query: 481 KVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAED 540
VP INVCFD+D NGIL V+AEDKT G+K +ITITN +GRLS EE+ +M++DAE+YKAED
Sbjct: 475 GVPQINVCFDIDANGILNVSAEDKTAGVKNQITITNDKGRLSKEEIEKMVQDAEKYKAED 534
Query: 541 EEVRKKVKAKNLFENYVYEMRERVK--------------KLEKVVEESIEWFERNQLAEI 586
E+V+KKV+AKN ENY Y MR +K K+EK ++E+IEW E NQLAE+
Sbjct: 535 EQVKKKVEAKNSLENYAYNMRNTIKDEKLAQKLTQEDKQKIEKAIDETIEWIEGNQLAEV 594
Query: 587 DEFEFKKQELE 597
DEFE+K +ELE
Sbjct: 595 DEFEYKLKELE 605
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 793 bits (2048), Expect = 0.0
Identities = 405/605 (66%), Positives = 482/605 (78%), Gaps = 20/605 (3%)
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTD+ERLIGDAAKNQ+A P N
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVN 68
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRFSD +VQ D+KLWPFK+ DKPMI V YKG+EK F+ +EISSMVL
Sbjct: 69 TVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPADKPMIYVEYKGEEKEFAAEEISSMVL 128
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE YLG + NAV+TVPAYFN+SQRQAT DAG IAG NVMRIINEPTAAAIAYG
Sbjct: 129 IKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAIAYG 188
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD I+ N +AL RLR++CE+AKR LSS++QT IE+DSL GID + T+TRA
Sbjct: 249 QEFKRKSKKD--ITGNPRALRRLRTSCERAKRTLSSTAQTTIEIDSLYEGIDFYSTITRA 306
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ KS VH+ VLVGGST IPK+QQLL++ F
Sbjct: 307 RFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQDFF----NG 362
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQ AIL+ EG++K++DLLLLDV P SLG+ET GGVM+ LIP+
Sbjct: 363 KELCKSINPDEAVAYGAAVQGAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTTLIPR 422
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE VFS DNQ +LI+VYEGE A+T+DN LLGKFELSG PR VP I V
Sbjct: 423 NTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVPQITV 482
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS +E+ +M+++AE+YK+EDEE +KKV
Sbjct: 483 CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEKYKSEDEEHKKKV 542
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
+AKN ENY Y MR + KK+E +E++I+W E NQLAE DEFE K
Sbjct: 543 EAKNALENYAYNMRNTIQDEKIGEKLPAADKKKIEDSIEQAIQWLEGNQLAEADEFEDKM 602
Query: 594 QELEN 598
+ELE+
Sbjct: 603 KELES 607
>At3g12580 putative protein
Length = 650
Score = 793 bits (2048), Expect = 0.0
Identities = 407/605 (67%), Positives = 483/605 (79%), Gaps = 20/605 (3%)
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTD+ERLIGDAAKNQ+A P N
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPTN 68
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRR+SD +VQ D WPFKVV +KPMIVVN+KG+EK FS +EISSMVL
Sbjct: 69 TVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQFSAEEISSMVL 128
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE +LG V NAV+TVPAYFN+SQRQAT DAG I+G NVMRIINEPTAAAIAYG
Sbjct: 129 IKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIINEPTAAAIAYG 188
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 189 LDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD I+ N +AL RLR+ACE+AKR LSS++QT IE+DSL GID + T+TRA
Sbjct: 249 QEFKRKNKKD--ITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGIDFYTTITRA 306
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ KS VH+ VLVGGST IPK+QQLL++ F
Sbjct: 307 RFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQLLQDFF----NG 362
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLIP+
Sbjct: 363 KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIPR 422
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE +FS DNQ +LI+VYEGE A+T+DN LLGKFELSG PR VP I V
Sbjct: 423 NTTIPTKKEQIFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVPQITV 482
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS EE+ +M+++AE+YKAEDEE +KKV
Sbjct: 483 CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKAEDEEHKKKV 542
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
AKN ENY Y MR + KK+E ++++IEW + NQLAE DEFE K
Sbjct: 543 DAKNALENYAYNMRNTIKDEKIASKLDAADKKKIEDAIDQAIEWLDGNQLAEADEFEDKM 602
Query: 594 QELEN 598
+ELE+
Sbjct: 603 KELES 607
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 786 bits (2030), Expect = 0.0
Identities = 404/605 (66%), Positives = 479/605 (78%), Gaps = 20/605 (3%)
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTD+ERLIGDAAKNQ+A P N
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPIN 68
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRF+D +VQ DIKLWPF + +KPMIVVNYKG++K FS +EISSM+L
Sbjct: 69 TVFDAKRLIGRRFTDSSVQSDIKLWPFTLKSGPAEKPMIVVNYKGEDKEFSAEEISSMIL 128
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE YLG + NAV+TVPAYFN+SQRQAT DAG IAG NVMRIINEPTAAAIAYG
Sbjct: 129 IKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIINEPTAAAIAYG 188
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD IS N +AL RLR+ACE+AKR LSS++QT IE+DSL GID + +TRA
Sbjct: 249 QEFKRKNKKD--ISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFDGIDFYAPITRA 306
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ K+ + + VLVGGST IPK+QQLL + F
Sbjct: 307 RFEELNIDLFRKCMEPVEKCLRDAKMDKNSIDDVVLVGGSTRIPKVQQLLVDFF----NG 362
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+VLI +
Sbjct: 363 KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTVLIQR 422
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE VFS DNQ +LI+VYEGE A+T+DN LLGKFELSG PR VP I V
Sbjct: 423 NTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVPQITV 482
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS +E+ +M+++AE+YK+EDEE +KKV
Sbjct: 483 CFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEKYKSEDEEHKKKV 542
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
AKN ENY Y MR + KK+E +E +IEW E NQLAE DEFE K
Sbjct: 543 DAKNALENYAYNMRNTIRDEKIGEKLAGDDKKKIEDSIEAAIEWLEANQLAECDEFEDKM 602
Query: 594 QELEN 598
+ELE+
Sbjct: 603 KELES 607
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 780 bits (2014), Expect = 0.0
Identities = 399/605 (65%), Positives = 479/605 (78%), Gaps = 20/605 (3%)
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTD+ERLIGDAAKNQ+A P N
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVN 68
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRFSD +VQ D +LWPF ++ +KPMIVV YKG+EK F+ +EISSMVL
Sbjct: 69 TVFDAKRLIGRRFSDASVQSDRQLWPFTIISGTAEKPMIVVEYKGEEKQFAAEEISSMVL 128
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE +LG V NAV+TVPAYFN+SQRQAT DAG IAG NV+RIINEPTAAAIAYG
Sbjct: 129 IKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK GEKNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 189 LDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK K+D I+ +AL RLR+ACE+AKR LSS++QT IE+DSL GG D + +TRA
Sbjct: 249 QEFKRKNKQD--ITGQPRALRRLRTACERAKRTLSSTAQTTIEIDSLYGGADFYSPITRA 306
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME VEKCL + K+ KS VHE VLVGGST IPK+QQLL++ F
Sbjct: 307 RFEEMNMDLFRKCMEPVEKCLRDAKMDKSTVHEIVLVGGSTRIPKVQQLLQDFF----NG 362
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+ LI +
Sbjct: 363 KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTTLIQR 422
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IPTKKE VFS DNQ +LI+V+EGE A+T+DN LLGKFELSG PR VP I V
Sbjct: 423 NTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIPPAPRGVPQITV 482
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDKT G K KITITN +GRLS E++ +M+++AE+YK+EDEE +KKV
Sbjct: 483 CFDIDANGILNVSAEDKTTGKKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEHKKKV 542
Query: 548 KAKNLFENYVYEMRERV--------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
+AKN ENY Y MR + KK+E +EE+I+W + NQL E DEFE K
Sbjct: 543 EAKNALENYAYNMRNTIRDEKIGEKLPAADKKKVEDSIEEAIQWLDGNQLGEADEFEDKM 602
Query: 594 QELEN 598
+ELE+
Sbjct: 603 KELES 607
>At1g56410 hypothetical protein
Length = 617
Score = 766 bits (1978), Expect = 0.0
Identities = 393/603 (65%), Positives = 474/603 (78%), Gaps = 18/603 (2%)
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
AIGIDLGT+YSC+ VW+++RVEII NDQGNR TPSYVAFTD+ERLIGDAAKNQ+A P N
Sbjct: 9 AIGIDLGTTYSCVGVWQHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAAKNQVAMNPVN 68
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TVFDAKRLIGRRFSD +VQ D+K WPFKV P DKPMI VNYKG+EK F+ +EISSMVL
Sbjct: 69 TVFDAKRLIGRRFSDASVQSDMKFWPFKVTPGQADKPMIFVNYKGEEKQFAAEEISSMVL 128
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
K++E+AE YLG + NAV+TVPAYFN+SQRQAT DAG IAG NV+RIINEPTAAAIAYG
Sbjct: 129 IKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIINEPTAAAIAYG 188
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
LDKK G KNVL+FDLGGGTFDVSL+TI+EG+F+VKAT GDTHLGG DFDN +VN V
Sbjct: 189 LDKKATSVGIKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGEDFDNRMVNHFV 248
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
+ F RK KKD IS + +AL RLR+ACE+AKR LSS++QT +E+DSL GID + +TRA
Sbjct: 249 QEFKRKNKKD--ISGDARALRRLRTACERAKRTLSSTAQTTVEVDSLFEGIDFYSPITRA 306
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEE+N DLF+KCME V KCL + K+ KS VH+ VLVGGST IPK+QQLL++ F
Sbjct: 307 KFEEMNMDLFRKCMEPVMKCLRDSKMDKSMVHDVVLVGGSTRIPKVQQLLQDFF----NG 362
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPK 427
KE CKSINPDEAVAYGAAVQAAIL+ EG++K++DLLLLDV P SLG+ET GGVM+ LI +
Sbjct: 363 KELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGIETIGGVMTTLIQR 422
Query: 428 NTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINV 487
NT IP KKE F+ DNQ +LI+VYEGE A+T DN +LG+F LSG PR +P V
Sbjct: 423 NTTIPAKKEQEFTTTVDNQPDVLIQVYEGERARTIDNNILGQFVLSGIPPAPRGIPQFTV 482
Query: 488 CFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKV 547
CFD+D NGIL V+AEDK G K KITITN +GRLS +++ +M+++AE+YK+EDEE +KKV
Sbjct: 483 CFDIDSNGILNVSAEDKATGKKNKITITNDKGRLSKDDIEKMVQEAEKYKSEDEEHKKKV 542
Query: 548 KAKNLFENYVY-------EMRERV-----KKLEKVVEESIEWFERNQLAEIDEFEFKKQE 595
+AKN ENY Y +M E++ KK E +EE I+W + NQLAE DEFE K +E
Sbjct: 543 EAKNGLENYAYNVGNTLRDMGEKLPAADKKKFEDSIEEVIQWLDDNQLAEADEFEHKMKE 602
Query: 596 LEN 598
LE+
Sbjct: 603 LES 605
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 660 bits (1702), Expect = 0.0
Identities = 344/612 (56%), Positives = 451/612 (73%), Gaps = 26/612 (4%)
Query: 2 ATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQL 61
AT+ IGIDLGT+YSC+ V++N VEII NDQGNR+TPS+V FTD+ERLIG+AAKNQ
Sbjct: 31 ATKLGSVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQA 90
Query: 62 AKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQ 120
A P TVFD KRLIGR+F D+ VQ+D KL P+++V N KP I V K G+ K FSP+
Sbjct: 91 AVNPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIV-NKDGKPYIQVKIKDGETKVFSPE 149
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EIS+M+L+K+KE AE YLG ++ +AV+TVPAYFN++QRQAT DAG IAG NV RIINEPT
Sbjct: 150 EISAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 209
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V +T GDTHLGG DFD+
Sbjct: 210 AAAIAYGLDKK---GGEKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDH 266
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
++ ++L +K++KD IS++ KALG+LR CE+AKR LSS Q +E++SL G+DL
Sbjct: 267 RIMEYFIKLIKKKHQKD--ISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDL 324
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+TRA FEE+N DLF+K M V+K + + + KSQ+ E VLVGGST IPK+QQLLK+
Sbjct: 325 SEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDF 384
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F E KEP K +NPDEAVAYGAAVQ IL+ EG + +D+LLLDV P +LG+ET GGV
Sbjct: 385 F----EGKEPNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGV 440
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPR 480
M+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D LLGKF+L+G PR
Sbjct: 441 MTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLTGVPPAPR 500
Query: 481 KVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAED 540
P I V F+VD NGIL V AEDK G +KITITN++GRLS EE+ RM+++AE + ED
Sbjct: 501 GTPQIEVTFEVDANGILNVKAEDKASGKSEKITITNEKGRLSQEEIDRMVKEAEEFAEED 560
Query: 541 EEVRKKVKAKNLFENYVYEMRERV---------------KKLEKVVEESIEWFERNQLAE 585
++V++K+ A+N E YVY M+ +V +K+E +E++EW + NQ +E
Sbjct: 561 KKVKEKIDARNALETYVYNMKNQVSDKDKLADKLEGDEKEKIEAATKEALEWLDENQNSE 620
Query: 586 IDEFEFKKQELE 597
+E++ K +E+E
Sbjct: 621 KEEYDEKLKEVE 632
>At1g09080 putative luminal binding protein
Length = 678
Score = 658 bits (1698), Expect = 0.0
Identities = 341/604 (56%), Positives = 442/604 (72%), Gaps = 25/604 (4%)
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHNT 68
IGIDLGT+YSC+ V+ N VEII NDQGNR+TPS+VAFTDTERLIG+AAKNQ AK P T
Sbjct: 53 IGIDLGTTYSCVGVYHNKHVEIIANDQGNRITPSWVAFTDTERLIGEAAKNQAAKNPERT 112
Query: 69 VFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLS 128
+FD KRLIGR+F D VQ+DIK P+KVV N KP I V KG+EK FSP+EIS+M+L+
Sbjct: 113 IFDPKRLIGRKFDDPDVQRDIKFLPYKVV-NKDGKPYIQVKVKGEEKLFSPEEISAMILT 171
Query: 129 KLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGL 188
K+KE AE +LG ++ +AVITVPAYFN++QRQAT DAG IAG NV+RIINEPT AAIAYGL
Sbjct: 172 KMKETAEAFLGKKIKDAVITVPAYFNDAQRQATKDAGAIAGLNVVRIINEPTGAAIAYGL 231
Query: 189 DKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLVE 248
DKK GE N+LV+DLGGGTFDVS++TID G+F+V +T GDTHLGG DFD+ +++ ++
Sbjct: 232 DKK---GGESNILVYDLGGGTFDVSILTIDNGVFEVLSTSGDTHLGGEDFDHRVMDYFIK 288
Query: 249 LFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRAF 308
L +KY KD IS++ KALG+LR CE AKR LS+ Q +E++SL G+D +TRA
Sbjct: 289 LVKKKYNKD--ISKDHKALGKLRRECELAKRSLSNQHQVRVEIESLFDGVDFSEPLTRAR 346
Query: 309 FEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEVK 368
FEE+N DLFKK ME V+K L + + KS + E VLVGGST IPK+QQ+LK+ F + K
Sbjct: 347 FEELNMDLFKKTMEPVKKALKDAGLKKSDIDEIVLVGGSTRIPKVQQMLKDFF----DGK 402
Query: 369 EPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSVLIPKN 428
EP K NPDEAVAYGAAVQ +L+ EG ++ +++LLLDV P SLG+ET GGVM+ +IP+N
Sbjct: 403 EPSKGTNPDEAVAYGAAVQGGVLSGEGGEETQNILLLDVAPLSLGIETVGGVMTNIIPRN 462
Query: 429 TMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVPNINVC 488
T+IPTKK VF+ D Q ++ I VYEGE + T+DN LGKF+L+G PR VP I V
Sbjct: 463 TVIPTKKSQVFTTYQDQQTTVTINVYEGERSMTKDNRELGKFDLTGILPAPRGVPQIEVT 522
Query: 489 FDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEVRKKVK 548
F+VD NGIL+V AEDK Q ITITN +GRL+ EE+ M+R+AE + ED+ +++K+
Sbjct: 523 FEVDANGILQVKAEDKVAKTSQSITITNDKGRLTEEEIEEMIREAEEFAEEDKIMKEKID 582
Query: 549 AKNLFENYVYEMRERV---------------KKLEKVVEESIEWFERNQLAEIDEFEFKK 593
A+N E YVY M+ V +K+E V++E++EW E N AE ++++ K
Sbjct: 583 ARNKLETYVYNMKSTVADKEKLAKKISDEDKEKMEGVLKEALEWLEENVNAEKEDYDEKL 642
Query: 594 QELE 597
+E+E
Sbjct: 643 KEVE 646
>At5g28540 luminal binding protein
Length = 669
Score = 658 bits (1697), Expect = 0.0
Identities = 343/612 (56%), Positives = 450/612 (73%), Gaps = 26/612 (4%)
Query: 2 ATRKSKAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQL 61
AT+ IGIDLGT+YSC+ V++N VEII NDQGNR+TPS+V FTD+ERLIG+AAKNQ
Sbjct: 31 ATKLGSVIGIDLGTTYSCVGVYKNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQA 90
Query: 62 AKYPHNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYK-GQEKHFSPQ 120
A P TVFD KRLIGR+F D+ VQ+D KL P+++V N KP I V K G+ K FSP+
Sbjct: 91 AVNPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIV-NKDGKPYIQVKIKDGETKVFSPE 149
Query: 121 EISSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPT 180
EIS+M+L+K+KE AE YLG ++ +AV+TVPAYFN++QRQAT DAG IAG NV RIINEPT
Sbjct: 150 EISAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPT 209
Query: 181 AAAIAYGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDN 240
AAAIAYGLDKK GEKN+LVFDLGGGTFDVS++TID G+F+V +T GDTHLGG DFD+
Sbjct: 210 AAAIAYGLDKK---GGEKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDH 266
Query: 241 NLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDL 300
++ ++L +K++KD IS++ KALG+LR CE+AKR LSS Q +E++SL G+D
Sbjct: 267 RVMEYFIKLIKKKHQKD--ISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDF 324
Query: 301 HFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNM 360
+TRA FEE+N DLF+K M V+K + + + KSQ+ E VLVGGST IPK+QQLLK+
Sbjct: 325 SEPLTRARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDF 384
Query: 361 FRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV 420
F E KEP K +NPDEAVAYGAAVQ IL+ EG + +D+LLLDV P +LG+ET GGV
Sbjct: 385 F----EGKEPNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGV 440
Query: 421 MSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPR 480
M+ LIP+NT+IPTKK VF+ D Q ++ I+V+EGE + T+D LLGKF+L+G PR
Sbjct: 441 MTKLIPRNTVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLNGIPPAPR 500
Query: 481 KVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAED 540
P I V F+VD NGIL V AEDK G +KITITN++GRLS EE+ RM+++AE + ED
Sbjct: 501 GTPQIEVTFEVDANGILNVKAEDKASGKSEKITITNEKGRLSQEEIDRMVKEAEEFAEED 560
Query: 541 EEVRKKVKAKNLFENYVYEMRERV---------------KKLEKVVEESIEWFERNQLAE 585
++V++K+ A+N E YVY M+ +V +K+E +E++EW + NQ +E
Sbjct: 561 KKVKEKIDARNALETYVYNMKNQVNDKDKLADKLEGDEKEKIEAATKEALEWLDENQNSE 620
Query: 586 IDEFEFKKQELE 597
+E++ K +E+E
Sbjct: 621 KEEYDEKLKEVE 632
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 471 bits (1212), Expect = e-133
Identities = 267/572 (46%), Positives = 364/572 (62%), Gaps = 26/572 (4%)
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDT-ERLIGDAAKNQLAKYPHN 67
IGIDLGT+ SC++V +I N +G+R TPS VA E L+G AK Q P N
Sbjct: 55 IGIDLGTTNSCVSVMEGKTARVIENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTNPTN 114
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
T+F +KRLIGRRF D Q+++K+ P+K+V + N GQ+ FSP +I + VL
Sbjct: 115 TIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEAN--GQK--FSPSQIGANVL 170
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
+K+KE AE YLG +N AV+TVPAYFN++QRQAT DAGKIAG +V RIINEPTAAA++YG
Sbjct: 171 TKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIINEPTAAALSYG 230
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
++ K E + VFDLGGGTFDVS++ I G+F+VKAT GDT LGG DFDN L+ LV
Sbjct: 231 MNNK-----EGVIAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTLLEYLV 285
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLC----GGIDLHFT 303
F R DL +++ AL RLR A EKAK LSS++QT I L + G L+ T
Sbjct: 286 NEFKRSDNIDL--TKDNLALQRLREAAEKAKIELSSTTQTEINLPFITADASGAKHLNIT 343
Query: 304 VTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRV 363
+TR+ FE + L ++ + CL + ++ +V E +LVGG T +PK+Q+++ +F
Sbjct: 344 LTRSKFEGLVGKLIERTRSPCQNCLKDAGVTIKEVDEVLLVGGMTRVPKVQEIVSEIFG- 402
Query: 364 NDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSV 423
K PCK +NPDEAVA GAA+Q IL GD ++DLLLLDV+P SLG+ET G V +
Sbjct: 403 ----KSPCKGVNPDEAVAMGAAIQGGILR--GD--VKDLLLLDVVPLSLGIETLGAVFTK 454
Query: 424 LIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVP 483
LIP+NT IPTKK VFS A DNQ + IKV +GE DN +LG+F+L G PR +P
Sbjct: 455 LIPRNTTIPTKKSQVFSTAADNQMQVGIKVLQGEREMAADNKVLGEFDLVGIPPAPRGMP 514
Query: 484 NINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEV 543
I V FD+D NGI V+A+DK G +Q ITI G LS +E+ RM+++AE +D+E
Sbjct: 515 QIEVTFDIDANGITTVSAKDKATGKEQNITI-RSSGGLSDDEINRMVKEAELNAQKDQEK 573
Query: 544 RKKVKAKNLFENYVYEMRERVKKLEKVVEESI 575
++ + +N + +Y + + + + + + I
Sbjct: 574 KQLIDLRNSADTTIYSVEKSLSEYREKIPAEI 605
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 462 bits (1190), Expect = e-130
Identities = 270/580 (46%), Positives = 371/580 (63%), Gaps = 34/580 (5%)
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAF-TDTERLIGDAAKNQLAKYPHN 67
IGIDLGT+ SC+AV ++I N +G R TPS VAF T E L+G AK Q P N
Sbjct: 60 IGIDLGTTNSCVAVMEGKNPKVIENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTNPTN 119
Query: 68 TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVL 127
TV KRLIGR+F D Q+++K+ P+K+V + N GQ+ +SP +I + +L
Sbjct: 120 TVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGDAWVEAN--GQQ--YSPSQIGAFIL 175
Query: 128 SKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYG 187
+K+KE AE YLG V AV+TVPAYFN++QRQAT DAG+IAG +V RIINEPTAAA++YG
Sbjct: 176 TKMKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIAGLDVERIINEPTAAALSYG 235
Query: 188 LDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
+ K E + VFDLGGGTFDVS++ I G+F+VKAT GDT LGG DFDN L++ LV
Sbjct: 236 MTNK-----EGLIAVFDLGGGTFDVSVLEISNGVFEVKATNGDTFLGGEDFDNALLDFLV 290
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLC----GGIDLHFT 303
F K + + ++++ AL RLR A EKAK LSS+SQT I L + G + T
Sbjct: 291 NEF--KTTEGIDLAKDRLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHFNIT 348
Query: 304 VTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRV 363
+TR+ FE + L ++ + + CL + IS +V E +LVGG T +PK+Q ++ +F
Sbjct: 349 LTRSRFETLVNHLIERTRDPCKNCLKDAGISAKEVDEVLLVGGMTRVPKVQSIVAEIFG- 407
Query: 364 NDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMSV 423
K P K +NPDEAVA GAA+Q IL GD +++LLLLDV P SLG+ET GGV +
Sbjct: 408 ----KSPSKGVNPDEAVAMGAALQGGILR--GD--VKELLLLDVTPLSLGIETLGGVFTR 459
Query: 424 LIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRKVP 483
LI +NT IPTKK VFS A DNQ + I+V +GE DN LLG+F+L G PR VP
Sbjct: 460 LITRNTTIPTKKSQVFSTAADNQTQVGIRVLQGEREMATDNKLLGEFDLVGIPPSPRGVP 519
Query: 484 NINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEV 543
I V FD+D NGI+ V+A+DKT G Q+ITI G LS +++++M+R+AE + +D+E
Sbjct: 520 QIEVTFDIDANGIVTVSAKDKTTGKVQQITI-RSSGGLSEDDIQKMVREAELHAQKDKER 578
Query: 544 RKKVKAKNLFENYVY-------EMRERV-KKLEKVVEESI 575
++ + KN + +Y E RE++ ++ K +E+++
Sbjct: 579 KELIDTKNTADTTIYSIEKSLGEYREKIPSEIAKEIEDAV 618
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 440 bits (1132), Expect = e-124
Identities = 261/604 (43%), Positives = 368/604 (60%), Gaps = 27/604 (4%)
Query: 7 KAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDT-ERLIGDAAKNQLAKYP 65
K +GIDLGT+ S +A + I+ N +G R TPS VA+T + +RL+G AK Q P
Sbjct: 79 KVVGIDLGTTNSAVAAMEGGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKRQAVVNP 138
Query: 66 HNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSM 125
NT F KR IGRR ++ V ++ K ++V+ + + ++ K F+ +EIS+
Sbjct: 139 ENTFFSVKRFIGRRMNE--VAEESKQVSYRVIKDENGN--VKLDCPAIGKQFAAEEISAQ 194
Query: 126 VLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIA 185
VL KL + A +L +V AVITVPAYFN+SQR AT DAG+IAG V+RIINEPTAA++A
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 186 YGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNR 245
YG ++K + +LVFDLGGGTFDVS++ + +G+F+V +T GDTHLGG DFD +V+
Sbjct: 255 YGFERK----SNETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDW 310
Query: 246 LVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGID----LH 301
L F K + + + ++ +AL RL A EKAK LSS +QT + L + D +
Sbjct: 311 LASTF--KKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKHIE 368
Query: 302 FTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMF 361
T+TR FEE+ DL + VE L + K+S + E +LVGGST IP +Q L++ +
Sbjct: 369 TTLTRGKFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQDLVRKLT 428
Query: 362 RVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVM 421
KEP S+NPDE VA GAAVQA +L+ GD + D++LLDV P SLG+ET GGVM
Sbjct: 429 G-----KEPNVSVNPDEVVALGAAVQAGVLS--GD--VSDIVLLDVTPLSLGLETLGGVM 479
Query: 422 SVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVPRK 481
+ +IP+NT +PT K VFS A D Q S+ I V +GE DN +G F L G PR
Sbjct: 480 TKIIPRNTTLPTSKSEVFSTAADGQTSVEINVLQGEREFVRDNKSIGSFRLDGIPPAPRG 539
Query: 482 VPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDE 541
VP I V FD+D NGIL V+A DK G KQ ITIT L +E+ M+++AER+ ED+
Sbjct: 540 VPQIEVKFDIDANGILSVSASDKGTGKKQDITITG-ASTLPKDEVDTMVQEAERFAKEDK 598
Query: 542 EVRKKVKAKNLFENYVYEMRERVKKLEKVVEESIEWFERNQLAEIDE--FEFKKQELENS 599
E R + KN ++ VY+ +++K+L + + ++ +L E+ E QE++++
Sbjct: 599 EKRDAIDTKNQADSVVYQTEKQLKELGEKIPGPVKEKVEAKLQELKEKIASGSTQEIKDT 658
Query: 600 MKFL 603
M L
Sbjct: 659 MAAL 662
>At4g24280 hsp 70-like protein
Length = 718
Score = 440 bits (1132), Expect = e-124
Identities = 267/610 (43%), Positives = 375/610 (60%), Gaps = 39/610 (6%)
Query: 7 KAIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDT-ERLIGDAAKNQLAKYP 65
K +GIDLGT+ S +A + I+ N +G R TPS VA+T + +RL+G AK Q P
Sbjct: 79 KVVGIDLGTTNSAVAAMEGGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKRQAVVNP 138
Query: 66 HNTVFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSM 125
NT F KR IGR+ ++ V ++ K ++VV + + + + K F+ +EIS+
Sbjct: 139 ENTFFSVKRFIGRKMNE--VDEESKQVSYRVVRDENNN--VKLECPAINKQFAAEEISAQ 194
Query: 126 VLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIA 185
VL KL + A +L +V AVITVPAYFN+SQR AT DAG+IAG V+RIINEPTAA++A
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 186 YGLDKKMWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNR 245
YG D+K + +LVFDLGGGTFDVS++ + +G+F+V +T GDTHLGG DFD R
Sbjct: 255 YGFDRK----ANETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDK----R 306
Query: 246 LVELFHRKYKKD--LKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGID---- 299
+V+ ++KKD + + ++ +AL RL A EKAK LSS +QT + L + D
Sbjct: 307 VVDWLAAEFKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKH 366
Query: 300 LHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKN 359
+ T+TRA FEE+ DL + VE L + K+S + E +LVGGST IP +Q+L++
Sbjct: 367 IETTLTRAKFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQELVRK 426
Query: 360 MFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGG 419
+ KEP ++NPDE VA GAAVQA +L GD + D++LLDV P S+G+ET GG
Sbjct: 427 VTG-----KEPNVTVNPDEVVALGAAVQAGVL--AGD--VSDIVLLDVTPLSIGLETLGG 477
Query: 420 VMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGCSLVP 479
VM+ +IP+NT +PT K VFS A D Q S+ I V +GE DN LG F L G P
Sbjct: 478 VMTKIIPRNTTLPTSKSEVFSTAADGQTSVEINVLQGEREFVRDNKSLGSFRLDGIPPAP 537
Query: 480 RKVPNINVCFDVDVNGILEVTAEDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAE 539
R VP I V FD+D NGIL V+A DK G KQ ITIT L +E+ +M+++AER+ +
Sbjct: 538 RGVPQIEVKFDIDANGILSVSAVDKGTGKKQDITITG-ASTLPKDEVDQMVQEAERFAKD 596
Query: 540 DEEVRKKVKAKNLFENYVYEMRERVKKL-EKVVEESIEWFERNQLAEIDEFEFK-----K 593
D+E R + KN ++ VY+ +++K+L EK+ E E E A++ E + K
Sbjct: 597 DKEKRDAIDTKNQADSVVYQTEKQLKELGEKIPGEVKEKVE----AKLQELKDKIGSGST 652
Query: 594 QELENSMKFL 603
QE++++M L
Sbjct: 653 QEIKDAMAAL 662
>At2g32120 70kD heat shock protein
Length = 563
Score = 249 bits (637), Expect = 2e-66
Identities = 165/506 (32%), Positives = 260/506 (50%), Gaps = 34/506 (6%)
Query: 8 AIGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHN 67
A+GID+GTS IAVW ++V I+ N + ++ S+V F D + NQLA
Sbjct: 30 ALGIDIGTSQCSIAVWNGSQVHILRNTRNQKLIKSFVTFKD--EVPAGGVSNQLAHEQEM 87
Query: 68 ----TVFDAKRLIGRRFSDQTVQQDIKLWPFKVVP-NHKDKPMIVVNYKGQEKHFSPQEI 122
+F+ KRL+GR +D V L PF V + +P I + +P+E+
Sbjct: 88 LTGAAIFNMKRLVGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNAWRSTTPEEV 146
Query: 123 SSMVLSKLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAA 182
++ L +L+ +AE L V N V+TVP F+ Q A +AG +V+R++ EPTA
Sbjct: 147 LAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQLTRFERACAMAGLHVLRLMPEPTAI 206
Query: 183 AIAYGLDKKMWRE------GEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGI 236
A+ Y ++M E+ ++F++G G DV++ G+ Q+KA G +GG
Sbjct: 207 ALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDVAVTATAGGVSQIKALAGSP-IGGE 265
Query: 237 DFDNNLVNRLVELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCG 296
D N + + N +A G LR A + A L+ IE+D L
Sbjct: 266 DILQNTIRHIAP-------------PNEEASGLLRVAAQDAIHRLTDQENVQIEVD-LGN 311
Query: 297 GIDLHFTVTRAFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQL 356
G + + R FEE+N+ +F++C V +CL + +++ + + ++VGG + IPK++ +
Sbjct: 312 GNKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGGDIDDLIMVGGCSYIPKVRTI 371
Query: 357 LKNMFRVNDEVKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIE-DLLLLDVMPFSLGVE 415
+KN+ + DE+ K +NP EA GAA++ A+ + D DLL + P ++GV
Sbjct: 372 IKNVCK-KDEIY---KGVNPLEAAVRGAALEGAVTSGIHDPFGSLDLLTIQATPLAVGVR 427
Query: 416 TKGGVMSVLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTEDNFLLGKFELSGC 475
G +IP+NTM+P +K+ F+ DNQ LI +YEGE E+N LLG F+L G
Sbjct: 428 ANGNKFIPVIPRNTMVPARKDLFFTTVQDNQKEALIIIYEGEGETVEENHLLGYFKLVGI 487
Query: 476 SLVPRKVPNINVCFDVDVNGILEVTA 501
P+ VP INVC D+D + L V A
Sbjct: 488 PPAPKGVPEINVCMDIDASNALRVFA 513
>At1g79930 putative heat-shock protein
Length = 831
Score = 249 bits (637), Expect = 2e-66
Identities = 148/444 (33%), Positives = 234/444 (52%), Gaps = 22/444 (4%)
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHNT 68
+G D G +AV R ++++ ND+ NR TP+ V F D +R IG A P N+
Sbjct: 4 VGFDFGNENCLVAVARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 63
Query: 69 VFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLS 128
+ KRLIGR+FSD +Q+DIK PF V P+I NY G+++ F+P ++ M+LS
Sbjct: 64 ISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEKRAFTPTQVMGMMLS 123
Query: 129 KLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGL 188
LK +AE L V + I +P YF + QR+A DA IAG + +R+I+E TA A+AYG+
Sbjct: 124 NLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLRLIHETTATALAYGI 183
Query: 189 DKKMWREGEK-NVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
K E ++ NV D+G + V + +G ++ + D LGG DFD L N
Sbjct: 184 YKTDLPESDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNHFA 243
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
F +YK D +S+N KA RLR+ CEK K++LS++ + ++ L D+ + R
Sbjct: 244 AKFKDEYKID--VSQNAKASLRLRATCEKLKKVLSANPLAPLNIECLMDEKDVRGVIKRE 301
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEEI+ + ++ +EK L + ++ VH ++G + +P + ++L F
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVIGSGSRVPAMIKILTEFFG----- 356
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV------- 420
KEP +++N E V+ G A+Q AIL+ K+ + + + PFS+ + KG
Sbjct: 357 KEPRRTMNASECVSRGCALQCAILSP--TFKVREFQVHESFPFSISLAWKGAASEAQNGG 414
Query: 421 -----MSVLIPKNTMIPTKKENVF 439
+++ PK IP+ K F
Sbjct: 415 AENQQSTIVFPKGNPIPSVKALTF 438
Score = 32.7 bits (73), Expect = 0.60
Identities = 26/109 (23%), Positives = 50/109 (45%), Gaps = 9/109 (8%)
Query: 497 LEVTAEDKTIGLKQKITITNKE------GRLSPEEMRRMMRDAERYKAEDEEVRKKVKAK 550
+++ + K K+K+ TN G L E+ + + +D + + K
Sbjct: 564 VQMETDSKAEAPKKKVKKTNVPLSELVYGALKTVEVEKAVEKEFEMALQDRVMEETKDRK 623
Query: 551 NLFENYVYEMRERV--KKLEKVVEESIEWFERNQLAEIDEFEFKKQELE 597
N E+YVY+MR ++ K E + + E F N L E++++ ++ E E
Sbjct: 624 NAVESYVYDMRNKLSDKYQEYITDSEREAFLAN-LQEVEDWLYEDGEDE 671
>At1g11660 heat-shock protein like
Length = 773
Score = 249 bits (635), Expect = 4e-66
Identities = 172/662 (25%), Positives = 316/662 (46%), Gaps = 86/662 (12%)
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHNT 68
+G D+G IAV + ++++ ND+ NR P+ V+F + +R +G AA +P +T
Sbjct: 4 VGFDVGNENCVIAVAKQRGIDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMHPKST 63
Query: 69 VFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLS 128
+ KRLIGR+F + VQ D++L+PF+ + I + Y G+ + FSP +I M+LS
Sbjct: 64 ISQLKRLIGRKFREPDVQNDLRLFPFETSEDSDGGIQIRLRYMGEIQSFSPVQILGMLLS 123
Query: 129 KLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGL 188
LK++AE L V++ VI +P+YF NSQR A DA IAG +R++++ TA A+ YG+
Sbjct: 124 HLKQIAEKSLKTPVSDCVIGIPSYFTNSQRLAYLDAAAIAGLRPLRLMHDSTATALGYGI 183
Query: 189 DKK--MWREGEKNVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRL 246
K + ++ D+G V + + + G +V++ D +LGG DFD L N
Sbjct: 184 YKTDLVANSSPTYIVFIDIGHCDTQVCVASFESGSMRVRSHAFDRNLGGRDFDEVLFNHF 243
Query: 247 VELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTR 306
F KY D + N KA RLR++CEK K++LS++++ + ++ L D+ + R
Sbjct: 244 ALEFKEKYNID--VYTNTKACVRLRASCEKVKKVLSANAEAQLNIECLMEEKDVRSFIKR 301
Query: 307 AFFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDE 366
FE+++ L ++ + +K L + +S Q+H LVG + IP I ++L ++F+
Sbjct: 302 EEFEQLSAGLLERLIVPCQKALADSGLSLDQIHSVELVGSGSRIPAISKMLSSLFK---- 357
Query: 367 VKEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGVMS---- 422
+E +++N E VA G A+Q A+L+ ++ D + D PF++G + G ++
Sbjct: 358 -RELGRTVNASECVARGCALQCAMLSPV--FRVRDYEVQDSYPFAIGFSSDKGPINTPSN 414
Query: 423 -VLIPKNTMIPTKKENVFSIACDNQDSLLIKVYEGEHAKTE-------DNFLLGKFELSG 474
+L PK + P+ K + +++ ++ + H + +F++G F +S
Sbjct: 415 ELLFPKGQIFPSVK----VLTLHRENTFQLEAFYANHNELSPDIPTQISSFMIGPFHIS- 469
Query: 475 CSLVPRKVPNINVCFDVDVNGILEVTA--------------------------------- 501
+ + V ++++GI+ + +
Sbjct: 470 ----HGEAARVKVRVQLNLHGIVTIDSATLESKLSVSEQLIEYHKENITSEEMISEENHQ 525
Query: 502 ------------------EDKTIGLKQKITITNKEGRLSPEEMRRMMRDAERYKAEDEEV 543
E K I + + N G L+ +E+ + +D ++
Sbjct: 526 SSAMKDGSLDPSSGSIGNEPKAIKRMEIPVVANVSGALTKDELSEAKQRENSLVEQDLKM 585
Query: 544 RKKVKAKNLFENYVYEMRERVKKLEK--VVEESIEWFERNQLAEIDEFEFKKQELENSMK 601
KN E++VYEMR+++ + E E RN L E +E+ ++ + E+
Sbjct: 586 ESTKDKKNALESFVYEMRDKMLNTYRNTATESERECIARN-LQETEEWLYEDGDDESENA 644
Query: 602 FL 603
++
Sbjct: 645 YI 646
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 248 bits (632), Expect = 9e-66
Identities = 148/444 (33%), Positives = 232/444 (51%), Gaps = 22/444 (4%)
Query: 9 IGIDLGTSYSCIAVWRNNRVEIIPNDQGNRVTPSYVAFTDTERLIGDAAKNQLAKYPHNT 68
+G D G +AV R ++++ ND+ NR TP+ V F D +R IG A P N+
Sbjct: 4 VGFDFGNENCLVAVARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPKNS 63
Query: 69 VFDAKRLIGRRFSDQTVQQDIKLWPFKVVPNHKDKPMIVVNYKGQEKHFSPQEISSMVLS 128
+ KRLIGR+FSD +Q+DIK PF V P+I NY G+ + F+P ++ M+LS
Sbjct: 64 ISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEIRAFTPTQVMGMMLS 123
Query: 129 KLKEVAETYLGHEVNNAVITVPAYFNNSQRQATNDAGKIAGFNVMRIINEPTAAAIAYGL 188
LK +AE L V + I +P YF + QR+A DA IAG + + +I+E TA A+AYG+
Sbjct: 124 NLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLHLIHETTATALAYGI 183
Query: 189 DKKMWREGEK-NVLVFDLGGGTFDVSLVTIDEGMFQVKATLGDTHLGGIDFDNNLVNRLV 247
K E ++ NV D+G + V + +G ++ + D LGG DFD L N
Sbjct: 184 YKTDLPENDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNHFA 243
Query: 248 ELFHRKYKKDLKISENFKALGRLRSACEKAKRLLSSSSQTVIELDSLCGGIDLHFTVTRA 307
F +YK D +S+N KA RLR+ CEK K++LS++ + ++ L D+ + R
Sbjct: 244 AKFKDEYKID--VSQNAKASLRLRATCEKLKKVLSANPMAPLNIECLMAEKDVRGVIKRE 301
Query: 308 FFEEINKDLFKKCMETVEKCLVEVKISKSQVHEFVLVGGSTTIPKIQQLLKNMFRVNDEV 367
FEEI+ + ++ +EK L + ++ VH +VG + +P + ++L F
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVVGSGSRVPAMIKILTEFFG----- 356
Query: 368 KEPCKSINPDEAVAYGAAVQAAILNTEGDKKIEDLLLLDVMPFSLGVETKGGV------- 420
KEP +++N E V+ G A+Q AIL+ K+ + + + PFS+ + KG
Sbjct: 357 KEPRRTMNASECVSRGCALQCAILSP--TFKVREFQVHESFPFSISLAWKGAATDAQNGG 414
Query: 421 -----MSVLIPKNTMIPTKKENVF 439
+++ PK IP+ K F
Sbjct: 415 TENQQSTIVFPKGNPIPSVKALTF 438
>At5g61460 SMC-like protein (MIM)
Length = 1057
Score = 40.0 bits (92), Expect = 0.004
Identities = 28/95 (29%), Positives = 53/95 (55%), Gaps = 6/95 (6%)
Query: 513 TITNKEGRLS--PEEMRRMMRDAERY-KAEDEEVRKKVKAKNLFE---NYVYEMRERVKK 566
T+T K+ +++ +E M R+ E + ++ VR+K+ + F NYV ++++RV++
Sbjct: 296 TLTKKKAQVACLMDESTAMKREIESFHQSAKTAVREKIALQEEFNHKCNYVQKIKDRVRR 355
Query: 567 LEKVVEESIEWFERNQLAEIDEFEFKKQELENSMK 601
LE+ V + E +N AE E E K + LE ++
Sbjct: 356 LERQVGDINEQTMKNTQAEQSEIEEKLKYLEQEVE 390
>At4g11100 unknown protein
Length = 287
Score = 38.9 bits (89), Expect = 0.008
Identities = 32/101 (31%), Positives = 48/101 (46%), Gaps = 8/101 (7%)
Query: 503 DKTIGLKQKITITNKEGRLSPEE--MRRMMRDAERYKA-----EDEEVRKKVKAKNLFEN 555
+KT+ L+ +I N E L E R+ E Y+A E++ +V K L E
Sbjct: 6 EKTMTLEHEILKENHETLLKDYESLQERIKHAEEAYEAIKLHHENKAKELEVSNKRLLEE 65
Query: 556 YVYEMRERVKKLEKVVEESIEWFERNQLAEIDEFEFKKQEL 596
+ E RE+ K + K EE + E + A +DE + K QEL
Sbjct: 66 CMKERREKAK-VRKTFEEMKKTMESERTAIVDELKSKNQEL 105
>At3g27710 putative protein
Length = 537
Score = 36.2 bits (82), Expect = 0.054
Identities = 17/53 (32%), Positives = 33/53 (62%), Gaps = 1/53 (1%)
Query: 534 ERYKAEDEEVRKKVKAKNLFENYVYEMRERVKKLEKVVEESIEWFERNQLAEI 586
E +K E E +++K KNLFE+ ++ V+KL K++EE + ++ ++ E+
Sbjct: 422 ELFKDEMSEKEREIK-KNLFEDQQQQLEGNVEKLSKILEEPFDEYDHEKVVEM 473
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.135 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,143,735
Number of Sequences: 26719
Number of extensions: 575315
Number of successful extensions: 2267
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 2104
Number of HSP's gapped (non-prelim): 131
length of query: 603
length of database: 11,318,596
effective HSP length: 105
effective length of query: 498
effective length of database: 8,513,101
effective search space: 4239524298
effective search space used: 4239524298
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140547.5


