

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140033.2 - phase: 0
(352 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
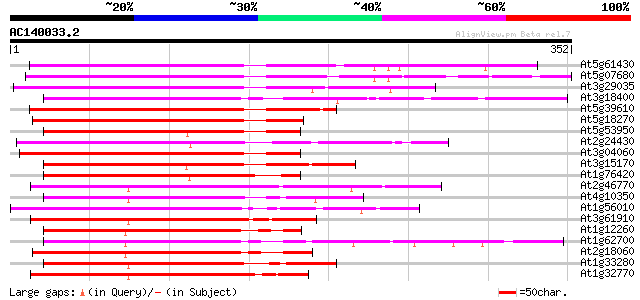
Score E
Sequences producing significant alignments: (bits) Value
At5g61430 NAM, no apical meristem, - like protein 255 3e-68
At5g07680 NAM (no apical meristem)-like protein 251 3e-67
At3g29035 NAM-like protein (No Apical Meristem) 233 9e-62
At3g18400 organ separation protein, putative 231 3e-61
At5g39610 NAM / CUC2 - like protein 228 5e-60
At5g18270 NAM (no apical meristem)-like protein 217 7e-57
At5g53950 CUC2 (dbj|BAA19529.1) 216 1e-56
At2g24430 NAM (no apical meristem)-like protein 213 2e-55
At3g04060 NAM-like protein (no apical meristem) 211 6e-55
At3g15170 NAM-like protein 207 7e-54
At1g76420 unknown protein 202 3e-52
At2g46770 NAM (no apical meristem)-like protein 189 2e-48
At4g10350 NAM/NAP like protein 184 6e-47
At1g56010 NAC1 181 4e-46
At3g61910 NAM-like protein 178 3e-45
At1g12260 unknown protein 177 1e-44
At1g62700 unknown protein 176 1e-44
At2g18060 putative NAM (no apical meristem)-like protein 175 3e-44
At1g33280 hypothetical protein 174 9e-44
At1g32770 OsNAC7 protein, putative 173 1e-43
>At5g61430 NAM, no apical meristem, - like protein
Length = 336
Score = 255 bits (651), Expect = 3e-68
Identities = 154/343 (44%), Positives = 196/343 (56%), Gaps = 41/343 (11%)
Query: 13 ENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGE 72
E E++ G RF PTDEELI YL KKV D SF A AI EVD+NK EPW+LP MAKMGE
Sbjct: 10 EEEQMDLPPGFRFHPTDEELITHYLHKKVLDTSFSAKAIGEVDLNKSEPWELPWMAKMGE 69
Query: 73 TEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRG 132
EWYFFCVRD+KYPTG RTNRAT AGYWKATGKDKEIY+G SL+GMKKTLVFY+GRAP+G
Sbjct: 70 KEWYFFCVRDRKYPTGLRTNRATEAGYWKATGKDKEIYRGKSLVGMKKTLVFYRGRAPKG 129
Query: 133 EKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGS 192
+K+NWVMHEYRLEG F HNL +EWV+ RVF+K GKK+ S
Sbjct: 130 QKTNWVMHEYRLEGK-------------FSAHNLPKTAKNEWVICRVFQKSAGGKKIPIS 176
Query: 193 KLGRSNSSREEPSNTNAASLWAPFLEFSPYNSENK---ITIPDFSNE--------FNSFT 241
L R S + + SL + SPYN + K + +P FSN+ N F+
Sbjct: 177 SLIRIGSLGTDFN----PSLLPSLTDSSPYNDKTKTEPVYVPCFSNQTDQNQGTTLNCFS 232
Query: 242 NP----------NQSEKPKTQYDNIVHNNETSILNISSSSKQMDVYPLAGATVADPNLTS 291
+P ++ +TQ + N ++ +L S + ++ +++
Sbjct: 233 SPVLNSIQADIFHRIPLYQTQSLQVSMNLQSPVLTQEHSVLHAMIENNRRQSLKTMSVSQ 292
Query: 292 MAGNSS--NFFFSQEFSFG-REFDADADISSVVYGNDMFQRWS 331
G S+ N S +F FG R FD+ D SS D+ W+
Sbjct: 293 ETGVSTDMNTDISSDFEFGKRRFDSQEDPSSSTGPVDLEPFWN 335
>At5g07680 NAM (no apical meristem)-like protein
Length = 329
Score = 251 bits (642), Expect = 3e-67
Identities = 155/354 (43%), Positives = 200/354 (55%), Gaps = 45/354 (12%)
Query: 11 KMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKM 70
K ++E++ G RF PTDEELI YL KKV D F A AI EVD+NK EPW+LP AK+
Sbjct: 9 KEDDEQMDLPPGFRFHPTDEELITHYLHKKVLDLGFSAKAIGEVDLNKAEPWELPYKAKI 68
Query: 71 GETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAP 130
GE EWYFFCVRD+KYPTG RTNRAT AGYWKATGKDKEI++G SL+GMKKTLVFY+GRAP
Sbjct: 69 GEKEWYFFCVRDRKYPTGLRTNRATQAGYWKATGKDKEIFRGKSLVGMKKTLVFYRGRAP 128
Query: 131 RGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMN 190
+G+K+NWVMHEYRL+G HNL +EWV+ RVF K GKK+
Sbjct: 129 KGQKTNWVMHEYRLDGK-------------LSAHNLPKTAKNEWVICRVFHKTAGGKKIP 175
Query: 191 GSKLGRSNSSREEPSNTNAASLWAPFLEFSPYNSENK---ITIPDFSNE-------FNSF 240
S L R S S P + SPYN + K + +P FSN+ N F
Sbjct: 176 ISTLIRIGS-------YGTGSSLPPLTDSSPYNDKTKTEPVYVPCFSNQAETRGTILNCF 228
Query: 241 TNPNQSEKPKTQYDNIVHNNETSILNISSSSKQMDVYPLAGATVADPNLTSMAGNSSNFF 300
+NP+ S + + ++ + LNIS SS + T L +M N+
Sbjct: 229 SNPSLS-SIQPDFLQMIPLYQPQSLNISESSNPV-------LTQEQSVLQAMMENNRRQN 280
Query: 301 FSQEFSFGREFD-ADADISSVV-YGNDMFQRWSGYQDLSPASTGLAANDSFWNF 352
F + S +E ++ D SSV +G F +Q++ S+G + FWN+
Sbjct: 281 F-KTLSISQETGVSNTDNSSVFEFGRKRFD----HQEVPSPSSGPVDLEPFWNY 329
>At3g29035 NAM-like protein (No Apical Meristem)
Length = 318
Score = 233 bits (595), Expect = 9e-62
Identities = 131/277 (47%), Positives = 164/277 (58%), Gaps = 26/277 (9%)
Query: 3 SNENVSNQKMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPW 62
S E V + ++EK+ G RF PTDEELI YL KV ++ F AIAI EVD+NK EPW
Sbjct: 8 SGEIVEGEVEDSEKIDLPPGFRFHPTDEELITHYLRPKVVNSFFSAIAIGEVDLNKVEPW 67
Query: 63 DLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTL 122
DLP AK+GE EWYFFCVRD+KYPTG RTNRAT AGYWKATGKDKEI+KG SL+GMKKTL
Sbjct: 68 DLPWKAKLGEKEWYFFCVRDRKYPTGLRTNRATKAGYWKATGKDKEIFKGKSLVGMKKTL 127
Query: 123 VFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK 182
VFYKGRAP+G K+NWVMHEYRLEG F NLS +E V++RVF
Sbjct: 128 VFYKGRAPKGVKTNWVMHEYRLEGK-------------FAIDNLSKTAKNECVISRVFHT 174
Query: 183 RNCGKK----------MNGSKLGRSNSSREEPSNTNAASLWAPFLEFSPYNSENKITIPD 232
R G K M+ S +S ++ + T L + FS +++K + D
Sbjct: 175 RTDGTKEHMSVGLPPLMDSSPYLKSR-GQDSLAGTTLGGLLSHVTYFSDQTTDDKSLVAD 233
Query: 233 FSNEF--NSFTNPNQSEKPKTQYDNIVHNNETSILNI 267
F + TN + +D + N +S+L +
Sbjct: 234 FKTTMFGSGSTNFLPNIGSLLDFDPLFLQNNSSVLKM 270
>At3g18400 organ separation protein, putative
Length = 314
Score = 231 bits (590), Expect = 3e-61
Identities = 144/333 (43%), Positives = 182/333 (54%), Gaps = 30/333 (9%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEELI YL +KV D F A+ +VD+NKCEPWDLP A MGE EWYFF R
Sbjct: 8 GFRFHPTDEELITHYLCRKVSDIGFTGKAVVDVDLNKCEPWDLPAKASMGEKEWYFFSQR 67
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWVMHE 141
D+KYPTG RTNRAT AGYWK TGKDKEIY+ L+GMKKTLVFYKGRAP+GEKSNWVMHE
Sbjct: 68 DRKYPTGLRTNRATEAGYWKTTGKDKEIYRSGVLVGMKKTLVFYKGRAPKGEKSNWVMHE 127
Query: 142 YRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLGRSNSSR 201
YRLE SK P + EWVV RVFEK KK + S S
Sbjct: 128 YRLE----SKQPFNPTNK------------EEWVVCRVFEKSTAAKKAQEQQPQSSQPSF 171
Query: 202 EEP--SNTNAASLWAPFLEFSPYNSENKITIPDFSNEFNSFTNPNQSEKPKTQYDNIVHN 259
P +N++ A+ + E NS N TI D++N + ++ N + T ++
Sbjct: 172 GSPCDANSSMANEFEDIDELPNLNS-NSSTI-DYNNHIHQYSQRNVYSEDNTTSTAGLNM 229
Query: 260 NETSILNISSSSKQMDVYPLAGATVADPNLTSMAGNSSNFFFSQEFSFGREFDADADISS 319
N +N++S++ Q L G ++ N + F +SF +E + SS
Sbjct: 230 N----MNMASTNLQSWTTSLLGPPLSPINSLLLKA----FQIRNSYSFPKEMIPSFNHSS 281
Query: 320 VVYG-NDMFQRWSGYQDLSPASTGLAAN-DSFW 350
+ G ++M Q S + P A N DS W
Sbjct: 282 LQQGVSNMIQNASSSSQVQPQPQEEAFNMDSIW 314
>At5g39610 NAM / CUC2 - like protein
Length = 285
Score = 228 bits (580), Expect = 5e-60
Identities = 116/193 (60%), Positives = 133/193 (68%), Gaps = 14/193 (7%)
Query: 13 ENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGE 72
+ E + G RF PTDEELI YL KV + F A AI EVD+NK EPWDLP AKMGE
Sbjct: 14 DEEHIDLPPGFRFHPTDEELITHYLKPKVFNTFFSATAIGEVDLNKIEPWDLPWKAKMGE 73
Query: 73 TEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRG 132
EWYFFCVRD+KYPTG RTNRAT AGYWKATGKDKEI+KG SL+GMKKTLVFYKGRAP+G
Sbjct: 74 KEWYFFCVRDRKYPTGLRTNRATEAGYWKATGKDKEIFKGKSLVGMKKTLVFYKGRAPKG 133
Query: 133 EKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGS 192
K+NWVMHEYRLEG + NL +EWV+ RVF+KR G K+ S
Sbjct: 134 VKTNWVMHEYRLEGK-------------YCIENLPQTAKNEWVICRVFQKRADGTKVPMS 180
Query: 193 KLGRSNSSREEPS 205
L + +R EP+
Sbjct: 181 MLD-PHINRMEPA 192
>At5g18270 NAM (no apical meristem)-like protein
Length = 335
Score = 217 bits (553), Expect = 7e-57
Identities = 105/171 (61%), Positives = 123/171 (71%), Gaps = 14/171 (8%)
Query: 15 EKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETE 74
E V G RF PTDEE+I YL +KV ++ F A+A+ E D+NKCEPWDLP+ AKMGE E
Sbjct: 17 ELVDLPPGFRFHPTDEEIITCYLKEKVLNSRFTAVAMGEADLNKCEPWDLPKRAKMGEKE 76
Query: 75 WYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNS-LIGMKKTLVFYKGRAPRGE 133
+YFFC RD+KYPTG RTNRAT +GYWKATGKDKEI+KG L+GMKKTLVFY+GRAP+GE
Sbjct: 77 FYFFCQRDRKYPTGMRTNRATESGYWKATGKDKEIFKGKGCLVGMKKTLVFYRGRAPKGE 136
Query: 134 KSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRN 184
K+NWVMHEYRLEG + +NL EWVV RVF K N
Sbjct: 137 KTNWVMHEYRLEGK-------------YSYYNLPKSARDEWVVCRVFHKNN 174
>At5g53950 CUC2 (dbj|BAA19529.1)
Length = 375
Score = 216 bits (551), Expect = 1e-56
Identities = 106/163 (65%), Positives = 121/163 (74%), Gaps = 15/163 (9%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEELI YL++KV D F + AIAEVD+NKCEPW LP AKMGE EWYFF +R
Sbjct: 20 GFRFHPTDEELITHYLLRKVLDGCFSSRAIAEVDLNKCEPWQLPGRAKMGEKEWYFFSLR 79
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIY--KGNSLIGMKKTLVFYKGRAPRGEKSNWVM 139
D+KYPTG RTNRAT AGYWKATGKD+EI+ K +L+GMKKTLVFYKGRAP+GEKSNWVM
Sbjct: 80 DRKYPTGLRTNRATEAGYWKATGKDREIFSSKTCALVGMKKTLVFYKGRAPKGEKSNWVM 139
Query: 140 HEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK 182
HEYRLEG F H +S EWV++RVF+K
Sbjct: 140 HEYRLEGK-------------FSYHFISRSSKDEWVISRVFQK 169
>At2g24430 NAM (no apical meristem)-like protein
Length = 316
Score = 213 bits (541), Expect = 2e-55
Identities = 125/273 (45%), Positives = 155/273 (55%), Gaps = 28/273 (10%)
Query: 5 ENVSNQKMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDL 64
E +Q+ + E+ G RF PTDEELI+ YLV K+ D +F AIA+VD+NK EPW+L
Sbjct: 2 EQGDHQQHKKEEEALPPGFRFHPTDEELISYYLVNKIADQNFTGKAIADVDLNKSEPWEL 61
Query: 65 PEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKG--NSLIGMKKTL 122
PE AKMG EWYFF +RD+KYPTG RTNRATN GYWK TGKDKEI+ + L+GMKKTL
Sbjct: 62 PEKAKMGGKEWYFFSLRDRKYPTGVRTNRATNTGYWKTTGKDKEIFNSTTSELVGMKKTL 121
Query: 123 VFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK 182
VFY+GRAPRGEK+ WVMHEYRL S + T EWVV RVF+K
Sbjct: 122 VFYRGRAPRGEKTCWVMHEYRLHSKS----------------SYRTSKQDEWVVCRVFKK 165
Query: 183 RNCGKKMNGSKLGRSNSSREEPSNTNAASLWAPFLEFSPYNSENKITIPDFSNEFNSFTN 242
KK + S+SS N + + +P S + + +P S N
Sbjct: 166 TEATKKY----ISTSSSSTSHHHNNHTRASILSTNNNNPNYSSDLLQLPPHLQPHPSL-N 220
Query: 243 PNQSEKPKTQYDNIVHNNETSILNISSSSKQMD 275
NQS N VH E S + +S+S MD
Sbjct: 221 INQS-----LMANAVHLAELSRVFRASTSTTMD 248
>At3g04060 NAM-like protein (no apical meristem)
Length = 338
Score = 211 bits (536), Expect = 6e-55
Identities = 105/177 (59%), Positives = 123/177 (69%), Gaps = 14/177 (7%)
Query: 7 VSNQKMENEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPE 66
V NQ + E V G RF PTDEE+I YL +KV + F A AI + D+NK EPWDLP+
Sbjct: 8 VVNQGGDQEVVDLPPGFRFHPTDEEIITHYLKEKVFNIRFTAAAIGQADLNKNEPWDLPK 67
Query: 67 MAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNS-LIGMKKTLVFY 125
+AKMGE E+YFFC RD+KYPTG RTNRAT +GYWKATGKDKEI++G L+GMKKTLVFY
Sbjct: 68 IAKMGEKEFYFFCQRDRKYPTGMRTNRATVSGYWKATGKDKEIFRGKGCLVGMKKTLVFY 127
Query: 126 KGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK 182
GRAP+GEK+NWVMHEYRL+G + HNL EWVV RVF K
Sbjct: 128 TGRAPKGEKTNWVMHEYRLDGK-------------YSYHNLPKTARDEWVVCRVFHK 171
>At3g15170 NAM-like protein
Length = 310
Score = 207 bits (527), Expect = 7e-54
Identities = 109/198 (55%), Positives = 133/198 (67%), Gaps = 16/198 (8%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEELI YL+KKV D++F AI++VD+NK EPW+LPE AKMGE EWYFF +R
Sbjct: 23 GFRFHPTDEELITYYLLKKVLDSNFSCAAISQVDLNKSEPWELPEKAKMGEKEWYFFTLR 82
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEI--YKGNSLIGMKKTLVFYKGRAPRGEKSNWVM 139
D+KYPTG RTNRAT AGYWKATGKD+EI K SL+GMKKTLVFYKGRAP+GEKS WVM
Sbjct: 83 DRKYPTGLRTNRATEAGYWKATGKDREIKSSKTKSLLGMKKTLVFYKGRAPKGEKSCWVM 142
Query: 140 HEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLGRSNS 199
HEYRL+G F H +S+ EWV+ +V K + + + S+S
Sbjct: 143 HEYRLDGK-------------FSYHYISSSAKDEWVLCKVCLKSGVVSR-ETNLISSSSS 188
Query: 200 SREEPSNTNAASLWAPFL 217
S ++A S AP +
Sbjct: 189 SAVTGEFSSAGSAIAPII 206
>At1g76420 unknown protein
Length = 334
Score = 202 bits (513), Expect = 3e-52
Identities = 97/163 (59%), Positives = 112/163 (68%), Gaps = 18/163 (11%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGETEWYFFCVR 81
G RF PTDEELI YL K+ I I+EVD+N+CEPW+LPEMAKMGE EWYF+ +R
Sbjct: 25 GFRFHPTDEELITFYLASKIFHGGLSGIHISEVDLNRCEPWELPEMAKMGEREWYFYSLR 84
Query: 82 DKKYPTGQRTNRATNAGYWKATGKDKEIYK--GNSLIGMKKTLVFYKGRAPRGEKSNWVM 139
D+KYPTG RTNRAT AGYWKATGKDKE++ G L+GMKKTLVFYKGRAPRG K+ WVM
Sbjct: 85 DRKYPTGLRTNRATTAGYWKATGKDKEVFSGGGGQLVGMKKTLVFYKGRAPRGLKTKWVM 144
Query: 140 HEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEK 182
HEYRLE + +H EWV+ RVF K
Sbjct: 145 HEYRLENDHSHRHTC----------------KEEWVICRVFNK 171
>At2g46770 NAM (no apical meristem)-like protein
Length = 365
Score = 189 bits (480), Expect = 2e-48
Identities = 105/266 (39%), Positives = 149/266 (55%), Gaps = 11/266 (4%)
Query: 14 NEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGET 73
N + + G RF PT+EEL+ YL KKV+ I +VD+NK EPWD+ EM K+G T
Sbjct: 11 NGQSQVPPGFRFHPTEEELLQYYLRKKVNSIEIDLDVIRDVDLNKLEPWDIQEMCKIGTT 70
Query: 74 ---EWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAP 130
+WYFF +DKKYPTG RTNRAT AG+WKATG+DK IY IGM+KTLVFYKGRAP
Sbjct: 71 PQNDWYFFSHKDKKYPTGTRTNRATAAGFWKATGRDKIIYSNGRRIGMRKTLVFYKGRAP 130
Query: 131 RGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMN 190
G+KS+W+MHEYRL+ N +S ++ + S WVV R+F+K+N K +N
Sbjct: 131 HGQKSDWIMHEYRLDDNIISPEDVTVHEVVSIIGEASQD--EGWVVCRIFKKKNLHKTLN 188
Query: 191 GSKLGRSNSSREEPSNTNAASLW-----APFLEFSPYNSENKITIPDFSNEFNSFTNPNQ 245
G S S + T ++ ++ FLE + + ++ + F + + +PN
Sbjct: 189 SPVGGASLSGGGDTPKTTSSQIFNEDTLDQFLELMGRSCKEELNLDPFM-KLPNLESPNS 247
Query: 246 SEKPKTQYDNIVHNNETSILNISSSS 271
+ N+ + N+ +S
Sbjct: 248 QAINNCHVSSPDTNHNIHVSNVVDTS 273
>At4g10350 NAM/NAP like protein
Length = 341
Score = 184 bits (467), Expect = 6e-47
Identities = 103/207 (49%), Positives = 125/207 (59%), Gaps = 21/207 (10%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGET---EWYFF 78
G RF PTDEEL++ YL KK+ F I EVD+NK EPWDL E K+G T EWYFF
Sbjct: 12 GFRFHPTDEELLHYYLKKKISYQKFEMEVIREVDLNKLEPWDLQERCKIGSTPQNEWYFF 71
Query: 79 CVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWV 138
+D+KYPTG RTNRAT+AG+WKATG+DK I IGM+KTLVFYKGRAP G+K++W+
Sbjct: 72 SHKDRKYPTGSRTNRATHAGFWKATGRDKCIRNSYKKIGMRKTLVFYKGRAPHGQKTDWI 131
Query: 139 MHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMN--GSKLGR 196
MHEYRLE P+ N S G WVV RVF K+N K +N S +
Sbjct: 132 MHEYRLEDAD------------DPQANPSEDG---WVVCRVFMKKNLFKVVNEGSSSINS 176
Query: 197 SNSSREEPSNTNAASLWAPFL-EFSPY 222
+ + SN N A F+ SPY
Sbjct: 177 LDQHNHDASNNNHALQARSFMHRDSPY 203
>At1g56010 NAC1
Length = 324
Score = 181 bits (460), Expect = 4e-46
Identities = 102/262 (38%), Positives = 151/262 (56%), Gaps = 23/262 (8%)
Query: 1 MGSNENVSNQKMENEKVKFDAGVRFFPTDEELINQYLVKK-VDDNSFCAIAIAEVDMNKC 59
M + E + + + K G RF P D+EL+ YL+++ + +N + + +VD+NKC
Sbjct: 1 METEEEMKESSISMVEAKLPPGFRFHPKDDELVCDYLMRRSLHNNHRPPLVLIQVDLNKC 60
Query: 60 EPWDLPEMAKMGETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMK 119
EPWD+P+MA +G +WYF+ RD+KY TG RTNRAT GYWKATGKD+ I + L+GM+
Sbjct: 61 EPWDIPKMACVGGKDWYFYSQRDRKYATGLRTNRATATGYWKATGKDRTILRKGKLVGMR 120
Query: 120 KTLVFYKGRAPRGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRV 179
KTLVFY+GRAPRG K++WVMHE+RL+G S H P H+LS+ +WV+ RV
Sbjct: 121 KTLVFYQGRAPRGRKTDWVMHEFRLQG---SHHP--------PNHSLSS-PKEDWVLCRV 168
Query: 180 FEKRNCGKKMNGSKLGRSNSSREEPSNTNAASLWAPFLEF----SPYNSENKITIPDFSN 235
F K G + S +E ++ + L P++ F S Y S++ I +
Sbjct: 169 FHKNTEGVICRDN----MGSCFDETASASLPPLMDPYINFDQEPSSYLSDDHHYI--INE 222
Query: 236 EFNSFTNPNQSEKPKTQYDNIV 257
F+N +Q++ + N V
Sbjct: 223 HVPCFSNLSQNQTLNSNLTNSV 244
>At3g61910 NAM-like protein
Length = 334
Score = 178 bits (452), Expect = 3e-45
Identities = 92/183 (50%), Positives = 119/183 (64%), Gaps = 7/183 (3%)
Query: 14 NEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGET 73
N + + G RF PT+EEL+ YL KK+ + I ++D+NK EPWD+ EM K+G T
Sbjct: 6 NGQSQVPPGFRFHPTEEELLKYYLRKKISNIKIDLDVIPDIDLNKLEPWDIQEMCKIGTT 65
Query: 74 ---EWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAP 130
+WYF+ +DKKYPTG RTNRAT G+WKATG+DK IY IGM+KTLVFYKGRAP
Sbjct: 66 PQNDWYFYSHKDKKYPTGTRTNRATTVGFWKATGRDKTIYTNGDRIGMRKTLVFYKGRAP 125
Query: 131 RGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSE-WVVTRVFEKRNCGKKM 189
G+KS+W+MHEYRL+ + L + +H++ E G E WVV RVF+K N K M
Sbjct: 126 HGQKSDWIMHEYRLDESVLI--SSCGDHDVNVE-TCDVIGSDEGWVVCRVFKKNNLCKNM 182
Query: 190 NGS 192
S
Sbjct: 183 ISS 185
>At1g12260 unknown protein
Length = 395
Score = 177 bits (448), Expect = 1e-44
Identities = 90/165 (54%), Positives = 111/165 (66%), Gaps = 20/165 (12%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMG---ETEWYFF 78
G RF PTDEEL++ YL KKV I ++D+ K EPWDL E+ K+G +++WYFF
Sbjct: 10 GFRFHPTDEELVDYYLRKKVASKRIEIDFIKDIDLYKIEPWDLQELCKIGHEEQSDWYFF 69
Query: 79 CVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWV 138
+DKKYPTG RTNRAT AG+WKATG+DK IY +SLIGM+KTLVFYKGRAP G+KS+W+
Sbjct: 70 SHKDKKYPTGTRTNRATKAGFWKATGRDKAIYLRHSLIGMRKTLVFYKGRAPNGQKSDWI 129
Query: 139 MHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKR 183
MHEYRLE + N P+ WVV RVF+KR
Sbjct: 130 MHEYRLE----------TDENGTPQE-------EGWVVCRVFKKR 157
>At1g62700 unknown protein
Length = 394
Score = 176 bits (447), Expect = 1e-44
Identities = 128/346 (36%), Positives = 175/346 (49%), Gaps = 46/346 (13%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMG---ETEWYFF 78
G RF PTDEEL++ YL KKV I +VD+ K EP DL E+ K+G ++EWYFF
Sbjct: 10 GFRFHPTDEELVDYYLRKKVASKRIEIDIIKDVDLYKIEPCDLQELCKIGNEEQSEWYFF 69
Query: 79 CVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWV 138
+DKKYPTG RTNRAT AG+WKATG+DK IY +SLIGM+KTLVFYKGRAP G+KS+W+
Sbjct: 70 SHKDKKYPTGTRTNRATKAGFWKATGRDKAIYIRHSLIGMRKTLVFYKGRAPNGQKSDWI 129
Query: 139 MHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLGRSN 198
MHEYRLE + N P+ WVV RVF+K+ K+G +
Sbjct: 130 MHEYRLE----TSENGTPQE-------------EGWVVCRVFKKKLAA---TVRKMGDYH 169
Query: 199 SSREEPSNTNAASLWA---------PFLEFSPYN-SENKITIPDFSNEFNSFTNPNQSEK 248
SS + + S A FL YN ++ T+P N FN+ NPN K
Sbjct: 170 SSPSQHWYDDQLSFMASEIISSSPRQFLPNHHYNRHHHQQTLPCGLNAFNN-NNPNLQCK 228
Query: 249 PKTQ--YDNIVHNNETSILNISSSSKQMDVY--PLAGATVADPNLTSMAGN---SSNFFF 301
+ + Y+ +V + + + S Q+ P + + N T N SSN
Sbjct: 229 QELELHYNQMVQHQQQNHHLRESMFLQLPQLESPTSNCNSDNNNNTRNISNLQKSSNISH 288
Query: 302 SQEFSFGREFDADADISSVVYGNDMFQRWSGYQDLSPASTGLAAND 347
++ G + SS+ Y + Q + ++ L +ND
Sbjct: 289 EEQLQQGNQ-----SFSSLYYDQGVEQMTTDWRVLDKFVASQLSND 329
>At2g18060 putative NAM (no apical meristem)-like protein
Length = 365
Score = 175 bits (444), Expect = 3e-44
Identities = 89/179 (49%), Positives = 109/179 (60%), Gaps = 20/179 (11%)
Query: 15 EKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMG--- 71
E G RF PTDEEL+ YL KK+ I ++D+ + EPWDL E ++G
Sbjct: 5 ESCSVPPGFRFHPTDEELVGYYLRKKIASQKIDLDVIRDIDLYRIEPWDLQEQCRIGYEE 64
Query: 72 ETEWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPR 131
+ EWYFF +DKKYPTG RTNRAT AG+WKATG+DK +Y LIGM+KTLVFYKGRAP
Sbjct: 65 QNEWYFFSHKDKKYPTGTRTNRATMAGFWKATGRDKAVYDKTKLIGMRKTLVFYKGRAPN 124
Query: 132 GEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMN 190
G+KS+W+MHEYRLE S N P+ WVV R F+KR G+ N
Sbjct: 125 GKKSDWIMHEYRLE----SDENAPPQE-------------EGWVVCRAFKKRATGQAKN 166
>At1g33280 hypothetical protein
Length = 305
Score = 174 bits (440), Expect = 9e-44
Identities = 93/187 (49%), Positives = 115/187 (60%), Gaps = 21/187 (11%)
Query: 22 GVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGET---EWYFF 78
G RF PTDEEL++ YL KK+ F I EVD+NK EPWDL + K+G T EWYFF
Sbjct: 11 GFRFHPTDEELLHYYLKKKISYEKFEMEVIKEVDLNKIEPWDLQDRCKIGSTPQNEWYFF 70
Query: 79 CVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAPRGEKSNWV 138
+D+KYPTG RTNRAT++G+WKATG+DK I IGM+KTLVFYKGRAP G+K++W+
Sbjct: 71 SHKDRKYPTGSRTNRATHSGFWKATGRDKCIRNSYKKIGMRKTLVFYKGRAPHGQKTDWI 130
Query: 139 MHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGKKMNGSKLGRSN 198
MHEYR+E E + G WVV RVF+K+N K N SN
Sbjct: 131 MHEYRIEDT---------------EDDPCEDG---WVVCRVFKKKNLFKVGNDVGSNISN 172
Query: 199 SSREEPS 205
+ E S
Sbjct: 173 NRLEARS 179
>At1g32770 OsNAC7 protein, putative
Length = 358
Score = 173 bits (438), Expect = 1e-43
Identities = 90/177 (50%), Positives = 115/177 (64%), Gaps = 8/177 (4%)
Query: 14 NEKVKFDAGVRFFPTDEELINQYLVKKVDDNSFCAIAIAEVDMNKCEPWDLPEMAKMGET 73
N + K G RF PT+EEL++ YL KKV+ I EVD+NK EPWD+ E ++G T
Sbjct: 11 NGQSKVPPGFRFHPTEEELLHYYLRKKVNSQKIDLDVIREVDLNKLEPWDIQEECRIGST 70
Query: 74 ---EWYFFCVRDKKYPTGQRTNRATNAGYWKATGKDKEIYKGNSLIGMKKTLVFYKGRAP 130
+WYFF +DKKYPTG RTNRAT AG+WKATG+DK I IG++KTLVFYKGRAP
Sbjct: 71 PQNDWYFFSHKDKKYPTGTRTNRATVAGFWKATGRDKIICSCVRRIGLRKTLVFYKGRAP 130
Query: 131 RGEKSNWVMHEYRLEGNSLSKHNIFPEHNLFPEHNLSTHGMSEWVVTRVFEKRNCGK 187
G+KS+W+MHEYRL+ +S ++ E +S + WVV RVF K+N K
Sbjct: 131 HGQKSDWIMHEYRLDDTPMSNGYA----DVVTEDPMS-YNEEGWVVCRVFRKKNYQK 182
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.313 0.130 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,697,027
Number of Sequences: 26719
Number of extensions: 390033
Number of successful extensions: 944
Number of sequences better than 10.0: 127
Number of HSP's better than 10.0 without gapping: 106
Number of HSP's successfully gapped in prelim test: 21
Number of HSP's that attempted gapping in prelim test: 682
Number of HSP's gapped (non-prelim): 134
length of query: 352
length of database: 11,318,596
effective HSP length: 100
effective length of query: 252
effective length of database: 8,646,696
effective search space: 2178967392
effective search space used: 2178967392
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC140033.2


