

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140546.3 + phase: 0
(266 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
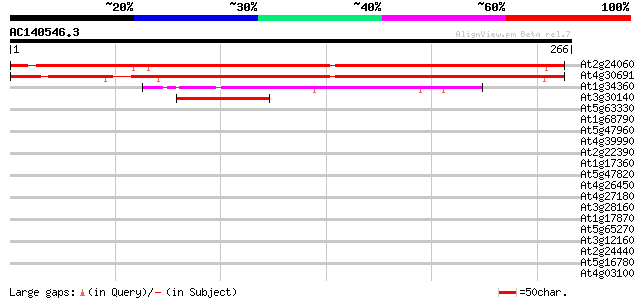
Score E
Sequences producing significant alignments: (bits) Value
At2g24060 putative chloroplast initiation factor 3 283 7e-77
At4g30691 unknown protein 282 1e-76
At1g34360 transcription factor like protein 70 2e-12
At3g30140 hypothetical protein 58 6e-09
At5g63330 unknown protein 32 0.35
At1g68790 putative nuclear matrix constituent protein 1 (NMCP1) 32 0.35
At5g47960 RAS superfamily GTP-binding protein-like 30 1.0
At4g39990 GTP-binding protein GB3 30 1.0
At2g22390 putative GTP-binding protein 30 1.0
At1g17360 COP1-Interacting Protein 7, putative 30 1.0
At5g47820 kinesin-like protein 30 1.3
At4g26450 unknown protein 30 1.3
At4g27180 heavy chain polypeptide of kinesin-like protein (katB) 29 3.0
At3g28160 non-LTR reverse transcriptase, putative 29 3.0
At1g17870 hypothetical protein 29 3.0
At5g65270 GTP-binding protein 28 3.9
At3g12160 hypothetical protein 28 3.9
At2g24440 unknown protein 28 5.1
At5g16780 putative protein 28 6.6
At4g03100 rac GTPase activating protein like 28 6.6
>At2g24060 putative chloroplast initiation factor 3
Length = 312
Score = 283 bits (724), Expect = 7e-77
Identities = 162/282 (57%), Positives = 200/282 (70%), Gaps = 24/282 (8%)
Query: 1 MAGITSTTVVPVKLKASHFFSNKLSIPHSLNLQSSPSFTLPSHATVRYGVNLRPSAY--- 57
MAGITS+TV A K HSL S +L T NL PS
Sbjct: 1 MAGITSSTV---GFNAVFTGITKTVSSHSLFSVDSKLCSLRLSKTELSFTNLTPSPRRAF 57
Query: 58 -------GGGGNFR-----RAPPEKDADDGQALDLSTLSSNTVRLIDQSQNMVGVVSLDQ 105
GGGG +R R K+A+ +ALD+S++ S TVRLID QNM+G+VS D+
Sbjct: 58 AVTCRFGGGGGGYRFSGDNRRGRPKEAEIDEALDISSIRSATVRLIDGQQNMLGLVSKDE 117
Query: 106 AIRMAEDAELDLVIVSAEADPPVVRIMNYSKYRYELQKKKRDQQKKSAASRMDLKELKMG 165
A+RMA+DAELDLVI+S +ADPPVV++M+YSKYRYE QK+K+DQQKK+ +RMDLKELKMG
Sbjct: 118 AVRMADDAELDLVILSPDADPPVVKMMDYSKYRYEQQKRKKDQQKKT--TRMDLKELKMG 175
Query: 166 YNIDQHDYSVRLKAARKFLSDGDKVKVIVNLKGREKEFRNNAIELIRRFQNDVGKLATEE 225
YNIDQHDYSVRL+AA+KFL DGDKVKVIV++KGRE EFRN AIEL+RRFQ ++G+LATEE
Sbjct: 176 YNIDQHDYSVRLRAAQKFLQDGDKVKVIVSMKGRENEFRNIAIELLRRFQTEIGELATEE 235
Query: 226 AKNFRDKNIFITLIPNKTEVQKPQETPT----KAATDEVSVS 263
+KNFRD+N+FI L+PNK ++KPQE PT K A +E S S
Sbjct: 236 SKNFRDRNMFIILVPNKEMIRKPQEPPTRKKKKTAENEASAS 277
>At4g30691 unknown protein
Length = 281
Score = 282 bits (721), Expect = 1e-76
Identities = 156/273 (57%), Positives = 196/273 (71%), Gaps = 23/273 (8%)
Query: 1 MAGITSTTVVPVKLKASHFFSNKLSIPHSLNLQSSPSFTLPSHA-TVRYGVNLRPSAYGG 59
+AG T T PVK K F +L +P + SP A T RYG GG
Sbjct: 14 LAGATKTVSHPVKSK---LFGLRLCVPEFSIVSLSPYHHRRCPAITCRYG--------GG 62
Query: 60 GGNFRRAPPE-----KDADDGQALDLSTLSSNTVRLIDQSQNMVGVVSLDQAIRMAEDAE 114
GG R P + K+++D +LD+S + S TVRLID QNM+G+VS ++A+R AEDAE
Sbjct: 63 GGGGSRFPGDRRGRQKESEDDDSLDISAIRSATVRLIDDQQNMIGLVSKEEAVRRAEDAE 122
Query: 115 LDLVIVSAEADPPVVRIMNYSKYRYELQKKKRDQQKKSAASRMDLKELKMGYNIDQHDYS 174
LDLVI+S +ADPPVVR+M+YSKYRYE QK+K++QQKK+ +RMDLKELKMGYNIDQHDYS
Sbjct: 123 LDLVILSPDADPPVVRMMDYSKYRYEQQKRKKEQQKKT--TRMDLKELKMGYNIDQHDYS 180
Query: 175 VRLKAARKFLSDGDKVKVIVNLKGREKEFRNNAIELIRRFQNDVGKLATEEAKNFRDKNI 234
VR++AARKFL DGDKVKVIVN+KGRE EFRN AIEL+RRFQ ++G+L TEE+KNFRD+N+
Sbjct: 181 VRMRAARKFLQDGDKVKVIVNMKGRENEFRNIAIELLRRFQTEIGELGTEESKNFRDRNL 240
Query: 235 FITLIPNKTEVQKPQETP----TKAATDEVSVS 263
FI L+PNK ++K QE P K A D+VS +
Sbjct: 241 FIVLVPNKEVIRKVQEPPPKKKKKPADDKVSAA 273
>At1g34360 transcription factor like protein
Length = 520
Score = 69.7 bits (169), Expect = 2e-12
Identities = 54/165 (32%), Positives = 88/165 (52%), Gaps = 8/165 (4%)
Query: 64 RRAPPEKDADDGQALDLSTLSSNTVRLIDQSQNMVGVVSLDQAIRMAEDAELDLVIVSAE 123
R+ E D+D G L+ ++ + VRL+ + + V VSL +A+R A++ + DLV V +
Sbjct: 73 RKKEAEVDSD-GPRLN-EKITGDYVRLVSEEGHCV--VSLREALRRAKELQCDLVEVQRD 128
Query: 124 ADPPVVRIMNYSKYRYELQK--KKRDQQKKSAASRMDLKELKMGYNIDQHDYSVRLKAAR 181
A PPV +I+ YS Y+ K K+R + K++ A R D+KE++ I+ D + A
Sbjct: 129 AKPPVCKIVKYSLELYKKAKVGKERAKAKRAEAIRPDIKEIRFTPKIEAKDLKFKSDQAL 188
Query: 182 KFLSDGDKVKVI-VNLKGREKEFR-NNAIELIRRFQNDVGKLATE 224
K + G +VK + V K + KE +EL+ RF +G E
Sbjct: 189 KLMESGYRVKCLAVPDKDKHKELEPEKLLELLFRFTCFIGDALVE 233
>At3g30140 hypothetical protein
Length = 140
Score = 57.8 bits (138), Expect = 6e-09
Identities = 29/44 (65%), Positives = 36/44 (80%)
Query: 80 LSTLSSNTVRLIDQSQNMVGVVSLDQAIRMAEDAELDLVIVSAE 123
+ + SS TVRLID QNM+G+VS D+A+RMA+DAELDLV VS E
Sbjct: 73 VQSYSSATVRLIDGQQNMLGLVSKDEAVRMADDAELDLVCVSIE 116
>At5g63330 unknown protein
Length = 477
Score = 32.0 bits (71), Expect = 0.35
Identities = 21/95 (22%), Positives = 43/95 (45%), Gaps = 15/95 (15%)
Query: 126 PPVVRIMNYSKYRYELQKKKRDQQKKSAASRMDLKELKMGYNIDQHDYSVRLKAARKFLS 185
PPV+ + + + E+ + +KK AA +D +R++ A+ ++
Sbjct: 271 PPVIPLTSSASLESEIPFEVAPMRKKEAA---------------MNDNKLRVEPAKLVMT 315
Query: 186 DGDKVKVIVNLKGREKEFRNNAIELIRRFQNDVGK 220
DG+K K+ +L E++F +L+R G+
Sbjct: 316 DGEKKKLGQDLMALEEDFPQKIADLLREQSGSDGQ 350
>At1g68790 putative nuclear matrix constituent protein 1 (NMCP1)
Length = 1085
Score = 32.0 bits (71), Expect = 0.35
Identities = 26/97 (26%), Positives = 49/97 (49%), Gaps = 11/97 (11%)
Query: 130 RIMNYSKYRYELQKKKRDQQKKSAASRMDLKELKMGYNIDQHDYSVRLKAARKFLSDGDK 189
R+M + + +K + Q+K + ++ +L E + I +D S++ K D +
Sbjct: 270 RVMENERTIEKKEKILENLQQKISVAKSELTEKEESIKIKLNDISLKEK-------DFEA 322
Query: 190 VKVIVNLKGRE-KEFRNNAIELIRRFQNDVGKLATEE 225
+K V++K +E EF N IE R Q ++GKL ++
Sbjct: 323 MKAKVDIKEKELHEFEENLIE---REQMEIGKLLDDQ 356
>At5g47960 RAS superfamily GTP-binding protein-like
Length = 223
Score = 30.4 bits (67), Expect = 1.0
Identities = 15/35 (42%), Positives = 21/35 (59%)
Query: 224 EEAKNFRDKNIFITLIPNKTEVQKPQETPTKAATD 258
EE + DKNI I LI NKT++ + PT+ A +
Sbjct: 111 EELRGHADKNIVIMLIGNKTDLGTLRAVPTEDAKE 145
>At4g39990 GTP-binding protein GB3
Length = 224
Score = 30.4 bits (67), Expect = 1.0
Identities = 14/35 (40%), Positives = 22/35 (62%)
Query: 224 EEAKNFRDKNIFITLIPNKTEVQKPQETPTKAATD 258
EE + DKNI I LI NK++++ + PT+ A +
Sbjct: 113 EELRAHADKNIVIILIGNKSDLEDQRAVPTEDAKE 147
>At2g22390 putative GTP-binding protein
Length = 176
Score = 30.4 bits (67), Expect = 1.0
Identities = 14/35 (40%), Positives = 21/35 (60%)
Query: 224 EEAKNFRDKNIFITLIPNKTEVQKPQETPTKAATD 258
EE + DKNI I LI NKT+++ + P + A +
Sbjct: 80 EELRVHADKNIVIILIGNKTDLENQRSVPVEDAKE 114
>At1g17360 COP1-Interacting Protein 7, putative
Length = 1061
Score = 30.4 bits (67), Expect = 1.0
Identities = 24/95 (25%), Positives = 47/95 (49%), Gaps = 5/95 (5%)
Query: 139 YELQKKKRDQQKKSAASRMDLKELKMGYNIDQHDYSVRLKAARKFLSDGDKVKVIVNLKG 198
YE KKRD + + S + K M +DQ ++ K + + D + + +
Sbjct: 556 YEEYMKKRDAKLREEWSSKESKLKSMQEALDQSRTEMKAKFSAASMKRQDSIS---STRQ 612
Query: 199 REKEFRN-NAIELIRRFQNDVGKLATEEAKNFRDK 232
R ++FR+ N+ +++Q+ + L +EE +N +DK
Sbjct: 613 RAEKFRSFNSRTSSKKYQHPISSLQSEE-ENEKDK 646
>At5g47820 kinesin-like protein
Length = 1035
Score = 30.0 bits (66), Expect = 1.3
Identities = 21/93 (22%), Positives = 46/93 (48%), Gaps = 2/93 (2%)
Query: 136 KYRYELQKKKRDQQKKSAASRM--DLKELKMGYNIDQHDYSVRLKAARKFLSDGDKVKVI 193
K ++Q K+ Q+ AA R+ +++ +K QH + R++ + +K +
Sbjct: 598 KQESQVQLLKQKQKSDDAARRLQDEIQSIKAQKVQLQHRMKQEAEQFRQWKASREKELLQ 657
Query: 194 VNLKGREKEFRNNAIELIRRFQNDVGKLATEEA 226
+ +GR+ E+ + ++ + + Q V + TEEA
Sbjct: 658 LRKEGRKSEYERHKLQALNQRQKMVLQRKTEEA 690
>At4g26450 unknown protein
Length = 489
Score = 30.0 bits (66), Expect = 1.3
Identities = 24/106 (22%), Positives = 48/106 (44%), Gaps = 14/106 (13%)
Query: 73 DDGQALDLSTLSSNTVRLIDQSQNMVGVVSLDQAIRMAEDAELDLVIVSAEADPPVVRIM 132
DDG++LD + S +V+L+ S ++ + + D A PV++
Sbjct: 28 DDGRSLDNGSCSDESVKLL----------STSNSVELGKPMSFDSPGDGGGAYSPVLKGQ 77
Query: 133 NYSKYRYELQKKKRDQQKKSAASRMDLKELKMGYNIDQHDYSVRLK 178
K+R + +RD K ++A+ + K LK G + H + +++
Sbjct: 78 GLRKWR----RIRRDLVKDTSANMENSKALKRGLSGVAHSHGKQMQ 119
>At4g27180 heavy chain polypeptide of kinesin-like protein (katB)
Length = 745
Score = 28.9 bits (63), Expect = 3.0
Identities = 28/103 (27%), Positives = 46/103 (44%), Gaps = 14/103 (13%)
Query: 130 RIMNYSKYRYELQKKKRDQQKKSAASRMDL-----KELKMGYNIDQHDYSVRLKAARKFL 184
RI SKY Y K+R + R+ L +EL++ Y +Q ++ K
Sbjct: 46 RIKYKSKYNY----KERCENTMDYVKRLRLCIRWFQELELDYAFEQEKLKNAMEMNEKHC 101
Query: 185 SDGDKVKVIVNLKGREKEFRNNAIELIRRFQNDVGKLATEEAK 227
+D + VNLK +E+E EL + F + +LA E+ +
Sbjct: 102 ADLE-----VNLKVKEEELNMVIDELRKNFASVQVQLAKEQTE 139
>At3g28160 non-LTR reverse transcriptase, putative
Length = 630
Score = 28.9 bits (63), Expect = 3.0
Identities = 38/169 (22%), Positives = 71/169 (41%), Gaps = 8/169 (4%)
Query: 80 LSTLSSNTVRLIDQSQNMVGVVSLDQAIRM--AEDAELDLVIVSAEADPPVVRIMNYSKY 137
L +L + L + QN ++LD +R +++ E D A+ + + MN SK
Sbjct: 454 LMSLPNQFSELKETMQNSGISLTLDNVMRAIYSKEQEFDQFWEQAKERKVLDQRMNLSKL 513
Query: 138 RYELQKKKRDQQKKSAASRMDLKELKMGYNIDQHDYSVRLKAARKFLSDGDKVK---VIV 194
R +K+K+ + K+ ++ + K G++ Q ++ KA K K V
Sbjct: 514 RANQEKEKKMNKSKAKGKKICWTDHKEGHSYKQCNH---CKAENKLGEQSHKSVCKFVGS 570
Query: 195 NLKGREKEFRNNAIELIRRFQNDVGKLATEEAKNFRDKNIFITLIPNKT 243
N E +N+ L R + + + EEA + K + + N+T
Sbjct: 571 NRLKAELMVKNDVSHLNERLEEKLVLCSEEEALSQSIKELKKEQVANET 619
>At1g17870 hypothetical protein
Length = 573
Score = 28.9 bits (63), Expect = 3.0
Identities = 20/107 (18%), Positives = 50/107 (46%), Gaps = 3/107 (2%)
Query: 66 APPEKDADDGQALDLSTLSSNTVRLIDQSQNMVGVVSLDQAIRMAE---DAELDLVIVSA 122
APP ++ + + S + ++ + +G S++ AI++ + D +L + +
Sbjct: 84 APPSPESSEEEEEKKSKQQEMDWKTDEEFKKFMGNPSIEAAIKLEKTRTDRKLKELNKES 143
Query: 123 EADPPVVRIMNYSKYRYELQKKKRDQQKKSAASRMDLKELKMGYNID 169
++ P++ I N ++K+R ++ + +DL +LK + D
Sbjct: 144 NSENPIIGIYNSLARDSLTKEKERLEKAEETFKALDLNKLKSCFGFD 190
>At5g65270 GTP-binding protein
Length = 226
Score = 28.5 bits (62), Expect = 3.9
Identities = 14/35 (40%), Positives = 21/35 (60%)
Query: 224 EEAKNFRDKNIFITLIPNKTEVQKPQETPTKAATD 258
EE + DKNI I LI NK+++ + PT+ A +
Sbjct: 113 EELRAHADKNIVIILIGNKSDLVDQRAIPTEDAKE 147
>At3g12160 hypothetical protein
Length = 220
Score = 28.5 bits (62), Expect = 3.9
Identities = 14/35 (40%), Positives = 20/35 (57%)
Query: 224 EEAKNFRDKNIFITLIPNKTEVQKPQETPTKAATD 258
EE + DKNI I LI NK ++ + PT+ A +
Sbjct: 109 EELRGHADKNIVIMLIGNKCDLGSLRAVPTEDAQE 143
>At2g24440 unknown protein
Length = 183
Score = 28.1 bits (61), Expect = 5.1
Identities = 16/71 (22%), Positives = 36/71 (50%)
Query: 140 ELQKKKRDQQKKSAASRMDLKELKMGYNIDQHDYSVRLKAARKFLSDGDKVKVIVNLKGR 199
E +K+K + K A++ +K+ ++ I++ + A ++ D DK K+++ +
Sbjct: 48 EKKKRKTTKAKNVGAAKKKVKKEEVAVKIEKEEEEDDDAAEKEEDDDSDKKKIVIEHCKQ 107
Query: 200 EKEFRNNAIEL 210
K F+ A E+
Sbjct: 108 CKSFKERANEV 118
>At5g16780 putative protein
Length = 820
Score = 27.7 bits (60), Expect = 6.6
Identities = 24/90 (26%), Positives = 38/90 (41%), Gaps = 2/90 (2%)
Query: 147 DQQKKSAASRMDLKELKMGYNIDQHDYSVRLKAARKFLSDGDKVKVIVNLKGREKEFRNN 206
D +A D KE+ NI + L A K L D +K V GR + + +
Sbjct: 597 DTDMDAAEDSSDTKEITPDENIHEVAVGKGLSGALKLLKDRGTLKEKVEWGGRNMDKKKS 656
Query: 207 AIELIRRFQNDVGKLATEEAKNFRDKNIFI 236
+L+ +D GK + ++ R K+I I
Sbjct: 657 --KLVGIVDDDGGKESKDKESKDRFKDIRI 684
>At4g03100 rac GTPase activating protein like
Length = 430
Score = 27.7 bits (60), Expect = 6.6
Identities = 18/64 (28%), Positives = 32/64 (49%), Gaps = 6/64 (9%)
Query: 185 SDGDKVKVIVNLKGREKEFRNNAIELIRRFQNDVGKLATEEAKNFRDKNIFITLIPNKTE 244
++ + V++I LK E N A++L+ DV + EE+ +NI + PN T+
Sbjct: 223 TEDESVELIKQLKPTESALLNWAVDLMA----DV--VEEEESNKMNARNIAMVFAPNMTQ 276
Query: 245 VQKP 248
+ P
Sbjct: 277 MTDP 280
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.350
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,659,725
Number of Sequences: 26719
Number of extensions: 235209
Number of successful extensions: 665
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 10
Number of HSP's that attempted gapping in prelim test: 650
Number of HSP's gapped (non-prelim): 22
length of query: 266
length of database: 11,318,596
effective HSP length: 98
effective length of query: 168
effective length of database: 8,700,134
effective search space: 1461622512
effective search space used: 1461622512
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140546.3


