

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140547.14 - phase: 0
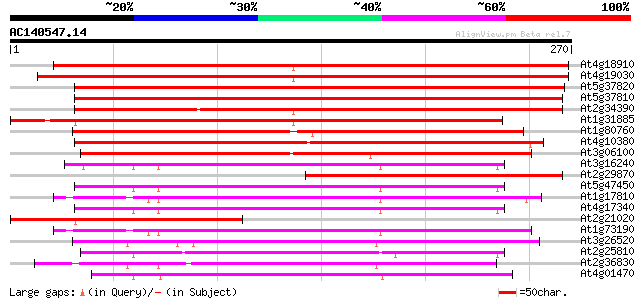
(270 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g18910 major intrinsic protein (MIP)- like 361 e-100
At4g19030 nodulin-26 like protein 348 1e-96
At5g37820 Membrane integral protein (MIP) -like 288 2e-78
At5g37810 Membrane integral protein (MIP) -like 286 8e-78
At2g34390 putative aquaporin (plasma membrane intrinsic protein) 259 1e-69
At1g31885 major intrinsic protein - like 258 2e-69
At1g80760 nodulin-like protein 223 6e-59
At4g10380 major intrinsic protein (MIP) - like 213 9e-56
At3g06100 putative major intrinsic protein 174 6e-44
At3g16240 delta tonoplast integral protein (delta-TIP) 127 6e-30
At2g29870 putative aquaporin (plasma membrane intrinsic protein) 122 2e-28
At5g47450 membrane channel protein-like; aquaporin (tonoplast in... 120 6e-28
At1g17810 putative protein 119 1e-27
At4g17340 membrane channel like protein 119 2e-27
At2g21020 putative major intrinsic (channel) protein 111 5e-25
At1g73190 tonoplast intrinsic protein, alpha (alpha-TIP) 111 5e-25
At3g26520 salt-stress induced tonoplast intrinsic protein 109 2e-24
At2g25810 putative aquaporin (tonoplast intrinsic protein) 108 4e-24
At2g36830 putative aquaporin (tonoplast intrinsic protein gamma) 105 2e-23
At4g01470 putative water channel protein 100 8e-22
>At4g18910 major intrinsic protein (MIP)- like
Length = 294
Score = 361 bits (927), Expect = e-100
Identities = 176/255 (69%), Positives = 215/255 (84%), Gaps = 7/255 (2%)
Query: 22 KCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMV 81
K D+ + VP LQKL+AEV+GT+FLIFAGCAAV VN +DK VTLPGI+IVWGL VMV
Sbjct: 38 KKQDSLLSISVPFLQKLMAEVLGTYFLIFAGCAAVAVNTQHDKAVTLPGIAIVWGLTVMV 97
Query: 82 LVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIF------ 135
LVYS+GHISGAHFNPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L+F
Sbjct: 98 LVYSLGHISGAHFNPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRLLFGLDQDV 157
Query: 136 -SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILN 194
SGK + F GTLP+GS+LQ+FV+EFIITF+LMF+ISGVATDNRAIGELAGLAVGSTV+LN
Sbjct: 158 CSGKHDVFVGTLPSGSNLQSFVIEFIITFYLMFVISGVATDNRAIGELAGLAVGSTVLLN 217
Query: 195 VLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRE 254
V+ AGP++GASMNP RSLGPA+V+ YRG+WIY+VSPI+GA++G W Y +R T+KP+RE
Sbjct: 218 VIIAGPVSGASMNPGRSLGPAMVYSCYRGLWIYIVSPIVGAVSGAWVYNMVRYTDKPLRE 277
Query: 255 LTKSSSFLKAVSKGA 269
+TKS SFLK V G+
Sbjct: 278 ITKSGSFLKTVRNGS 292
>At4g19030 nodulin-26 like protein
Length = 296
Score = 348 bits (894), Expect = 1e-96
Identities = 169/263 (64%), Positives = 212/263 (80%), Gaps = 7/263 (2%)
Query: 14 NINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISI 73
+I++ K D+ + VP LQKL+AE +GT+FL+F GCA+VVVN+ ND VVTLPGI+I
Sbjct: 33 DIHNPRPLKKQDSLLSVSVPFLQKLIAEFLGTYFLVFTGCASVVVNMQNDNVVTLPGIAI 92
Query: 74 VWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKL 133
VWGL +MVL+YS+GHISGAH NPAVTIA + GRFPLKQ+PAY+I+QV+GSTLA+ L+L
Sbjct: 93 VWGLTIMVLIYSLGHISGAHINPAVTIAFASCGRFPLKQVPAYVISQVIGSTLAAATLRL 152
Query: 134 IF-------SGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLA 186
+F SGK + F G+ P GSDLQAF +EFI+TF+LMFIISGVATDNRAIGELAGLA
Sbjct: 153 LFGLDHDVCSGKHDVFIGSSPVGSDLQAFTMEFIVTFYLMFIISGVATDNRAIGELAGLA 212
Query: 187 VGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLR 246
+GSTV+LNVL A P++ ASMNP RSLGPA+V+ Y+GIWIY+V+P LGA+AG W Y +R
Sbjct: 213 IGSTVLLNVLIAAPVSSASMNPGRSLGPALVYGCYKGIWIYLVAPTLGAIAGAWVYNTVR 272
Query: 247 ITNKPVRELTKSSSFLKAVSKGA 269
T+KP+RE+TKS SFLK V G+
Sbjct: 273 YTDKPLREITKSGSFLKTVRIGS 295
>At5g37820 Membrane integral protein (MIP) -like
Length = 283
Score = 288 bits (738), Expect = 2e-78
Identities = 142/236 (60%), Positives = 178/236 (75%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V L QKL+AE++GT+F+IF+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIIFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AHFNPAVT+ RFP Q+P YI AQ+ GS LAS L+L+F+ F GT P S
Sbjct: 99 AHFNPAVTVTFAVFRRFPWYQVPLYIGAQLTGSLLASLTLRLMFNVTPKAFFGTTPTDSS 158
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
QA V E II+F LMF+ISGVATD+RA GELAG+AVG T+ILNV AGPI+GASMNPARS
Sbjct: 159 GQALVAEIIISFLLMFVISGVATDSRATGELAGIAVGMTIILNVFVAGPISGASMNPARS 218
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVSK 267
LGPAIV Y+GIW+Y+V P +G AG + Y F+R T+KP+RELTKS+SFL++V++
Sbjct: 219 LGPAIVMGRYKGIWVYIVGPFVGIFAGGFVYNFMRFTDKPLRELTKSASFLRSVAQ 274
>At5g37810 Membrane integral protein (MIP) -like
Length = 283
Score = 286 bits (732), Expect = 8e-78
Identities = 139/235 (59%), Positives = 177/235 (75%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V L QKL+AE++GT+F++F+GC VVVN+ +T PGI + WGL VMV++YS GHISG
Sbjct: 39 VCLTQKLIAEMIGTYFIVFSGCGVVVVNVLYGGTITFPGICVTWGLIVMVMIYSTGHISG 98
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AHFNPAVT+ RFP Q+P YI AQ GS LAS L+L+F F GT PA S
Sbjct: 99 AHFNPAVTVTFAIFRRFPWHQVPLYIGAQFAGSLLASLTLRLMFKVTPEAFFGTTPADSP 158
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
+A V E II+F LMF+ISGVATDNRA+GELAG+AVG T+++NV AGPI+GASMNPARS
Sbjct: 159 ARALVAEIIISFLLMFVISGVATDNRAVGELAGIAVGMTIMVNVFVAGPISGASMNPARS 218
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKAVS 266
LGPA+V Y+ IW+Y+V P+LG ++G + Y +R T+KP+RELTKS+SFL+AVS
Sbjct: 219 LGPALVMGVYKHIWVYIVGPVLGVISGGFVYNLIRFTDKPLRELTKSASFLRAVS 273
>At2g34390 putative aquaporin (plasma membrane intrinsic protein)
Length = 288
Score = 259 bits (661), Expect = 1e-69
Identities = 133/242 (54%), Positives = 176/242 (71%), Gaps = 8/242 (3%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V LQKL+AE+VGT++LIFAGCAA+ VN ++ VVTL GI++VWG+ +MVLVY +GH+S
Sbjct: 44 VHFLQKLLAELVGTYYLIFAGCAAIAVNAQHNHVVTLVGIAVVWGIVIMVLVYCLGHLS- 102
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIF-------SGKENQFAG 144
AHFNPAVT+A ++ RFPL Q+PAYI QV+GSTLAS L+L+F S K + F G
Sbjct: 103 AHFNPAVTLALASSQRFPLNQVPAYITVQVIGSTLASATLRLLFDLNNDVCSKKHDVFLG 162
Query: 145 TLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGA 204
+ P+GSDLQAFV+EFIIT FLM ++ V T R EL GL +G+TV LNV+FAG ++GA
Sbjct: 163 SSPSGSDLQAFVMEFIITGFLMLVVCAVTTTKRTTEELEGLIIGATVTLNVIFAGEVSGA 222
Query: 205 SMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFLKA 264
SMNPARS+GPA+V Y+GIWIY+++P LGA++G + L E +K+ S K
Sbjct: 223 SMNPARSIGPALVWGCYKGIWIYLLAPTLGAVSGALIHKMLPSIQNAEPEFSKTGSSHKR 282
Query: 265 VS 266
V+
Sbjct: 283 VT 284
>At1g31885 major intrinsic protein - like
Length = 294
Score = 258 bits (659), Expect = 2e-69
Identities = 130/250 (52%), Positives = 182/250 (72%), Gaps = 15/250 (6%)
Query: 1 MGDISNGNLDVVMNINDDATKKCDDTTIDD------HVPLLQKLVAEVVGTFFLIFAGCA 54
+ DI+ VV++I + + DD+ D V +QKL+ E VGTF +IFAGC+
Sbjct: 4 ISDITTQTQTVVLDIEN--YQSIDDSRSSDLSAPLVSVSFVQKLIGEFVGTFTMIFAGCS 61
Query: 55 AVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLP 114
A+VVN K VTLPGI++VWGL V V++YSIGH+SGAHFNPAV+IA ++ +FP Q+P
Sbjct: 62 AIVVNETYGKPVTLPGIALVWGLVVTVMIYSIGHVSGAHFNPAVSIAFASSKKFPFNQVP 121
Query: 115 AYIIAQVVGSTLASGVLKLIF-------SGKENQFAGTLPAGSDLQAFVVEFIITFFLMF 167
YI AQ++GSTLA+ VL+L+F S K + + GT P+ S+ +FV+EFI TF LMF
Sbjct: 122 GYIAAQLLGSTLAAAVLRLVFHLDDDVCSLKGDVYVGTYPSNSNTTSFVMEFIATFNLMF 181
Query: 168 IISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIY 227
+IS VATD RA G AG+A+G+T++L++LF+GPI+GASMNPARSLGPA++ Y+ +W+Y
Sbjct: 182 VISAVATDKRATGSFAGIAIGATIVLDILFSGPISGASMNPARSLGPALIWGCYKDLWLY 241
Query: 228 MVSPILGALA 237
+VSP++GAL+
Sbjct: 242 IVSPVIGALS 251
>At1g80760 nodulin-like protein
Length = 305
Score = 223 bits (569), Expect = 6e-59
Identities = 119/219 (54%), Positives = 150/219 (68%), Gaps = 5/219 (2%)
Query: 31 HVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHIS 90
+V L +KL AE VGT LIFAG A +VN D TL G + GLAVM+++ S GHIS
Sbjct: 75 NVSLYRKLGAEFVGTLILIFAGTATAIVNQKTDGAETLIGCAASAGLAVMIVILSTGHIS 134
Query: 91 GAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAG--TLPA 148
GAH NPAVTIA FP K +P YI AQV+ S A+ LK +F E +G T+P
Sbjct: 135 GAHLNPAVTIAFAALKHFPWKHVPVYIGAQVMASVSAAFALKAVF---EPTMSGGVTVPT 191
Query: 149 GSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNP 208
QAF +EFII+F LMF+++ VATD RA+GELAG+AVG+TV+LN+L AGP T ASMNP
Sbjct: 192 VGLSQAFALEFIISFNLMFVVTAVATDTRAVGELAGIAVGATVMLNILIAGPATSASMNP 251
Query: 209 ARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRI 247
R+LGPAI + YR IW+Y+ +PILGAL G TYT +++
Sbjct: 252 VRTLGPAIAANNYRAIWVYLTAPILGALIGAGTYTIVKL 290
>At4g10380 major intrinsic protein (MIP) - like
Length = 304
Score = 213 bits (542), Expect = 9e-56
Identities = 115/232 (49%), Positives = 152/232 (64%), Gaps = 7/232 (3%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISG 91
V L +KL AE VGTF LIF A +VN D TL G + GLAVM+++ S GHISG
Sbjct: 74 VSLTRKLGAEFVGTFILIFTATAGPIVNQKYDGAETLIGNAACAGLAVMIIILSTGHISG 133
Query: 92 AHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSD 151
AH NP++TIA FP +PAYI AQV S AS LK +F + T+P+ S
Sbjct: 134 AHLNPSLTIAFAALRHFPWAHVPAYIAAQVSASICASFALKGVFHPFMSGGV-TIPSVSL 192
Query: 152 LQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARS 211
QAF +EFIITF L+F+++ VATD RA+GELAG+AVG+TV+LN+L AGP TG SMNP R+
Sbjct: 193 GQAFALEFIITFILLFVVTAVATDTRAVGELAGIAVGATVMLNILVAGPSTGGSMNPVRT 252
Query: 212 LGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITN------KPVRELTK 257
LGPA+ YR +W+Y+V+P LGA++G YT +++ + +PVR +
Sbjct: 253 LGPAVASGNYRSLWVYLVAPTLGAISGAAVYTGVKLNDSVTDPPRPVRSFRR 304
>At3g06100 putative major intrinsic protein
Length = 275
Score = 174 bits (440), Expect = 6e-44
Identities = 92/218 (42%), Positives = 136/218 (62%), Gaps = 2/218 (0%)
Query: 35 LQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHF 94
L+ ++AE+VGTF L+F+ C + + V L ++ GL+V+V+VYSIGHISGAH
Sbjct: 45 LRIVMAELVGTFILMFSVCGVISSTQLSGGHVGLLEYAVTAGLSVVVVVYSIGHISGAHL 104
Query: 95 NPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQA 154
NP++TIA G FP Q+P YI AQ +G+T A+ V ++ G T PA S + A
Sbjct: 105 NPSITIAFAVFGGFPWSQVPLYITAQTLGATAATLVGVSVY-GVNADIMATKPALSCVSA 163
Query: 155 FVVEFIITFFLMFIISGV-ATDNRAIGELAGLAVGSTVILNVLFAGPITGASMNPARSLG 213
F VE I T ++F+ S + ++ +G L G +G+ + L VL GPI+G SMNPARSLG
Sbjct: 164 FFVELIATSIVVFLASALHCGPHQNLGNLTGFVIGTVISLGVLITGPISGGSMNPARSLG 223
Query: 214 PAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKP 251
PA+V ++ +WIYM +P++GA+ G TY + + +P
Sbjct: 224 PAVVAWDFEDLWIYMTAPVIGAIIGVLTYRSISLKTRP 261
>At3g16240 delta tonoplast integral protein (delta-TIP)
Length = 250
Score = 127 bits (319), Expect = 6e-30
Identities = 80/222 (36%), Positives = 118/222 (53%), Gaps = 10/222 (4%)
Query: 27 TIDDHVPL--LQKLVAEVVGTFFLIFAGCAAVVV--NLNNDKVVTLPG---ISIVWGLAV 79
+ DD L L+ +AE + T +FAG + + L +D + PG I++ G A+
Sbjct: 8 SFDDSFSLASLRAYLAEFISTLLFVFAGVGSAIAYAKLTSDAALDTPGLVAIAVCHGFAL 67
Query: 80 MVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKE 139
V V +ISG H NPAVT G+ + Y IAQ++GST A +LK + G
Sbjct: 68 FVAVAIGANISGGHVNPAVTFGLAVGGQITVITGVFYWIAQLLGSTAACFLLKYVTGGLA 127
Query: 140 NQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNR--AIGELAGLAVGSTVILNVLF 197
++ V+E IITF L++ + A D + ++G +A LA+G V N+L
Sbjct: 128 VPTHSVAAGLGSIEGVVMEIIITFALVYTVYATAADPKKGSLGTIAPLAIGLIVGANILA 187
Query: 198 AGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPIL-GALAG 238
AGP +G SMNPARS GPA+ ++ G W+Y V P++ G LAG
Sbjct: 188 AGPFSGGSMNPARSFGPAVAAGDFSGHWVYWVGPLIGGGLAG 229
>At2g29870 putative aquaporin (plasma membrane intrinsic protein)
Length = 139
Score = 122 bits (307), Expect = 2e-28
Identities = 62/124 (50%), Positives = 85/124 (68%)
Query: 143 AGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGELAGLAVGSTVILNVLFAGPIT 202
+G+ P+GSDLQAFV+EFIIT FLM ++ V T R EL GL +G+TV LNV+F G ++
Sbjct: 12 SGSSPSGSDLQAFVMEFIITGFLMLVVCAVTTTKRTTEELEGLIIGATVTLNVIFVGEVS 71
Query: 203 GASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVRELTKSSSFL 262
GASMNPARS+GPA+V Y+GIWIY+++P LGA++ + L + +K+ S
Sbjct: 72 GASMNPARSIGPALVWGCYKGIWIYLLAPTLGAVSRALIHKMLPSIPNAEPKFSKTGSSH 131
Query: 263 KAVS 266
K VS
Sbjct: 132 KRVS 135
Score = 34.7 bits (78), Expect = 0.055
Identities = 28/100 (28%), Positives = 46/100 (46%), Gaps = 6/100 (6%)
Query: 35 LQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHF 94
LQ V E + T FL+ CA ++ L G+ I G V + V +G +SGA
Sbjct: 21 LQAFVMEFIITGFLMLVVCAVTTTKRTTEE---LEGLII--GATVTLNVIFVGEVSGASM 75
Query: 95 NPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLI 134
NPA +I K + Y++A +G+ + + K++
Sbjct: 76 NPARSIGPALVWGC-YKGIWIYLLAPTLGAVSRALIHKML 114
>At5g47450 membrane channel protein-like; aquaporin (tonoplast
intrinsic protein)-like
Length = 250
Score = 120 bits (302), Expect = 6e-28
Identities = 78/215 (36%), Positives = 115/215 (53%), Gaps = 8/215 (3%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVV--NLNNDKVVTLPG---ISIVWGLAVMVLVYSI 86
V L+ ++E + T +FAG + V L +D + G I+I A+ V V
Sbjct: 15 VSSLKAYLSEFIATLLFVFAGVGSAVAFAKLTSDGALDPAGLVAIAIAHAFALFVGVSIA 74
Query: 87 GHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTL 146
+ISG H NPAVT+ G L Y IAQ +GS +A +L + +GK G
Sbjct: 75 ANISGGHLNPAVTLGLAIGGNITLITGFFYWIAQCLGSIVACLLLVFVTNGKSVPTHGVS 134
Query: 147 PAGSDLQAFVVEFIITFFLMFIISGVATDNR--AIGELAGLAVGSTVILNVLFAGPITGA 204
++ V+E ++TF L++ + A D + ++G +A +A+G V N+L AGP +G
Sbjct: 135 AGLGAVEGVVMEIVVTFALVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSGG 194
Query: 205 SMNPARSLGPAIVHHEYRGIWIYMVSPIL-GALAG 238
SMNPARS GPA+V + IWIY V P++ GALAG
Sbjct: 195 SMNPARSFGPAVVSGDLSQIWIYWVGPLVGGALAG 229
>At1g17810 putative protein
Length = 267
Score = 119 bits (299), Expect = 1e-27
Identities = 81/252 (32%), Positives = 127/252 (50%), Gaps = 23/252 (9%)
Query: 22 KCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV---------VTLPG-- 70
+ D+ T H ++ +AE + TF +FAG ++ L DK+ PG
Sbjct: 13 RADEAT---HPDSIRATLAEFLSTFVFVFAGEGSI---LALDKLYWDTAAHTGTNTPGGL 66
Query: 71 --ISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLAS 128
+++ LA+ V + ++SG H NPAVT A GR + + Y +AQ++G+ LA
Sbjct: 67 VLVALAHALALFAAVSAAINVSGGHVNPAVTFAALIGGRISVIRAIYYWVAQLIGAILAC 126
Query: 129 GVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNR--AIGELAGLA 186
+L+L +G S+L ++E I+TF L++++ A D + +IG +A LA
Sbjct: 127 LLLRLATNGLRPVGFHVASGVSELHGLLMEIILTFALVYVVYSTAIDPKRGSIGIIAPLA 186
Query: 187 VGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLR 246
+G V N+L GP GASMNPAR+ GPA+V + WIY V P +G Y ++
Sbjct: 187 IGLIVGANILVGGPFDGASMNPARAFGPALVGWRWSNHWIYWVGPFIGGALAALIYEYMI 246
Query: 247 I--TNKPVRELT 256
I N+P T
Sbjct: 247 IPSVNEPPHHST 258
>At4g17340 membrane channel like protein
Length = 250
Score = 119 bits (298), Expect = 2e-27
Identities = 73/215 (33%), Positives = 116/215 (53%), Gaps = 8/215 (3%)
Query: 32 VPLLQKLVAEVVGTFFLIFAGCAAVVV--NLNNDKVVTLPG---ISIVWGLAVMVLVYSI 86
V L+ ++E + T +FAG + + L +D + G +++ A+ V V
Sbjct: 15 VASLKAYLSEFIATLLFVFAGVGSALAFAKLTSDAALDPAGLVAVAVAHAFALFVGVSIA 74
Query: 87 GHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTL 146
+ISG H NPAVT+ G + Y IAQ +GS +A +L + +G+ G
Sbjct: 75 ANISGGHLNPAVTLGLAVGGNITVITGFFYWIAQCLGSIVACLLLVFVTNGESVPTHGVA 134
Query: 147 PAGSDLQAFVVEFIITFFLMFIISGVATDNR--AIGELAGLAVGSTVILNVLFAGPITGA 204
++ V+E ++TF L++ + A D + ++G +A +A+G V N+L AGP +G
Sbjct: 135 AGLGAIEGVVMEIVVTFALVYTVYATAADPKKGSLGTIAPIAIGFIVGANILAAGPFSGG 194
Query: 205 SMNPARSLGPAIVHHEYRGIWIYMVSPIL-GALAG 238
SMNPARS GPA+V ++ IWIY V P++ GALAG
Sbjct: 195 SMNPARSFGPAVVSGDFSQIWIYWVGPLVGGALAG 229
>At2g21020 putative major intrinsic (channel) protein
Length = 262
Score = 111 bits (277), Expect = 5e-25
Identities = 56/116 (48%), Positives = 78/116 (66%), Gaps = 4/116 (3%)
Query: 1 MGDISNGNLDVVMNINDDATKKCDDTTIDD----HVPLLQKLVAEVVGTFFLIFAGCAAV 56
+ DI+ VV +I +D + +++ V +QKL+ E VGTF +IFAGC+A+
Sbjct: 15 ISDITTQTQTVVSDIENDRSIDDSQSSVLSGPLVSVSFVQKLIGEFVGTFSMIFAGCSAI 74
Query: 57 VVNLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQ 112
VVN K VTLPGI++VWGL V V++YSIGH+SGAHFNPAV+IA ++ +FP Q
Sbjct: 75 VVNETYGKPVTLPGIALVWGLVVTVMIYSIGHVSGAHFNPAVSIAFASSKKFPFNQ 130
Score = 33.1 bits (74), Expect = 0.16
Identities = 24/84 (28%), Positives = 41/84 (48%), Gaps = 5/84 (5%)
Query: 134 IFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNRAIGE---LAGLAV--G 188
I + + +G L + S +Q + EF+ TF ++F N G+ L G+A+ G
Sbjct: 35 IDDSQSSVLSGPLVSVSFVQKLIGEFVGTFSMIFAGCSAIVVNETYGKPVTLPGIALVWG 94
Query: 189 STVILNVLFAGPITGASMNPARSL 212
V + + G ++GA NPA S+
Sbjct: 95 LVVTVMIYSIGHVSGAHFNPAVSI 118
>At1g73190 tonoplast intrinsic protein, alpha (alpha-TIP)
Length = 268
Score = 111 bits (277), Expect = 5e-25
Identities = 74/245 (30%), Positives = 121/245 (49%), Gaps = 21/245 (8%)
Query: 22 KCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKV---------VTLPG-- 70
+ D+ T H ++ +AE + TF +FA ++ L+ DK+ PG
Sbjct: 13 RADEAT---HPDSIRATLAEFLSTFVFVFAAEGSI---LSLDKLYWEHAAHAGTNTPGGL 66
Query: 71 --ISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLAS 128
+++ A+ V + ++SG H NPAVT GR + Y IAQ++G+ LA
Sbjct: 67 ILVALAHAFALFAAVSAAINVSGGHVNPAVTFGALVGGRVTAIRAIYYWIAQLLGAILAC 126
Query: 129 GVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATDNR--AIGELAGLA 186
+L+L +G + V+E I+TF L++++ D + ++G +A LA
Sbjct: 127 LLLRLTTNGMRPVGFRLASGVGAVNGLVLEIILTFGLVYVVYSTLIDPKRGSLGIIAPLA 186
Query: 187 VGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLR 246
+G V N+L GP +GASMNPAR+ GPA+V + WIY V P +G+ Y ++
Sbjct: 187 IGLIVGANILVGGPFSGASMNPARAFGPALVGWRWHDHWIYWVGPFIGSALAALIYEYMV 246
Query: 247 ITNKP 251
I +P
Sbjct: 247 IPTEP 251
>At3g26520 salt-stress induced tonoplast intrinsic protein
Length = 253
Score = 109 bits (272), Expect = 2e-24
Identities = 74/232 (31%), Positives = 110/232 (46%), Gaps = 7/232 (3%)
Query: 31 HVPLLQKLVAEVVGTFFLIFAGCAA-VVVNLNNDKVVTLPGISIVWGLAV---MVLVYSI 86
H L+ +AE + T +FAG + + N D T P + LA + + S+
Sbjct: 17 HPNALRAALAEFISTLIFVFAGSGSGIAFNKITDNGATTPSGLVAAALAHAFGLFVAVSV 76
Query: 87 G-HISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGT 145
G +ISG H NPAVT G L + Y IAQ++GS A +L G+ G
Sbjct: 77 GANISGGHVNPAVTFGVLLGGNITLLRGILYWIAQLLGSVAACFLLSFATGGEPIPAFGL 136
Query: 146 LPAGSDLQAFVVEFIITFFLMFIISGVATD--NRAIGELAGLAVGSTVILNVLFAGPITG 203
L A V E ++TF L++ + A D N ++G +A +A+G V N+L G +G
Sbjct: 137 SAGVGSLNALVFEIVMTFGLVYTVYATAVDPKNGSLGTIAPIAIGFIVGANILAGGAFSG 196
Query: 204 ASMNPARSLGPAIVHHEYRGIWIYMVSPILGALAGTWTYTFLRITNKPVREL 255
ASMNPA + GPA+V + W+Y P++G Y F+ I +L
Sbjct: 197 ASMNPAVAFGPAVVSWTWTNHWVYWAGPLIGGGLAGIIYDFVFIDENAHEQL 248
>At2g25810 putative aquaporin (tonoplast intrinsic protein)
Length = 249
Score = 108 bits (269), Expect = 4e-24
Identities = 71/210 (33%), Positives = 106/210 (49%), Gaps = 8/210 (3%)
Query: 35 LQKLVAEVVGTFFLIFAGCAAVVV--NLNNDKVVTLPGISIVWGLAVMVLVYSIGHISGA 92
++ L+ E + TF +FAG + + +L + +V L +++ V V++ S GHISG
Sbjct: 18 IKALIVEFITTFLFVFAGVGSAMATDSLVGNTLVGLFAVAVAHAFVVAVMI-SAGHISGG 76
Query: 93 HFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDL 152
H NPAVT+ G + + Y I Q++ S+ A +L + G S
Sbjct: 77 HLNPAVTLGLLLGGHISVFRAFLYWIDQLLASSAACFLLSYLTGGMGTPVHTLASGVSYT 136
Query: 153 QAFVVEFIITFFLMFIISGVATDNRAIGELAG---LAVGSTVILNVLFAGPITGASMNPA 209
Q + E I+TF L+F + D + G L G L G V N+L G +GASMNPA
Sbjct: 137 QGIIWEIILTFSLLFTVYATIVDPKK-GSLDGFGPLLTGFVVGANILAGGAFSGASMNPA 195
Query: 210 RSLGPAIVHHEYRGIWIYMVSPIL-GALAG 238
RS GPA+V + W+Y V P++ G LAG
Sbjct: 196 RSFGPALVSGNWTDHWVYWVGPLIGGGLAG 225
Score = 29.3 bits (64), Expect = 2.3
Identities = 22/72 (30%), Positives = 35/72 (48%), Gaps = 5/72 (6%)
Query: 27 TIDDHVPLLQKLVAEVVGTFFLIFAGCAAVVVNLNNDKVVTLPGIS-IVWGLAVMVLVYS 85
T+ V Q ++ E++ TF L+F A +V + K +L G ++ G V + +
Sbjct: 128 TLASGVSYTQGIIWEIILTFSLLFTVYATIV----DPKKGSLDGFGPLLTGFVVGANILA 183
Query: 86 IGHISGAHFNPA 97
G SGA NPA
Sbjct: 184 GGAFSGASMNPA 195
>At2g36830 putative aquaporin (tonoplast intrinsic protein gamma)
Length = 251
Score = 105 bits (263), Expect = 2e-23
Identities = 76/231 (32%), Positives = 114/231 (48%), Gaps = 14/231 (6%)
Query: 13 MNINDDATKKCDDTTIDDHVPLLQKLVAEVVGTFFLIFAGCAA-VVVNLNNDKVVTLPG- 70
M I + A + D+ T D L+ +AE + T + AG + + N + T P
Sbjct: 1 MPIRNIAIGRPDEATRPD---ALKAALAEFISTLIFVVAGSGSGMAFNKLTENGATTPSG 57
Query: 71 -----ISIVWGLAVMVLVYSIGHISGAHFNPAVTIAHTTTGRFPLKQLPAYIIAQVVGST 125
++ +GL V V V + +ISG H NPAVT G L + Y IAQ++GS
Sbjct: 58 LVAAAVAHAFGLFVAVSVGA--NISGGHVNPAVTFGAFIGGNITLLRGILYWIAQLLGSV 115
Query: 126 LASGVLKLIFSGKENQFAGTLPAGSDLQAFVVEFIITFFLMFIISGVATD--NRAIGELA 183
+A +LK G G L AFV E ++TF L++ + A D N ++G +A
Sbjct: 116 VACLILKFATGGLAVPAFGLSAGVGVLNAFVFEIVMTFGLVYTVYATAIDPKNGSLGTIA 175
Query: 184 GLAVGSTVILNVLFAGPITGASMNPARSLGPAIVHHEYRGIWIYMVSPILG 234
+A+G V N+L G +GASMNPA + GPA+V + W+Y P++G
Sbjct: 176 PIAIGFIVGANILAGGAFSGASMNPAVAFGPAVVSWTWTNHWVYWAGPLVG 226
>At4g01470 putative water channel protein
Length = 252
Score = 100 bits (249), Expect = 8e-22
Identities = 70/210 (33%), Positives = 99/210 (46%), Gaps = 7/210 (3%)
Query: 40 AEVVGTFFLIFAGCAAVVV--NLNNDKVVTLPGI---SIVWGLAVMVLVYSIGHISGAHF 94
AE +FAG + + L D T G+ S+ A+ V V ++SG H
Sbjct: 25 AEFFSMVIFVFAGQGSGMAYGKLTGDGPATPAGLVAASLSHAFALFVAVSVGANVSGGHV 84
Query: 95 NPAVTIAHTTTGRFPLKQLPAYIIAQVVGSTLASGVLKLIFSGKENQFAGTLPAGSDLQA 154
NPAVT G L + Y IAQ++G+ +A +LK+ G E + A
Sbjct: 85 NPAVTFGAFIGGNITLLRAILYWIAQLLGAVVACLLLKVSTGGMETAAFSLSYGVTPWNA 144
Query: 155 FVVEFIITFFLMFIISGVATDNRA--IGELAGLAVGSTVILNVLFAGPITGASMNPARSL 212
V E ++TF L++ + A D + IG +A LA+G V N+L G GASMNPA S
Sbjct: 145 VVFEIVMTFGLVYTVYATAVDPKKGDIGIIAPLAIGLIVGANILVGGAFDGASMNPAVSF 204
Query: 213 GPAIVHHEYRGIWIYMVSPILGALAGTWTY 242
GPA+V + W+Y V P +GA Y
Sbjct: 205 GPAVVSWIWTNHWVYWVGPFIGAAIAAIVY 234
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.139 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,508,013
Number of Sequences: 26719
Number of extensions: 218494
Number of successful extensions: 617
Number of sequences better than 10.0: 38
Number of HSP's better than 10.0 without gapping: 36
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 485
Number of HSP's gapped (non-prelim): 52
length of query: 270
length of database: 11,318,596
effective HSP length: 98
effective length of query: 172
effective length of database: 8,700,134
effective search space: 1496423048
effective search space used: 1496423048
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140547.14


