

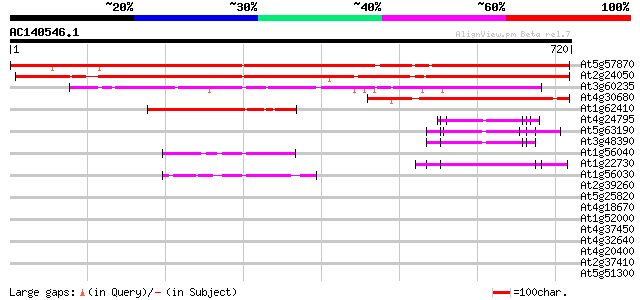
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140546.1 - phase: 0
(720 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g57870 eukaryotic initiation factor 4, eIF4-like protein 909 0.0
At2g24050 putative eukaryotic initiation factor 4, eIF4 808 0.0
At3g60235 initiation factor eIF-4 gamma like 240 2e-63
At4g30680 translation initiation factor-like protein 231 8e-61
At1g62410 hypothetical protein 181 1e-45
At4g24795 unknown protein 64 4e-10
At5g63190 topoisomerase-like protein 62 1e-09
At3g48390 putative protein 60 6e-09
At1g56040 hypothetical protein 55 1e-07
At1g22730 putative topoisomerase 54 3e-07
At1g56030 hypothetical protein 45 1e-04
At2g39260 unknown protein 42 0.002
At5g25820 putative protein 34 0.25
At4g18670 extensin-like protein 34 0.25
At1g52000 34 0.25
At4g37450 arabinogalactan protein AGP18 33 0.43
At4g32640 putative protein 33 0.43
At4g20400 unknown protein 33 0.43
At2g37410 translocase like protein 33 0.43
At5g51300 unknown protein 33 0.73
>At5g57870 eukaryotic initiation factor 4, eIF4-like protein
Length = 780
Score = 909 bits (2350), Expect = 0.0
Identities = 467/733 (63%), Positives = 578/733 (78%), Gaps = 28/733 (3%)
Query: 2 GDSQFESRERVRYTKEELLHIRETLEETPEDILKLRHDIDAELFGEDQSWGR----VENN 57
GDS+F+ RERV+YT+E+LL ++ET + + E IL+++ + AELFGE+ +W R V N
Sbjct: 60 GDSRFDGRERVKYTREQLLELKETTQLSDE-ILRVQRETAAELFGEEGTWARGESVVSNL 118
Query: 58 PPTQIQNRYSEPDNRDWRGRSAQPPANADERSWDNIKENRE--FGNTSQVNRQDQPRT-- 113
P Q +R+SEPD+RDWR RS QPP + +ERSWDN++E ++ + SQ NRQDQP +
Sbjct: 119 VPVQSASRFSEPDSRDWRSRSTQPPPSGEERSWDNLREAKDSRYVEASQYNRQDQPNSQF 178
Query: 114 -------NQGGGPAPTLVKAEVPWSARRGTLSDKDRVLKTVKGILNKLTPEKFDLLKGQL 166
NQGGGPAP LVKAEVPWSARRG LS+ DRVLKTVKGILNKLTPEK+DLLKGQL
Sbjct: 179 SRANISSNQGGGPAPVLVKAEVPWSARRGNLSENDRVLKTVKGILNKLTPEKYDLLKGQL 238
Query: 167 IDSGITSADILKGVISLIFDKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKEITFKR 226
I+SGITSADILKGVI+LIFDKAVLEPTFCPMYA+LCSD+N++LP+FP E G KEITFKR
Sbjct: 239 IESGITSADILKGVITLIFDKAVLEPTFCPMYAKLCSDINDQLPTFPPAEPGDKEITFKR 298
Query: 227 LLLDNCQEAFEGAGKLREELAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVP 286
+LL+ CQEAFEGA +LREEL QM++P+QE ER DK++L+K++TLGNIRLIGELLKQKMVP
Sbjct: 299 VLLNICQEAFEGASQLREELRQMSAPDQEAERNDKEKLLKLKTLGNIRLIGELLKQKMVP 358
Query: 287 ERIVHHIVQELLGAADSNVCPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKEL 346
E+IVHHIVQELLG AD VCPAEENVEAICHFF TIGKQLD + KS+RIND+YF RL+ L
Sbjct: 359 EKIVHHIVQELLG-ADEKVCPAEENVEAICHFFKTIGKQLDGNVKSKRINDVYFKRLQAL 417
Query: 347 STNPQLAPRMKFMVRDVIDLRASNWVPRREEIKAKTISEIHDEAEKNLGLRPGATAGMRN 406
S NPQL R++FMV+++ID+R++ WVPRREE+KA+TI+EIH EAEKNLGLRPGATA MR
Sbjct: 418 SKNPQLELRLRFMVQNIIDMRSNGWVPRREEMKARTITEIHTEAEKNLGLRPGATANMRR 477
Query: 407 TRVTGVQGNTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDMS 466
V+ + +P RPG GGLMPGMPG R+MPGMPG+DNDNWEVPRTRSM R D
Sbjct: 478 GMVSSGGPVSPGPVYPGGRPGAGGLMPGMPGTRRMPGMPGVDNDNWEVPRTRSMSRRDGP 537
Query: 467 GAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPA 526
G SP +SK + +N++LLPQGSSG++SGK SAL+ G G+ S + VS PA
Sbjct: 538 GPL-----HSPAVSKSASMNTRLLPQGSSGIMSGKTSALLQGSGSVSRPVT--VSAERPA 590
Query: 527 PQIPSPVKPVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAP 586
+ PV EKPQ KL+ + L RKT SLLEEYFNVRLL EALQCVEEL P
Sbjct: 591 QSVAPLTVPVPV---EKPQPSGPKLSEEVLQRKTKSLLEEYFNVRLLGEALQCVEELGLP 647
Query: 587 TYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGID 646
+YHPE VKEAISL L+KSPP VEP+A L+EYL +KK++ +D+ TG LL+ ++LDDIGID
Sbjct: 648 SYHPEFVKEAISLSLEKSPPVVEPIATLLEYLLSKKVVAPKDLETGFLLYGAMLDDIGID 707
Query: 647 LPKAPNNFGEIIGKLVLSAGLDFKVVKEILKKVGDDYFQKAIFNSAVQVI-SSASGQAVL 705
LPKAPNNFGEI+GKL+L+ G+DFK+V+EI+ K+ DD FQK + ++AV+++ SS G+++L
Sbjct: 708 LPKAPNNFGEIVGKLILAGGVDFKLVREIIGKMEDDRFQKMVVDAAVRIVESSEQGKSLL 767
Query: 706 DSQASDIEACQAL 718
SQA+DIEAC+ L
Sbjct: 768 ASQAADIEACRNL 780
>At2g24050 putative eukaryotic initiation factor 4, eIF4
Length = 747
Score = 808 bits (2088), Expect = 0.0
Identities = 421/722 (58%), Positives = 542/722 (74%), Gaps = 39/722 (5%)
Query: 8 SRERVRYTKEELLHIRETLEETPEDILKLRHDIDAELFGEDQSWGR--VENNPPTQIQNR 65
S ERVR+++EE+L RE+++ + E IL+ +I ELFGE+QSWG E+ Q+R
Sbjct: 53 SGERVRFSREEILQHRESVQVSDE-ILRRCKEIAVELFGEEQSWGNHAAESKITNHTQHR 111
Query: 66 YSEPDNRDWRGRSAQPPANADERSWDNIKENREFGNTSQVNRQDQPRTNQGGGPAPTLVK 125
++E DNRDW RS Q P + E D+ +E + T QG GP P L+K
Sbjct: 112 HTETDNRDWHSRS-QIPTSGKEWLRDDPREAKS--------------TWQGSGPTPVLIK 156
Query: 126 AEVPWSARRGTLSDKDRVLKTVKGILNKLTPEKFDLLKGQLIDSGITSADILKGVISLIF 185
AEVPWSA+RG LSDKDRV+K+VKGILNKLTPEK++LLKGQLID+GITSADILK VI LIF
Sbjct: 157 AEVPWSAKRGALSDKDRVVKSVKGILNKLTPEKYELLKGQLIDAGITSADILKEVIQLIF 216
Query: 186 DKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKEITFKRLLLDNCQEAFEGAGKLREE 245
+ A+L+PTFC MYA LC D+N +LPSFPSEE GGKEITFKR+LL+NCQEAFEGAGKL+EE
Sbjct: 217 ENAILQPTFCEMYALLCFDINGQLPSFPSEEPGGKEITFKRVLLNNCQEAFEGAGKLKEE 276
Query: 246 LAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLGAADSNV 305
+ QMT+P+QE ER DK+++ K+RTLGNIRLIGELLKQKMVPE+IVHHIVQELLG D+
Sbjct: 277 IRQMTNPDQEMERMDKEKMAKLRTLGNIRLIGELLKQKMVPEKIVHHIVQELLG-DDTKA 335
Query: 306 CPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKFMVRDVID 365
CPAE +VEA+C FF TIGKQLD+SP+SR IND YFGRLKEL+ +PQL R++FMV++V+D
Sbjct: 336 CPAEGDVEALCQFFITIGKQLDDSPRSRGINDTYFGRLKELARHPQLELRLRFMVQNVVD 395
Query: 366 LRASNWVPRREEIKAKTISEIHDEAEKNLGLRPGATAGMRNTR-----VTGVQGNTGAGG 420
LRA+ WVPRREE+KAK I+EIH EAE+NLG+RPGA A MRN V+G G G
Sbjct: 396 LRANKWVPRREEVKAKKINEIHSEAERNLGMRPGAMASMRNNNNNRAAVSGAADGMGLGN 455
Query: 421 FPIARPGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDMSGAQTGGRGQSPYLS 480
+ RPGTGG+MPGMPG R MP +D D WE+ RTRSMPRG+ Q P ++
Sbjct: 456 I-LGRPGTGGMMPGMPGTRVMP----MDEDGWEMARTRSMPRGNRQTVQQPRFQPPPAIN 510
Query: 481 KPSVINSKLLPQGSSGLIS--GKNSALVLGGGTPSALPSNIVSGTEPAPQIPSPVKPVSA 538
K +NS+LLPQGS GL++ G+ S L+ G G+ SA ++P P + P +P S
Sbjct: 511 KSLSVNSRLLPQGSGGLLNGGGRPSPLLQGNGSSSA-----PQASKPIPTVEKP-QPRSQ 564
Query: 539 ASPEKPQAP-AVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAI 597
P+ AP A LN +L RKT SLLEEYF++RL+DEALQCVEELK+P+YHPE+VKE I
Sbjct: 565 PQPQPQAAPLANSLNAGELERKTKSLLEEYFSIRLVDEALQCVEELKSPSYHPELVKETI 624
Query: 598 SLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEI 657
SLGL+K+PP VEP+A L+++L +K +LT++D+G GCLL+ S+LDDIGIDLPKAPN+FGE
Sbjct: 625 SLGLEKNPPLVEPIAKLLKHLISKNVLTSKDLGAGCLLYGSMLDDIGIDLPKAPNSFGEF 684
Query: 658 IGKLVLSAGLDFKVVKEILKKVGDDYFQKAIFNSAVQVISSA-SGQAVLDSQASDIEACQ 716
+G+LV + LDF++V+++LKK+ D++F+K + N+ ++ + SGQ+VLDSQA ++EACQ
Sbjct: 685 LGELVSAKVLDFELVRDVLKKMEDEWFRKTVLNAVIKSVRECPSGQSVLDSQAVEVEACQ 744
Query: 717 AL 718
+L
Sbjct: 745 SL 746
>At3g60235 initiation factor eIF-4 gamma like
Length = 1544
Score = 240 bits (613), Expect = 2e-63
Identities = 198/658 (30%), Positives = 312/658 (47%), Gaps = 74/658 (11%)
Query: 77 RSAQP--PANADERSWDNIKENREFGNTSQVNRQDQPRTNQGGGPAPTLVKAEVPWSARR 134
R QP P R+ +++ + N Q P T P + KAE +
Sbjct: 850 RPMQPVGPMGGMGRNTPDLERWQRGSNFQQKGLFPSPHT-----PMQVMHKAERKYQV-- 902
Query: 135 GTLSDKDRVL-KTVKGILNKLTPEKFDLLKGQLIDSGITSADILKGVISLIFDKAVLEPT 193
GT++D+++ + +K ILNKLTP+ F+ L Q+ I +A L GVIS IFDKA++EPT
Sbjct: 903 GTIADEEQAKQRQLKSILNKLTPQNFEKLFEQVKSVNIDNAVTLSGVISQIFDKALMEPT 962
Query: 194 FCPMYAQLCSDLNEKLPSFPSEESGGKEITFKRLLLDNCQEAFEGAGKLREELAQMTSPE 253
FC MYA C L+ LP F G++ITFKRLLL+ CQE FE K EE +++
Sbjct: 963 FCEMYADFCFHLSGALPDF---NENGEKITFKRLLLNKCQEEFERGEKEEEEASRVAEEG 1019
Query: 254 Q----ETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLGAADSNVCPAE 309
Q E ER +K V+ R LGNIRLIGEL K++M+ E+I+H +Q+LLG N P E
Sbjct: 1020 QVEQTEEEREEKRLQVRRRMLGNIRLIGELYKKRMLTEKIMHACIQKLLG---YNQDPHE 1076
Query: 310 ENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKFMVRDVIDLRAS 369
EN+EA+C +TIG +D + K++ D YF ++K LS +L+ R++FM+ + IDLR +
Sbjct: 1077 ENIEALCKLMSTIGVMIDHN-KAKFQMDGYFEKMKMLSCKQELSSRVRFMLINAIDLRKN 1135
Query: 370 NWVPRREEIKAKTISEIHDEAEKNLGLRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTG 429
W R + K I E+H +A + T R +R + + G + P G
Sbjct: 1136 KWQERMKVEGPKKIEEVHRDAAQE-----RQTQANRLSRGPSMNSSGRRGHMEFSSPRGG 1190
Query: 430 GLMPGMPGARK-----MPGMPGIDNDNWEV-------PRTRSMPRGDMS------GAQTG 471
G M P A+ P G N + PR MP+ + G Q G
Sbjct: 1191 GGMLSPPAAQMGSYHGPPQGRGFSNQDIRFDDRPSYEPRMVPMPQRSVCEEPITLGPQ-G 1249
Query: 472 GRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGG--GTPSALPSNIVSGTEPAPQ- 528
G GQ + +P+V ++ Q + G +S GG G S P++ V+ +PQ
Sbjct: 1250 GLGQGMSIRRPAVASNTY--QSDATQAGGGDSRRPAGGLNGFGSHRPASPVTHGRSSPQE 1307
Query: 529 --------------IPSPVKPVSAASPEKPQAPAVKLNI--------DDLHRKTVSLLEE 566
S + P +++ + Q P+ +N + L ++S ++E
Sbjct: 1308 RGTAYVHREFASLSRASDLSPEVSSARQVLQGPSATVNSPRENALSEEQLENLSLSAIKE 1367
Query: 567 YFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEYLF--TKKIL 624
Y++ R +E C++++ +P YHP ++ ++ ++ + +A L+ L L
Sbjct: 1368 YYSARDENEIGMCMKDMNSPAYHPTMISLWVTDSFERKDKERDLLAKLLVNLVKSADNAL 1427
Query: 625 TARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLSAGLDFKVVKEILKKVGDD 682
+ G L+D D PKA G I GK V + + ++++ G++
Sbjct: 1428 NEVQLVKGFESVLKTLEDAVNDAPKAAEFLGRIFGKSVTEKVVTLTEIGRLIQEGGEE 1485
>At4g30680 translation initiation factor-like protein
Length = 263
Score = 231 bits (590), Expect = 8e-61
Identities = 129/273 (47%), Positives = 179/273 (65%), Gaps = 25/273 (9%)
Query: 460 MPRGD--MSGAQTGGRGQSPYLSKPSVINSK-----------LLPQGSSGLISGKNSALV 506
M +GD +S GGRG S + ++S +GS GL++ + SALV
Sbjct: 1 MQQGDSMLSLRPGGGRGGSRRFAPRFTLSSSSDLTNGGDAPSFAVKGSGGLLNDRPSALV 60
Query: 507 LGGGTPSALPSNIVSGTEPAPQIPSPVKPVSAASPEKPQAPAV-KLNIDDLHRKTVSLLE 565
G G+ P P+P + KP P++ P LN +L RKT SLLE
Sbjct: 61 QGNGSQQPKPV-------PSPTRQTVEKPKPQPQPQEVAPPTTTSLNTVELSRKTNSLLE 113
Query: 566 EYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILT 625
EYFNVRLLDEALQC+EELK P+YHPE+VKEAISLGL+K+PP VEPVA L+E+L +K +LT
Sbjct: 114 EYFNVRLLDEALQCIEELKTPSYHPELVKEAISLGLEKNPPCVEPVAKLLEHLVSKNVLT 173
Query: 626 ARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLSAGLDFKVVKEILKKVGDDYFQ 685
+D+ GCLL+ S+LDDIGIDLPKAPNNFGEI+G LV++ D ++VKEIL K+GD++F+
Sbjct: 174 PKDLRNGCLLYGSMLDDIGIDLPKAPNNFGEILGSLVMAKASDSELVKEILMKMGDEWFK 233
Query: 686 KAIFNSAVQVISSASGQAVLDSQASDIEACQAL 718
KA+ + ++ +S +++L ++A ++EAC+ L
Sbjct: 234 KAVLEAVMRSVS----ESLLTTEAVEVEACRGL 262
>At1g62410 hypothetical protein
Length = 223
Score = 181 bits (459), Expect = 1e-45
Identities = 95/192 (49%), Positives = 133/192 (68%), Gaps = 3/192 (1%)
Query: 177 LKGVISLIFDKAVLEPTFCPMYAQLCSDLNEKLPSFPSEESGGKEITFKRLLLDNCQEAF 236
L+ + +LIFDKAVLEPTFCPMYAQLC D+ K+P FP EI+FKR+LL+ CQ+ F
Sbjct: 10 LQRLTTLIFDKAVLEPTFCPMYAQLCFDIRHKMPRFPPSAPKTDEISFKRVLLNTCQKVF 69
Query: 237 EGAGKLREELAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQE 296
E L EE+ +M +P+QE ER D+ RL+ +RTLGN+R GEL ++M+ E++V I Q+
Sbjct: 70 ERTDDLSEEIRKMNAPDQEAEREDEVRLLNLRTLGNLRFCGELFLKRMLTEKVVLAIGQK 129
Query: 297 LLGAADSNVCPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRM 356
LL A+ +CP+EE + AIC F NT+GK+LD S S+ +N++ RLK LS +PQL +
Sbjct: 130 LLEDAE-QMCPSEEKIIAICLFLNTVGKKLD-SLNSKLMNEI-LRRLKNLSNHPQLVMSL 186
Query: 357 KFMVRDVIDLRA 368
+ MV +I L +
Sbjct: 187 RLMVGKIIHLHS 198
>At4g24795 unknown protein
Length = 702
Score = 63.5 bits (153), Expect = 4e-10
Identities = 31/106 (29%), Positives = 62/106 (58%), Gaps = 2/106 (1%)
Query: 553 IDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVA 612
++D K +LLEEY + L+ EA +C+ EL P ++ EVVK+A+ +G++K ++ +
Sbjct: 575 VEDAKDKISNLLEEYESSGLVSEACKCIHELGMPFFNHEVVKKALVMGMEKKKDKM--ML 632
Query: 613 NLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEII 658
+L++ F++ ++T + G L+D+ +D+P A F + +
Sbjct: 633 DLLQESFSEGLITTNQMTKGFTRVKDGLEDLALDIPNAKEKFNDYV 678
Score = 60.8 bits (146), Expect = 3e-09
Identities = 34/119 (28%), Positives = 57/119 (47%), Gaps = 1/119 (0%)
Query: 550 KLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVE 609
+ ++++ +K +L EY EA +CV EL +H EVVK A+ L+
Sbjct: 275 RTTVEEVKKKIADILNEYVETGETYEACRCVRELGVSFFHHEVVKRALVTALENHAAEA- 333
Query: 610 PVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLSAGLD 668
PV L+ ++ ++++ + G LDD+ +D+P A FG I+ K V LD
Sbjct: 334 PVLKLLNEAASENLISSSQMVKGFSRLRESLDDLALDIPSARTKFGLIVPKAVSGGWLD 392
Score = 53.5 bits (127), Expect = 4e-07
Identities = 32/119 (26%), Positives = 61/119 (50%), Gaps = 1/119 (0%)
Query: 561 VSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFT 620
V+++ EYFN + E ++ +E+L AP Y+P +K+ I+L LD+ E + L+ L
Sbjct: 420 VTIIHEYFNSDDIPELIRSLEDLGAPEYNPIFLKKLITLALDRKNHEKEMASVLLSSLHI 479
Query: 621 KKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLSAGLDFKVVKEILKKV 679
++ T D+ G ++ +D +D+ A N + + V+ L ++EI K+
Sbjct: 480 -EMFTTEDVADGFVMLLESAEDTALDILDASNELALFLARAVIDDVLAPFNLEEISSKL 537
Score = 53.1 bits (126), Expect = 5e-07
Identities = 29/111 (26%), Positives = 53/111 (47%), Gaps = 1/111 (0%)
Query: 553 IDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVA 612
+DD + S++ EYF+ +D A + EL + YHP +K +S+ +D+ E +
Sbjct: 114 LDDYKKAAASIINEYFSTGDVDVAAADLIELGSSEYHPYFIKRLVSVAMDRHDKEKEMAS 173
Query: 613 NLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVL 663
L+ L+ ++ I G +L DD +D+P A N + + V+
Sbjct: 174 VLLSALYA-DVINPNQIRDGFVLLLESADDFVVDIPDAVNVLALFLARAVV 223
>At5g63190 topoisomerase-like protein
Length = 729
Score = 62.0 bits (149), Expect = 1e-09
Identities = 40/142 (28%), Positives = 67/142 (47%), Gaps = 3/142 (2%)
Query: 535 PVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVK 594
P A EK + +++ +K +L+EY EA +C+ EL +H EVVK
Sbjct: 293 PHHAELVEKKWGGSTHTTVEETKKKISEILKEYVENGDTYEACRCIRELGVSFFHHEVVK 352
Query: 595 EAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNF 654
A+ L +D SP V L++ + ++++ + G A LDD+ +D+P A F
Sbjct: 353 RALVLAMD-SPTAESLVLKLLKETAEEGLISSSQMVKGFFRVAESLDDLALDIPSAKKLF 411
Query: 655 GEIIGKLVLSAGLD--FKVVKE 674
I+ K + LD FK+ +
Sbjct: 412 DSIVPKAISGGWLDDSFKITSD 433
Score = 60.1 bits (144), Expect = 4e-09
Identities = 33/102 (32%), Positives = 54/102 (52%), Gaps = 3/102 (2%)
Query: 553 IDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVA 612
++D K LLEEY + EA QC+ +L P ++ EVVK+A+ + ++K R +
Sbjct: 608 VEDAKDKISKLLEEYETGGVTSEACQCIRDLGMPFFNHEVVKKALVMAMEKQNDR---LL 664
Query: 613 NLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNF 654
NL+E F + ++T + G LDD+ +D+P A F
Sbjct: 665 NLLEECFGEGLITTNQMTKGFGRVNDSLDDLSLDIPNAKEKF 706
Score = 49.3 bits (116), Expect = 8e-06
Identities = 35/151 (23%), Positives = 75/151 (49%), Gaps = 2/151 (1%)
Query: 558 RKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEY 617
+ TV++++EYF + E ++ +++L AP Y+P +K I+L LD+ R + +A+++
Sbjct: 450 KDTVNIIQEYFLSDDIPELIRSLQDLGAPEYNPVFLKRLITLALDRK-NREKEMASVLLS 508
Query: 618 LFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLSAGLDFKVVKEILK 677
++ + D G ++ +D +D+ A N + + V+ L +++I
Sbjct: 509 ALHMELFSTEDFINGFIMLLESAEDTALDIMDASNELALFLARAVIDDVLAPLNLEDIST 568
Query: 678 KVGDDYFQKAIFNSAVQVISSA-SGQAVLDS 707
K+ SA +IS+ +G+ +L S
Sbjct: 569 KLPPKSTGTETVRSARSLISARHAGERLLRS 599
Score = 48.9 bits (115), Expect = 1e-05
Identities = 28/111 (25%), Positives = 54/111 (48%), Gaps = 1/111 (0%)
Query: 553 IDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVA 612
++D + VS+++EYF+ + A + EL + YHP K +S+ +D+ E +
Sbjct: 147 LNDYKKSVVSIIDEYFSTGDVKVAASDLRELGSSEYHPYFTKRLVSMAMDRHDKEKEMAS 206
Query: 613 NLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVL 663
L+ L+ IL + I G + +DD+ +D+ A N I + ++
Sbjct: 207 VLLSALYADVILPDQ-IRDGFIRLLRSVDDLAVDILDAVNVLALFIARAIV 256
>At3g48390 putative protein
Length = 633
Score = 59.7 bits (143), Expect = 6e-09
Identities = 32/106 (30%), Positives = 56/106 (52%), Gaps = 3/106 (2%)
Query: 553 IDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVA 612
++D K LLEEY ++ EA +C+ +L P ++ EVVK+A+ + ++K R +
Sbjct: 512 VEDAKDKIWKLLEEYEVGGVISEACRCIRDLGMPFFNHEVVKKALVMAMEKKNDR---ML 568
Query: 613 NLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEII 658
NL++ F + I+T + G LDD+ +D+P A F +
Sbjct: 569 NLLQECFAEGIITTNQMTKGFGRVKDSLDDLSLDIPNAEEKFNSYV 614
Score = 54.7 bits (130), Expect = 2e-07
Identities = 36/141 (25%), Positives = 63/141 (44%), Gaps = 3/141 (2%)
Query: 535 PVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVK 594
P A E + + +++ RK L EY EA +C+ EL +H E+VK
Sbjct: 200 PHHAELVETKWGGSTHITVEETKRKISEFLNEYVENGDTREACRCIRELGVSFFHHEIVK 259
Query: 595 EAISLGLDKSPPRVEP-VANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNN 653
+ L ++ EP + L++ + ++++ + G A LDD+ +D+P A
Sbjct: 260 SGLVLVMESRTS--EPLILKLLKEATEEGLISSSQMAKGFSRVADSLDDLSLDIPSAKTL 317
Query: 654 FGEIIGKLVLSAGLDFKVVKE 674
F I+ K ++ LD KE
Sbjct: 318 FESIVPKAIIGGWLDEDSFKE 338
Score = 50.8 bits (120), Expect = 3e-06
Identities = 27/111 (24%), Positives = 56/111 (50%), Gaps = 1/111 (0%)
Query: 553 IDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVA 612
++D R+ VS+++EYF+ ++ A + +L YHP VK +S+ +D+ E +
Sbjct: 54 LEDYKREVVSIIDEYFSSGDVEVAASDLMDLGLSEYHPYFVKRLVSMAMDRGNKEKEKAS 113
Query: 613 NLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVL 663
L+ L+ +++ I G + + D+ +D+P A N I + ++
Sbjct: 114 VLLSRLYA-LVVSPDQIRVGFIRLLESVGDLALDIPDAVNVLALFIARAIV 163
Score = 41.2 bits (95), Expect = 0.002
Identities = 32/157 (20%), Positives = 72/157 (45%), Gaps = 2/157 (1%)
Query: 552 NIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPV 611
N+ + ++++EYF + E ++ +E+L P Y+P +K+ I+L +D+ E +
Sbjct: 348 NLRRFKKDAETIIQEYFLSDDIPELIRSLEDLGLPEYNPVFLKKLITLAMDRKNKEKE-M 406
Query: 612 ANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLSAGLDFKV 671
A++ ++ + D G ++ +D +D+ A + + + V+ L
Sbjct: 407 ASVFLASLHMEMFSTEDFINGFIMLLESAEDTALDILAASDELALFLARAVIDDVLAPLN 466
Query: 672 VKEILKKVGDDYFQKAIFNSAVQVISSA-SGQAVLDS 707
++EI + SA +IS+ +G+ +L S
Sbjct: 467 LEEISNSLPPKSTGSETIRSARSLISARHAGERLLRS 503
>At1g56040 hypothetical protein
Length = 437
Score = 55.5 bits (132), Expect = 1e-07
Identities = 44/172 (25%), Positives = 77/172 (44%), Gaps = 15/172 (8%)
Query: 197 MYAQLCSDLNEKLPSFPSEESGGKEITFKRLLLDNCQEAFEGAGKLREELAQMTSPEQET 256
++A+LC +L +LP PS+ES G TF+RL L C+ E A E L P
Sbjct: 8 LFARLCVELFHELPPLPSDESHGDITTFERLFLRKCRIELENASPKTEHL-----PLVYV 62
Query: 257 ERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLGAADSNVCPAEENVEAIC 316
+ + R +K +R++ E+ K + E ++Q L+ + + P E +A+
Sbjct: 63 DETNYYRFIK-----TVRVLAEVYKNSKITETTRKSMIQVLM----NPILPPERITDAMN 113
Query: 317 HFFNTIGKQLDESPKSRRINDMY-FGRLKELSTNPQLAPRMKFMVRDVIDLR 367
F + IGK D + N + R+ EL N +++ D + ++
Sbjct: 114 LFRSIIGKLADFHFSDEKFNQLVRSSRVVELEGNYNEEVKLRKEAEDALAMK 165
>At1g22730 putative topoisomerase
Length = 693
Score = 53.9 bits (128), Expect = 3e-07
Identities = 37/181 (20%), Positives = 74/181 (40%), Gaps = 1/181 (0%)
Query: 535 PVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVK 594
P+ A EK +D+ + LL+EY EA +C++ LK P +H E+VK
Sbjct: 234 PLHAEVVEKRWGGTDNWTAEDVKARINDLLKEYVMSGDKKEAFRCIKGLKVPFFHHEIVK 293
Query: 595 EAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNF 654
A+ + +++ +V + +L++ ++ + + G ++D+ +D+P A
Sbjct: 294 RALIMAMERRKAQVR-LLDLLKETIEVGLINSTQVTKGFSRIIDSIEDLSLDIPDARRIL 352
Query: 655 GEIIGKLVLSAGLDFKVVKEILKKVGDDYFQKAIFNSAVQVISSASGQAVLDSQASDIEA 714
I K L +K + G+ + + N S + L S++
Sbjct: 353 QSFISKAASEGWLCASSLKSLSADAGEKLLENSSANVFKDKAKSIIREYFLSGDTSEVVH 412
Query: 715 C 715
C
Sbjct: 413 C 413
Score = 52.0 bits (123), Expect = 1e-06
Identities = 34/122 (27%), Positives = 59/122 (47%), Gaps = 1/122 (0%)
Query: 553 IDDLHRKTVSLLEEYFNVRLLDEALQCVEELKAPTYHPEVVKEAISLGLDKSPPRVEPVA 612
+ ++ K LLEEY + L EA +CV+EL P +H EVVK+++ + + + E +
Sbjct: 558 VKEVKEKIQILLEEYVSGGDLREASRCVKELGMPFFHHEVVKKSV-VRIIEEKENEERLW 616
Query: 613 NLIEYLFTKKILTARDIGTGCLLFASLLDDIGIDLPKAPNNFGEIIGKLVLSAGLDFKVV 672
L++ F ++T + G L+D+ +D+P A F + + L LD
Sbjct: 617 KLLKVCFDSGLVTIYQMTKGFKRVDESLEDLSLDVPDAAKKFSSCVERGKLEGFLDESFA 676
Query: 673 KE 674
E
Sbjct: 677 SE 678
Score = 44.3 bits (103), Expect = 2e-04
Identities = 29/161 (18%), Positives = 67/161 (41%), Gaps = 1/161 (0%)
Query: 522 GTEPAPQIPSPVKPVSAASPEKPQAPAVKLNIDDLHRKTVSLLEEYFNVRLLDEALQCVE 581
G E + P+ + ++ + +K ++EEYF + + ++
Sbjct: 57 GVEDDDDLTDPIFDTIEGNGHSDPTSCFDADLSEYKKKATVIVEEYFGTNDVVSVVNELK 116
Query: 582 ELKAPTYHPEVVKEAISLGLDKSPPRVEPVANLIEYLFTKKILTARDIGTGCLLFASLLD 641
EL Y VK+ +S+ +D+ E A L+ L+ ++ ++ G + D
Sbjct: 117 ELGMAEYRYYFVKKLVSMAMDRHDKEKEMAAFLLSTLYA-DVIDPPEVYRGFNKLVASAD 175
Query: 642 DIGIDLPKAPNNFGEIIGKLVLSAGLDFKVVKEILKKVGDD 682
D+ +D+P A + + + ++ L +K+ +K + D+
Sbjct: 176 DLSVDIPDAVDVLAVFVARAIVDDILPPAFLKKQMKLLPDN 216
>At1g56030 hypothetical protein
Length = 371
Score = 45.4 bits (106), Expect = 1e-04
Identities = 46/198 (23%), Positives = 81/198 (40%), Gaps = 30/198 (15%)
Query: 197 MYAQLCSDLNEKLPSFPSEESGGKEITFKRLLLDNCQEAFEGAGKLREELAQMTSPEQET 256
++A+LC +L P PS+ES G+ T +R L NCQ E L+ EL + E +
Sbjct: 8 LFARLCVEL----PPLPSDESHGEITTVERQFLRNCQMELENL-SLKTELPLVYVDESKY 62
Query: 257 ERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLGAADSNVCPAEENVEAIC 316
R K +R++ E+ I+Q L+ + + P+E + +A+
Sbjct: 63 WRFIK----------TVRVLAEVFNNMKTTRTTRKSIIQVLM----NPILPSERSTDAMN 108
Query: 317 HFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKFMVRDVIDLRASNWVPRRE 376
F +TI K D N ++ +L + +++ +R E
Sbjct: 109 LFLSTIEKLADLQFSDEDFNQLFVSSRLDLQLENKYNDKVEVKLR-----------KEAE 157
Query: 377 EIKAKTISEIHDEAEKNL 394
+ A+ I E+ D E+ L
Sbjct: 158 DALARKIKEVVDLTERLL 175
>At2g39260 unknown protein
Length = 1029
Score = 41.6 bits (96), Expect = 0.002
Identities = 39/149 (26%), Positives = 57/149 (38%), Gaps = 9/149 (6%)
Query: 246 LAQMTSPEQETERRDKDRLVKIRTLGNIRLIGELLKQKMVPERIVHHIVQELLGAADSNV 305
L QM E + KD++ + NIR IGEL K K+VP +V ++ L
Sbjct: 381 LVQMLEDEFNSLVHKKDQMNIETKIRNIRFIGELCKFKIVPAGLVFSCLKACLDEF---- 436
Query: 306 CPAEENVEAICHFFNTIGKQLDESPKSRRINDMYFGRLKELSTNPQLAPRMKFMVRDVID 365
N++ C+ T G+ L SP++ L L L PR +V +
Sbjct: 437 --THHNIDVACNLLETCGRFLYRSPETTLRMTNMLDILMRLKNVKNLDPRQSTLVENAYY 494
Query: 366 LRASNWVPRREEIKAKTISEIHDEAEKNL 394
L P R +K +H K L
Sbjct: 495 LCKP---PERSARISKVRPPLHQYVRKLL 520
>At5g25820 putative protein
Length = 654
Score = 34.3 bits (77), Expect = 0.25
Identities = 34/159 (21%), Positives = 57/159 (35%), Gaps = 16/159 (10%)
Query: 396 LRPGATAGMRNTRVTGVQGNTGAGGFPIARPGTGGLMPGMPGAR----------KMPGM- 444
+ P V + ++G P P T L P +P + K PG+
Sbjct: 65 IAPSPAGDEEEVEVDQIYDSSGNATAPAISPTTATLPPLLPILKENATAPTANAKAPGLN 124
Query: 445 PGIDNDNWEVPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSA 504
P + D+ P + P + G ++ PSV + LP + + +A
Sbjct: 125 PSLVKDHATAPSPSANPPAALPGLNPSLVKENATAPAPSVKSPVALPILNPSTVKENATA 184
Query: 505 LVLGGGTPSALPSNIVSGTEPAPQIPSPVKPVSAASPEK 543
V P ALPS P+P + + P ++ PE+
Sbjct: 185 PVASAKAPVALPS-----INPSPVMKNETLPTTSKVPER 218
>At4g18670 extensin-like protein
Length = 839
Score = 34.3 bits (77), Expect = 0.25
Identities = 39/145 (26%), Positives = 50/145 (33%), Gaps = 14/145 (9%)
Query: 416 TGAGGFPIARPGT---GGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDMSGAQTGG 472
T + G P P T GG P P G P + P P + GG
Sbjct: 462 TPSPGSPPTSPTTPTPGGSPPSSPTTPTPGGSPP---SSPTTPTPGGSPPSSPTTPSPGG 518
Query: 473 RGQSPYLSK------PSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPA 526
SP +S PS ++ P S S+ + TPS P+ I G
Sbjct: 519 SPPSPSISPSPPITVPSPPSTPTSPGSPPSPSSPTPSSPIPSPPTPSTPPTPISPGQNSP 578
Query: 527 PQIPSP--VKPVSAASPEKPQAPAV 549
P IPSP P +SP P P +
Sbjct: 579 PIIPSPPFTGPSPPSSPSPPLPPVI 603
>At1g52000
Length = 730
Score = 34.3 bits (77), Expect = 0.25
Identities = 36/128 (28%), Positives = 48/128 (37%), Gaps = 11/128 (8%)
Query: 395 GLRPGATAGMRNTRVTGVQG-NTGAGGFPIARPGTGGLMPGMPGARKMPGMPGIDNDNWE 453
G +PGA+ ++ G G N GA G GG P A PG + N E
Sbjct: 134 GAKPGASGIGSDSGSIGSAGTNPGADGTRETEKNAGGSKPSSGSAGTNPGASAVGNGETE 193
Query: 454 VPRTRSMP----RGDMSGAQTGGRGQSPYL---SKPSVINSKLLPQGSSGLISGKNSALV 506
S P G GA GG G++ SKPS + P ++G G
Sbjct: 194 KNAGGSKPSSGSAGTNPGASAGGNGETEKNVGGSKPSSGKAGTNPGANAG---GNGGTEK 250
Query: 507 LGGGTPSA 514
GG+ S+
Sbjct: 251 NAGGSKSS 258
>At4g37450 arabinogalactan protein AGP18
Length = 209
Score = 33.5 bits (75), Expect = 0.43
Identities = 23/82 (28%), Positives = 34/82 (41%), Gaps = 2/82 (2%)
Query: 468 AQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPAP 527
A T +SP ++ P+ +K +S + S + +P P S PAP
Sbjct: 37 APTTSPTKSPAVTSPTTAPAKTPTASASSPVESPKSPAPVSESSPPPTPVPESSPPVPAP 96
Query: 528 QIPSPVK--PVSAASPEKPQAP 547
+ SPV PV A + P AP
Sbjct: 97 MVSSPVSSPPVPAPVADSPPAP 118
>At4g32640 putative protein
Length = 1069
Score = 33.5 bits (75), Expect = 0.43
Identities = 38/143 (26%), Positives = 51/143 (35%), Gaps = 25/143 (17%)
Query: 411 GVQGNTGAGGFPIARPGTGGLMPGMPGARKMP--GMPGIDNDN-WEVPRTRSMPRGDMSG 467
G G+ AG P +RP G P + MP GM G N ++ + PRG
Sbjct: 153 GPSGSVAAGPPPGSRPMAFGSPPPVGSGMSMPPSGMIGGPVSNGHQMVGSGGFPRGTQFP 212
Query: 468 AQTGGRGQSPYLSKPSVINSKLLPQGSSGLISGKNSALVLGGGTPSALPSNIVSGTEPAP 527
Q+PY+ PS ++ PQ L S+ +SG P
Sbjct: 213 GAAVTTPQAPYVRPPSAPYARTPPQ---------------------PLGSHSLSGNPPLT 251
Query: 528 QIPSPVKPVSAASPEKPQA-PAV 549
+P P A P P PAV
Sbjct: 252 PFTAPSMPPPATFPGAPHGRPAV 274
>At4g20400 unknown protein
Length = 954
Score = 33.5 bits (75), Expect = 0.43
Identities = 31/114 (27%), Positives = 48/114 (41%), Gaps = 21/114 (18%)
Query: 79 AQPPANADERSWDNIKENREFGNTSQVNRQDQPRTNQGGGPAPTLVKAEVPWSARRGTLS 138
A P + E W N K + FG + ++ T +GG P+L
Sbjct: 862 ALDPKHQLEEYW-NQKAVKLFG-AEPIKEGEKDDTEKGGASDPSL--------------- 904
Query: 139 DKDRVLKTVKGILNKLTPEKFDLLKGQLIDSGITSADILKGVISLIFDKAVLEP 192
DR + ++G+L K TPE+ ++ G L G T LK +S + DK + P
Sbjct: 905 --DRDTRLLRGLLKKATPEELVMMHGLL--CGETRNTELKEELSTLVDKMEISP 954
>At2g37410 translocase like protein
Length = 243
Score = 33.5 bits (75), Expect = 0.43
Identities = 24/63 (38%), Positives = 31/63 (49%), Gaps = 6/63 (9%)
Query: 426 PGTGGLMPGMPGARKMPGMPGIDNDNWEVPRTRSMPRGDM-SGAQTGGRGQSPYLSKPSV 484
PG G MPGM G + MPGMPG+ +P + M G M S AQ Q+ + S
Sbjct: 148 PGMQG-MPGMQGMQGMPGMPGMQG----MPGMQGMQMGQMQSQAQIRSESQNQNTASSSS 202
Query: 485 INS 487
+S
Sbjct: 203 SSS 205
>At5g51300 unknown protein
Length = 804
Score = 32.7 bits (73), Expect = 0.73
Identities = 26/94 (27%), Positives = 37/94 (38%), Gaps = 5/94 (5%)
Query: 445 PGIDNDNWEVPRTRSMPRGDMSGAQTGGRGQSPYLSKPSVINS---KLLPQGSSGLISGK 501
PG+ D+ T S+P + T GQ PY+S PS N+ P +S +
Sbjct: 670 PGVQADSGAA--TSSIPPNVYGSSVTAMPGQPPYMSYPSYYNAVPPPTPPAPASSTDHSQ 727
Query: 502 NSALVLGGGTPSALPSNIVSGTEPAPQIPSPVKP 535
N + PS + G AP P+P P
Sbjct: 728 NMGNMPWANNPSVSTPDHSQGLVNAPWAPNPPMP 761
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,143,404
Number of Sequences: 26719
Number of extensions: 828991
Number of successful extensions: 2312
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 2176
Number of HSP's gapped (non-prelim): 113
length of query: 720
length of database: 11,318,596
effective HSP length: 106
effective length of query: 614
effective length of database: 8,486,382
effective search space: 5210638548
effective search space used: 5210638548
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140546.1


