

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140068.2 + phase: 0
(501 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
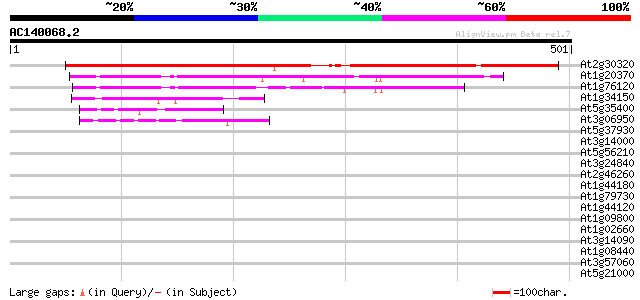
Score E
Sequences producing significant alignments: (bits) Value
At2g30320 putative pseudouridine synthase 490 e-139
At1g20370 putative tRNA pseudouridine synthase 118 7e-27
At1g76120 unknown protein 117 2e-26
At1g34150 pseudouridine synthase-like protein 55 7e-08
At5g35400 unknown protein 48 1e-05
At3g06950 putative tRNA pseudouridine synthase 48 1e-05
At5g37930 unknown protein (At5g37930) 31 1.4
At3g14000 unknown protein 31 1.8
At5g56210 unknown protein 30 2.4
At3g24840 phosphatidylinositol transfer protein like 30 2.4
At2g46260 unknown protein 30 2.4
At1g44180 putative aminoacylase 30 2.4
At1g79730 unknown protein 30 3.1
At1g44120 hypothetical protein 30 3.1
At1g09800 hypothetical protein 30 3.1
At1g02660 unknown protein 30 3.1
At3g14090 unknown protein 30 4.1
At1g08440 hypothetical protein 30 4.1
At3g57060 putative protein 29 5.3
At5g21000 hypothetical protein (fragment) 29 6.9
>At2g30320 putative pseudouridine synthase
Length = 510
Score = 490 bits (1262), Expect = e-139
Identities = 251/452 (55%), Positives = 320/452 (70%), Gaps = 39/452 (8%)
Query: 51 WQPFRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKI 110
W+ +RKKKVV+RI Y GT+YRGLQ+QRD S+ T+E ELE AI+KAGGIR+SN GDL KI
Sbjct: 57 WESYRKKKVVIRIGYVGTDYRGLQIQRDDPSIKTIEGELEVAIYKAGGIRDSNYGDLHKI 116
Query: 111 GWARSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQ 170
GWARSSRTDKGVHSLAT ++ KMEIPE +W +DP G LA ++ +LP +I+V S+LP+
Sbjct: 117 GWARSSRTDKGVHSLATSISLKMEIPETAWKDDPQGTVLAKCISKHLPENIRVFSVLPSN 176
Query: 171 RSFDPRKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTSR 230
R FDPR+EC LRKYSYLLP D++GI++ ++DEIDYH+++FN+ILKEFEG +PFHNYT R
Sbjct: 177 RRFDPRRECTLRKYSYLLPVDVLGIKNSFTSDEIDYHITDFNEILKEFEGEYPFHNYTQR 236
Query: 231 SRYRR-----------HIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQ 279
SRYRR PR P K E + E +EEE +VD
Sbjct: 237 SRYRRKSEQKIKQRNGRPPREPKSIKASESEFREENHIEIGEEEEEKEVD---------- 286
Query: 280 NQKSSGTSDSCEPVIETGNKGNLHDQSSSSVV-RAKWLYEPDEADRLNASHFRRVFRCSS 338
G SD E V+ D +S V RAKWLYEPDE D+++ +HFR+VFRC S
Sbjct: 287 -----GESD--EHVVTP-------DSDNSQVYSRAKWLYEPDETDKISGAHFRKVFRCRS 332
Query: 339 GKLEKSMGHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIILPL 398
GKLE S+G ++EI I GESFMLHQIRKM+ TAVAVKR+LLPRDII LSL+KF+RI+LPL
Sbjct: 333 GKLENSLGFGFVEISIWGESFMLHQIRKMIGTAVAVKRELLPRDIIRLSLNKFTRIVLPL 392
Query: 399 APSEVLLLRGNSFSMRTKPGNVTRPEMQTIVESEEINKVVDDFYRSVMLPQVSKFLDPSR 458
APSEVL+LRGNSF +R P RP M+ ESEE+ K +++FYR+VM+PQVS FLD +
Sbjct: 393 APSEVLILRGNSFEVRRLP---ERPGMEATGESEEVEKEIEEFYRAVMVPQVSIFLDSEK 449
Query: 459 VPWVEWIEKFDAHTSIPNDQLDEVRKARNLWK 490
PW EW++ D + + +++L++VRK WK
Sbjct: 450 SPWKEWVDHLDRNDGLIDEELEDVRKGWEEWK 481
>At1g20370 putative tRNA pseudouridine synthase
Length = 549
Score = 118 bits (296), Expect = 7e-27
Identities = 113/421 (26%), Positives = 176/421 (40%), Gaps = 52/421 (12%)
Query: 54 FRKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWA 113
+R++KV + A+ G Y+G+Q T+E ELE A+F AG + +++ WA
Sbjct: 55 YRRRKVAIVFAFCGVGYQGMQKNP---GAKTIEGELEEALFHAGAVPDADRNKPRNYEWA 111
Query: 114 RSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSF 173
RS+RTDKGV ++ +V+ + + DP GF +NS LP I+V SF
Sbjct: 112 RSARTDKGVSAVGQVVSGRFYV-------DPPGF--VERLNSKLPDQIRVFGYKRVAPSF 162
Query: 174 DPRKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMS-EFNDILKEFEGGHPF------HN 226
+K C R+Y YL+P + H + + + E+ ++ E G+ +
Sbjct: 163 SSKKFCDRRRYVYLIPVFALDPCVHGEAEMVRTDLGYEYVKCVECSEKGYKIPLGVMGKD 222
Query: 227 YTSRSRYRRHIPRRPSHSKCGEILSAYNSEQEDSD--EEENFKVDEALTG-NIECQNQKS 283
I S S C I + E S EN E L G ++ + K+
Sbjct: 223 TDCCDTKSLEIQSDISSSTCDAIGADVKCETLSSSVPSAENNLNSEVLDGADVSALSSKA 282
Query: 284 SGTSDSCEPVIETGNKGNLHDQSSSSVVRAKWLYEPDEADRLN-------ASH------- 329
G +S N G D + S +K+ Y E +R N SH
Sbjct: 283 EGMEESNTLAKVEMNNGEGEDMTKS---ESKFCYGEKEKERFNRILSYYVGSHNFHNFTT 339
Query: 330 ---------FRRVFRCSSGKLEKSMGHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLP 380
R + ++ + G +++ + G+SFMLHQIRKM+ AVA+ R P
Sbjct: 340 RTKAADPAANRYILSFNANTVINLDGKDFVKCEVVGQSFMLHQIRKMIGLAVAIMRNYAP 399
Query: 381 RDIITLSLSKFSRIILPLAPSEVLLLRGNSFSMRTKPGNVTRPEMQTIVESEEINKVVDD 440
+I S K RI +P+AP L L F+ K + E V EE +V ++
Sbjct: 400 ESLIEASFKKDVRINVPMAPEVGLYLDECFFTSYNKRFKGSHEE----VSMEEYKEVAEE 455
Query: 441 F 441
F
Sbjct: 456 F 456
>At1g76120 unknown protein
Length = 463
Score = 117 bits (292), Expect = 2e-26
Identities = 105/377 (27%), Positives = 167/377 (43%), Gaps = 53/377 (14%)
Query: 57 KKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARSS 116
+KV + A+ G Y+G+Q T+E ELE A+F AG + ES G + +ARS+
Sbjct: 22 QKVAIIFAFCGVGYQGMQKNP---GAKTIEGELEEALFHAGAVPESIRGKPKLYDFARSA 78
Query: 117 RTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDPR 176
RTDKGV ++ +V+ + + DP GF N +NS LP I++ SF +
Sbjct: 79 RTDKGVSAVGQVVSGRFIV-------DPLGFV--NRLNSNLPNQIRIFGYKHVTPSFSSK 129
Query: 177 KECVLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTSRSRYRRH 236
K C R+Y YLLP + SH + + + + +K FE S R
Sbjct: 130 KFCDRRRYVYLLPVFALDPISHRDRETVMASLGPGEEYVKCFEC----------SERGRK 179
Query: 237 IPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCEPVIET 296
IP G + + ++ D + + AL +I+ + SS + C +E
Sbjct: 180 IPPGLVGKWKG---TNFGTKSLDFQSDISSNNSSALRSDIKIE-ALSSNLAGLCSVDVEV 235
Query: 297 G----NKGNLHDQSSSSVVRAKWLYEPDEADRL-----------NASHF----------- 330
G + L+ SS + V++K+ Y E +R N +F
Sbjct: 236 GRIQEDSCKLNTNSSETKVKSKFCYGEKEKERFSRILSCYVGSYNFHNFTTRTKADDPTA 295
Query: 331 -RRVFRCSSGKLEKSMGHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLS 389
R++ ++ + G +I+ + G+SFMLHQIRKM+ AVA+ R +I + S
Sbjct: 296 NRQIISFTANTVINLDGIDFIKCEVLGKSFMLHQIRKMMGLAVAIMRNCASESLIQSAFS 355
Query: 390 KFSRIILPLAPSEVLLL 406
K I +P+AP L L
Sbjct: 356 KDVNITVPMAPEVGLYL 372
>At1g34150 pseudouridine synthase-like protein
Length = 446
Score = 55.5 bits (132), Expect = 7e-08
Identities = 47/183 (25%), Positives = 75/183 (40%), Gaps = 30/183 (16%)
Query: 56 KKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARS 115
K+ V ++I Y G + G + + +E +ES +FKA +GD + ++R
Sbjct: 89 KRYVALKIMYFGKRFYGFSAE------AQMEPSIESEVFKALERTRLLVGDKKDSCYSRC 142
Query: 116 SRTDKGVHSLATMVAF----KMEIPENSWNEDPFG-------FALANYVNSYLPCDIKVI 164
RTDKGV S ++A +++ P G + +N LP DI+VI
Sbjct: 143 GRTDKGVSSTGQVIALFLRSRLKSPPGDSKAQVNGRTGERPEYDYVRVLNRALPDDIRVI 202
Query: 165 SILPAQRSFDPRKECVLRKYSYLLPADIIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPF 224
PA F R C R+Y Y + ++S + K+F G H F
Sbjct: 203 GWSPAPIDFHARFSCYAREYKYFFWRQ-------------NLNLSAMDFAGKKFIGEHDF 249
Query: 225 HNY 227
N+
Sbjct: 250 RNF 252
>At5g35400 unknown protein
Length = 420
Score = 47.8 bits (112), Expect = 1e-05
Identities = 38/136 (27%), Positives = 61/136 (43%), Gaps = 19/136 (13%)
Query: 63 IAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWAR-------S 115
++Y G ++ G Q Q D L TV+ +E ++ GG + L K +
Sbjct: 99 LSYNGASFDGWQKQPD---LHTVQGVVEKSL---GGFVDERKAQLLKKKCKPLEGRVLVA 152
Query: 116 SRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDP 175
RTDKGV +L + +F +W +D A+ + +N ++V+SI RSF P
Sbjct: 153 GRTDKGVSALNQVCSFY------TWRKDIEPIAIEDAINKDASGKLRVVSISKVSRSFHP 206
Query: 176 RKECVLRKYSYLLPAD 191
R+Y Y+ P D
Sbjct: 207 NFSAKWRRYLYIFPLD 222
>At3g06950 putative tRNA pseudouridine synthase
Length = 298
Score = 47.8 bits (112), Expect = 1e-05
Identities = 51/173 (29%), Positives = 74/173 (42%), Gaps = 18/173 (10%)
Query: 63 IAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARSSRTDKGV 122
IAY GT + G Q Q S T++ LE A+ + I E +L+ IG + RTD GV
Sbjct: 22 IAYDGTRFAGWQY---QESPPTIQSMLEKALIQ---ITELGRKELQLIG---AGRTDAGV 72
Query: 123 HSLATMVAFKMEIPENSWNEDPFGFALANYVNSYLPCDIKVISILPAQRSFDPRKECVLR 182
H+ + F P N + D F AL N LP DI+V + A F R +
Sbjct: 73 HAWGQVAHF--VTPFNYTSLDSFHAAL----NGLLPKDIRVRELSAAVPEFHARFSASSK 126
Query: 183 KYSYLLPADII--GIQSH-SSNDEIDYHMSEFNDILKEFEGGHPFHNYTSRSR 232
Y Y + D Q H + + + S+ + F G H F + + +R
Sbjct: 127 VYRYQIYNDTFMDPFQRHWAYHCAYKLNASKMREAANLFVGKHDFSAFANATR 179
Score = 35.4 bits (80), Expect = 0.074
Identities = 15/49 (30%), Positives = 28/49 (56%)
Query: 348 SYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKFSRIIL 396
S +++ +EG F+ Q+R MVA + + ++ L DI+ + L R +L
Sbjct: 200 SLLQLEVEGSGFLYRQVRNMVALLIQIGKEALDSDIVPMILETKDRRVL 248
>At5g37930 unknown protein (At5g37930)
Length = 349
Score = 31.2 bits (69), Expect = 1.4
Identities = 31/122 (25%), Positives = 52/122 (42%), Gaps = 12/122 (9%)
Query: 226 NYTSRSRYRRHIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSSG 285
N +SR R+ +P + + E A +S +ED DE +N + E +++ S+
Sbjct: 17 NNNHQSRKRQRLPSIDENEEDAETSDAGSSGEEDEDETQNQGM------RPESEDRGSTS 70
Query: 286 TSDSCEPVIETGNKGNLHDQSSSSVVR----AKWLYEPDEADRLNASHFRR--VFRCSSG 339
E VIE G + SSS + + L +PD D + +F+C +G
Sbjct: 71 DDSDREVVIEERRFGKFVNSQSSSSSKDSPLSVTLLDPDVLDCPICCEPLKIPIFQCDNG 130
Query: 340 KL 341
L
Sbjct: 131 HL 132
>At3g14000 unknown protein
Length = 374
Score = 30.8 bits (68), Expect = 1.8
Identities = 22/75 (29%), Positives = 36/75 (47%), Gaps = 10/75 (13%)
Query: 246 CGEILSAYN--SEQEDSDEEENFKVDE--------ALTGNIECQNQKSSGTSDSCEPVIE 295
C + L+ N S++++ DEEE +V E +LT I+ K+SG SC+P
Sbjct: 7 CTKQLNTNNGGSKKQEEDEEEEDRVIETPRSKQIKSLTSQIKDMAVKASGAYKSCKPCSG 66
Query: 296 TGNKGNLHDQSSSSV 310
+ N+ + S V
Sbjct: 67 SSNQNKNRSYADSDV 81
>At5g56210 unknown protein
Length = 509
Score = 30.4 bits (67), Expect = 2.4
Identities = 16/48 (33%), Positives = 32/48 (66%), Gaps = 1/48 (2%)
Query: 252 AYNSEQEDSDEEENFKVDEALTGNIECQNQ-KSSGTSDSCEPVIETGN 298
A+ S+++D+ EEE + ++ G+ ++Q KS+G+SD+ +P+I N
Sbjct: 232 AHTSKRKDNVEEEEEETEDYSQGDCVEESQIKSNGSSDNLDPLIVAVN 279
>At3g24840 phosphatidylinositol transfer protein like
Length = 579
Score = 30.4 bits (67), Expect = 2.4
Identities = 43/160 (26%), Positives = 64/160 (39%), Gaps = 28/160 (17%)
Query: 298 NKGNLHDQSSSSVVRAK-WLYEPDEADRLNASHFRRVFRCSSGKLEKSM---GHSYIEIY 353
NKG +D +VR++ +Y+P E L ++F + S GH
Sbjct: 327 NKGPWNDPEIMKLVRSRDAMYKPKEMGLLENGEVAKLFSLRHVNTDMSSPDGGHVR---- 382
Query: 354 IEGESFMLHQIRKMV---ATAVAVKR-------KLLPRDI-----ITLSLSK----FSRI 394
E ES H R + A AV V R LP ++ +T SL K +R
Sbjct: 383 -ERESHPEHDKRAQLSNQAEAVGVGRMEQSDSTSPLPNNLAVERSLTTSLQKVASFLARF 441
Query: 395 ILPLAPSEVLLLRGNSFSMRTKPGNVTRPEMQTIVESEEI 434
IL L S L+ R + +P N RPE+ V +++
Sbjct: 442 ILQLLGSLCLMFRILGRLVNKQPENQLRPELSVSVSQQQV 481
>At2g46260 unknown protein
Length = 561
Score = 30.4 bits (67), Expect = 2.4
Identities = 57/251 (22%), Positives = 95/251 (37%), Gaps = 65/251 (25%)
Query: 198 HSSNDEIDYHM----SEFNDILKEFE--GG-----HPFHNYTSRSRYRRHIPRRPSHSK- 245
H S+ E D+ S F+D L E GG TS + + RH RR +K
Sbjct: 23 HGSSSEGDFGFAFNDSNFSDRLLRIEILGGPSDSRSDAEGCTSIADWARHRKRRREDNKK 82
Query: 246 -----------CGE--ILSAYNSEQEDS-------DEEENFKVDEALTGNIECQNQKSSG 285
C E IL+ N D D+E V+EAL+G+ + ++ + G
Sbjct: 83 DNGVAISDIVACAEEQILTDNNQPDMDDAPGGDNLDDEGEAMVEEALSGDDDASSEPNWG 142
Query: 286 TSDSCEPVIETGNKGNLHDQSSSSVVRAKWLYEPDEADRLNASHFRRVFRCSSGKLEKSM 345
S+VVR K L+ + F ++F S+G E
Sbjct: 143 I-------------------DCSTVVRVKELHISSPILAAKSPFFYKLF--SNGMRESEQ 181
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKLLPRDIITLSLSKF---------SRII- 395
H + I + E ++ + M + +++V D++ ++ KF SR++
Sbjct: 182 RHVTLRISAQEEGALMELLNFMYSNSLSVTTAPALLDVL-MAADKFEVASCMRYCSRLLR 240
Query: 396 -LPLAPSEVLL 405
+P+ P LL
Sbjct: 241 NMPMTPDSALL 251
>At1g44180 putative aminoacylase
Length = 435
Score = 30.4 bits (67), Expect = 2.4
Identities = 15/38 (39%), Positives = 21/38 (54%), Gaps = 1/38 (2%)
Query: 192 IIGIQSHSSNDEIDYHMSEFNDILKEFEGGHPFHNYTS 229
I +QSHS +E D ++ F L+ F HP NYT+
Sbjct: 16 IFSLQSHSKEEEEDTPITRFQQYLR-FNTAHPNPNYTA 52
>At1g79730 unknown protein
Length = 589
Score = 30.0 bits (66), Expect = 3.1
Identities = 46/236 (19%), Positives = 95/236 (39%), Gaps = 33/236 (13%)
Query: 229 SRSRYRRHIPRRPSHSKCGEILSA--YNSEQEDSDEEENFKVDEALTGNIECQNQKSSGT 286
S+ +R H+P HSK E ++E + ++ K + + + + Q K
Sbjct: 116 SKHHHRSHLP----HSKKIETEEERRLRKKRELEKQRQDEKHRQQMKNSHKSQMPKGHTE 171
Query: 287 SDSCEPVIETGNKGNLHDQSSSSVVRAKWLYE-PDEADRLNASHFRRVFRCSSGKLEKSM 345
P++ T N + ++ + + K+ E PD + +L +R +K
Sbjct: 172 EKKPTPLLTTDRVENRLKKPTTFICKLKFRNELPDPSAQLKLMTIKR---------DKDQ 222
Query: 346 GHSYIEIYIEGESFMLHQIRKMVATAVAVKRKL-LPRDIITLSLSKFSRIILPLAPSEVL 404
Y + + K+ + V+ L +P D++ LS+ ++ PLAP +
Sbjct: 223 FTKYT----------ITSLEKLWKPKIFVEPDLGIPLDLLDLSVYNPPKVKAPLAPEDEE 272
Query: 405 LLRGNSFSMRTKPGNVTRPE------MQTIVESEEINKVVDDFYRSVMLPQVSKFL 454
LLR + K + R E M +V+++ I+ + ++ R + + +K L
Sbjct: 273 LLRDDDAVTPIKKDGIRRKERPTDKGMSWLVKTQYISSINNESARQSLTEKQAKEL 328
>At1g44120 hypothetical protein
Length = 2114
Score = 30.0 bits (66), Expect = 3.1
Identities = 19/46 (41%), Positives = 27/46 (58%), Gaps = 2/46 (4%)
Query: 73 LQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWARSSRT 118
L+MQR Q ++T K L + + K GIR NL ++ +G RSS T
Sbjct: 1563 LEMQRFQEEITT--KNLIAPLVKLVGIRVRNLQEIALMGLERSSVT 1606
>At1g09800 hypothetical protein
Length = 372
Score = 30.0 bits (66), Expect = 3.1
Identities = 24/80 (30%), Positives = 36/80 (45%), Gaps = 12/80 (15%)
Query: 55 RKKKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGWAR 114
R ++ ++ I Y GT + G Q Q TV L+ A K G + +
Sbjct: 53 RVQRYLVAIEYIGTRFSGSQQQAKDR---TVVGVLQEAFHKFIG---------QPVKIFC 100
Query: 115 SSRTDKGVHSLATMVAFKME 134
SSRTD GVH+L+ + +E
Sbjct: 101 SSRTDAGVHALSNVCHVDVE 120
>At1g02660 unknown protein
Length = 713
Score = 30.0 bits (66), Expect = 3.1
Identities = 20/66 (30%), Positives = 32/66 (48%), Gaps = 10/66 (15%)
Query: 256 EQEDSDEEENFKVDEALTGNIECQNQKSSGTSDSCEPVIETGNKGN---LHDQSSSSVVR 312
E ED DE+E ++D+A+ + G D C + + GN+ N L +S S ++R
Sbjct: 130 EDEDGDEDEEVELDDAVV-------SEDDGGCDVCSVLEDDGNEANKFQLDRESFSKLLR 182
Query: 313 AKWLYE 318
L E
Sbjct: 183 RVTLPE 188
>At3g14090 unknown protein
Length = 623
Score = 29.6 bits (65), Expect = 4.1
Identities = 16/56 (28%), Positives = 29/56 (51%), Gaps = 6/56 (10%)
Query: 57 KKVVMRIAYTGTNYRGLQMQRDQNSLSTVEKELESAIFKAGGIRESNLGDLEKIGW 112
+ +V R+ G + +Q+ TV K AIFK GI + ++GD++++ W
Sbjct: 185 RSIVQRMVAAGYSRECIQVY------GTVRKSAMEAIFKQLGIVKISIGDVQRLEW 234
>At1g08440 hypothetical protein
Length = 501
Score = 29.6 bits (65), Expect = 4.1
Identities = 22/84 (26%), Positives = 40/84 (47%), Gaps = 4/84 (4%)
Query: 225 HNYTSRSRYRRHIPRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIECQNQKSS 284
H S ++++ ++PS S G I A ++ D D+ + + T N Q+Q
Sbjct: 390 HELASAAKFKNK--KKPSKSNSGSIGQAMPNKSHDDDDHVVTILGDVDTSNNVDQSQSHG 447
Query: 285 GTS-DSCEPV-IETGNKGNLHDQS 306
S DSC V I+ + ++HD++
Sbjct: 448 EISVDSCHHVTIKINDDDSIHDKN 471
>At3g57060 putative protein
Length = 1439
Score = 29.3 bits (64), Expect = 5.3
Identities = 25/82 (30%), Positives = 34/82 (40%), Gaps = 15/82 (18%)
Query: 238 PRRPSHSKCGEILSAYNSEQEDSDEEENFKVDEALTGNIEC-------QNQKSSGTSDSC 290
PR + G+ L SE+E SD EE E + +C NQK+SG
Sbjct: 1358 PRSLNRKTSGDNLIETESEEEQSDSEEEPSDSEEEPDSAQCGTTNPRSLNQKTSGG---- 1413
Query: 291 EPVIETGNKGNLHDQSSSSVVR 312
E G + +SSSS+ R
Sbjct: 1414 ----EEGESESKSTESSSSIRR 1431
>At5g21000 hypothetical protein (fragment)
Length = 151
Score = 28.9 bits (63), Expect = 6.9
Identities = 31/125 (24%), Positives = 54/125 (42%), Gaps = 18/125 (14%)
Query: 102 SNLGDLEKIGWARSSRTDKGVHSLATMVAFKMEIPENSWNEDPFGFALA--NYVNSYLPC 159
+NL + ++ + +S KG++S + F+M+ E+ FG +L + YLP
Sbjct: 33 ANLKGIRRLVLSPASLISKGIYSTQS---FQMD------EEEGFGRSLIVIDITTDYLPH 83
Query: 160 DIKVISILPAQRSFDPRKECVLRKYSYLLPADIIG-IQSHSSNDE------IDYHMSEFN 212
V + + F+ + + K + A + G IQ ND DYH+ FN
Sbjct: 84 LKAVALFIVSLLKFEKKLKSAAEKQCSEIIASVYGEIQLELENDRSVYTDRYDYHVLVFN 143
Query: 213 DILKE 217
I K+
Sbjct: 144 QITKK 148
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.132 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,824,374
Number of Sequences: 26719
Number of extensions: 531383
Number of successful extensions: 1652
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1627
Number of HSP's gapped (non-prelim): 34
length of query: 501
length of database: 11,318,596
effective HSP length: 103
effective length of query: 398
effective length of database: 8,566,539
effective search space: 3409482522
effective search space used: 3409482522
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC140068.2


