

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140032.3 - phase: 0 /pseudo
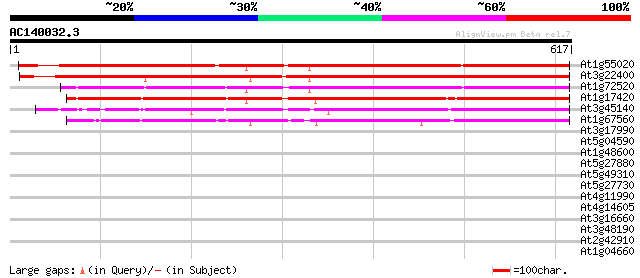
(617 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g55020 unknown protein 641 0.0
At3g22400 putative lipoxygenase 614 e-176
At1g72520 putative lipoxygenase 444 e-125
At1g17420 lipoxygenase 439 e-123
At3g45140 lipoxygenase (AtLox2) 429 e-120
At1g67560 putative lipoxygenase 391 e-109
At3g17990 unknown protein 35 0.16
At5g04590 sulphite reductase 33 0.47
At1g48600 phosphoethanolamine N-methyltransferase, putative 31 2.3
At5g27880 putative protein 30 3.0
At5g49310 importin alpha 30 5.2
At5g27730 unknown protein 30 5.2
At4g11990 unknown protein 30 5.2
At4g14605 unknown protein 29 6.8
At3g16660 unknown protein 29 6.8
At3g48190 ataxia-telangiectasia mutated protein AtATM 29 8.8
At2g42910 putative ribose phosphate pyrophosphokinase 29 8.8
At1g04660 unknown protein 29 8.8
>At1g55020 unknown protein
Length = 859
Score = 641 bits (1653), Expect = 0.0
Identities = 342/620 (55%), Positives = 427/620 (68%), Gaps = 43/620 (6%)
Query: 10 QKIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLGRNVSM 69
+K+KGTVVLM KNVLDFN + +D FLG +++
Sbjct: 19 KKVKGTVVLMKKNVLDFNDFNA----------------------SFLDRLHEFLGNKITL 56
Query: 70 QLISATKTDA-NG-KGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPGAFYIR 127
+L+S+ TD+ NG KG +GK L + + +L A + AF V F+Y+ +FG PGAF IR
Sbjct: 57 RLVSSDVTDSENGSKGKLGKAAHLEDWITTITSLTAGESAFKVTFDYETDFGYPGAFLIR 116
Query: 128 NYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAPLNH 187
N +EF L S+TLED+P V +ICNSW+Y K Y DR+FF+N TYLP +TPA L
Sbjct: 117 NSHFSEFLLKSLTLEDVPGHGRVHYICNSWIYPAKHYTTDRVFFSNKTYLPHETPATLLK 176
Query: 188 YREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRRVRS 247
YREEEL +LRG G GE KE DRVYDY YNDLG P RPVLGG+ YPYPRR R+
Sbjct: 177 YREEELVSLRGTGEGELKEWDRVYDYAYYNDLGVPPKNP---RPVLGGTQEYPYPRRGRT 233
Query: 248 GRKPTRKDPKSE------KPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIF 301
GRKPT++DP++E IYVPRDE FGHLK SDFL Y +K+++Q + P ++V
Sbjct: 234 GRKPTKEDPQTESRLPITSSLDIYVPRDERFGHLKMSDFLAYALKAIAQFIQPALEAVFD 293
Query: 302 DLNFTPNEFDSFDEVRGLFEGGIKLPT----DILSQISPLPALKEIFRTDGEQVLKFPPP 357
D TP EFDSF++V ++E GI LP D + + PL LKEIFRTDG++ LKFP P
Sbjct: 294 D---TPKEFDSFEDVLKIYEEGIDLPNQALIDSIVKNIPLEMLKEIFRTDGQKFLKFPVP 350
Query: 358 HVIKVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLAT 417
VIK K+AW TDEEF REM+AG+NP VI+LL+EFPPKS LD+ YG+QNSTITK H+
Sbjct: 351 QVIKEDKTAWRTDEEFAREMLAGLNPVVIQLLKEFPPKSKLDSESYGNQNSTITKSHIEH 410
Query: 418 NLGDITVEEALNGKKLFLLDYHDAFMPYLERIN-INAKAYATRTILFLKDDGTLKPIAIE 476
NL +TVEEAL ++LF+LD+HD MPYL R+N K YA+RT+LFLKDDGTLKP+ IE
Sbjct: 411 NLDGLTVEEALEKERLFILDHHDTLMPYLGRVNTTTTKTYASRTLLFLKDDGTLKPLVIE 470
Query: 477 LSLPHSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEP 536
LSLPH NG ++G+ S+V+ P EGV ++W LAKA V VNDS HQLISHW+ THA +EP
Sbjct: 471 LSLPHPNGDKFGAVSEVYTPG-EGVYDSLWQLAKAFVGVNDSGNHQLISHWMQTHASIEP 529
Query: 537 FIIATNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSI 596
F+IATNR LSVLHP+ KLL PHFRDT+NIN LARQ LIN GGI E T P ++E+SS
Sbjct: 530 FVIATNRQLSVLHPVFKLLEPHFRDTMNINALARQILINGGGIFEITVFPSKYAMEMSSF 589
Query: 597 VYRD-WVFTDQALPADLIKR 615
+Y++ W F DQALPA+L KR
Sbjct: 590 IYKNHWTFPDQALPAELKKR 609
>At3g22400 putative lipoxygenase
Length = 864
Score = 614 bits (1583), Expect = e-176
Identities = 326/623 (52%), Positives = 424/623 (67%), Gaps = 43/623 (6%)
Query: 11 KIKGTVVLMPKNVLDFNAITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLGRNVSMQ 70
KI+G VV+M KN+LDF ++ ++D LGR VS+
Sbjct: 12 KIEGEVVVMKKNLLDFK----------------------DVMASLLDRVNELLGRRVSLH 49
Query: 71 LISATKTD-ANGK-GLVGKETFLSKHLPQLPT-LGARQDAFSVFFEYDANFGIPGAFYIR 127
LIS+ + D AN K G +GK L K + ++ T + A + AF V F++D + G P AF I+
Sbjct: 50 LISSHQPDPANEKRGRLGKAAHLEKWVTKIKTSVTAEETAFGVTFDWDESMGPPAAFVIK 109
Query: 128 NYTQAEFFLVSVTLEDIPNRE----SVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPA 183
N+ ++F+L S+TL P+ E ++ FICNSW+Y Y+ DR+FF+N YLPS+TP
Sbjct: 110 NHHHSQFYLKSLTLRGFPDGEGGATAIHFICNSWIYPNHRYRSDRVFFSNKAYLPSETPE 169
Query: 184 PLNHYREEELQTLRGDGTG-ERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYP 242
+ REEEL+ LRG+ G E KE DRVYDY YNDLG PD G VRPVLGGS PYP
Sbjct: 170 LIKELREEELKNLRGNEKGGEFKEWDRVYDYAYYNDLGAPDKGPDSVRPVLGGSPELPYP 229
Query: 243 RRVRSGRKPTRKDPKSEKPGV-----IYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFK 297
RR ++GRK T+ DPKSE IYVPRDE F H+K SDFL Y +KS++Q ++P
Sbjct: 230 RRGKTGRKSTKSDPKSESRLALLNLNIYVPRDERFSHVKFSDFLAYALKSVTQVLVPEIA 289
Query: 298 SVIFDLNFTPNEFDSFDEVRGLFEGGIKLPT----DILSQISPLPALKEIFRTDGEQVLK 353
SV + T NEFDSF++V L++G IKL L + P +E+ R DGE+ LK
Sbjct: 290 SVC---DKTINEFDSFEDVFHLYDGSIKLANGHTISKLRDVIPWEMFRELVRNDGERFLK 346
Query: 354 FPPPHVIKVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKE 413
+P P ++K S+SAW TDEEF REM+AG+NP VI LQEFPPKS LD+ YG+Q+S+I E
Sbjct: 347 YPLPDILKESRSAWRTDEEFAREMLAGLNPVVISRLQEFPPKSCLDSAKYGNQHSSIRTE 406
Query: 414 HLATNLGDITVEEALNGKKLFLLDYHDAFMPYLERIN-INAKAYATRTILFLKDDGTLKP 472
H+ +N+ + V+EAL KL++LD+HDA MPYL RIN N K YATRT+L L+ DGTLKP
Sbjct: 407 HIESNMNGLNVQEALEQNKLYILDHHDALMPYLTRINSTNTKTYATRTLLLLQADGTLKP 466
Query: 473 IAIELSLPHSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHA 532
+AIELSLPH+ G YGS SKVF PA++GVE ++W LAKA+ VNDS YHQLISHWL THA
Sbjct: 467 LAIELSLPHAQGESYGSVSKVFTPAEKGVEGSVWQLAKAYAAVNDSGYHQLISHWLQTHA 526
Query: 533 VVEPFIIATNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVE 592
V+EPFIIA+NR LSV+HPI+KLL PHFRDT+NIN LAR LIN+ G++E+T P ++E
Sbjct: 527 VIEPFIIASNRQLSVVHPIHKLLHPHFRDTMNINALARHVLINSDGVLERTVFPSRYAME 586
Query: 593 ISSIVYRDWVFTDQALPADLIKR 615
+SS +Y++WVFT+QALP DL+KR
Sbjct: 587 MSSSIYKNWVFTEQALPKDLLKR 609
>At1g72520 putative lipoxygenase
Length = 926
Score = 444 bits (1142), Expect = e-125
Identities = 250/571 (43%), Positives = 345/571 (59%), Gaps = 20/571 (3%)
Query: 56 VDTATAFLGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYD 115
+D T +GRNV ++L+S T+ D + + K + A + ++ F D
Sbjct: 114 LDAFTDKIGRNVVLELMS-TQVDPKTNEPKKSKAAVLKDWSKKSNSKAERVHYTAEFTVD 172
Query: 116 ANFGIPGAFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDT 175
+ FG PGA + N Q EFFL S+T+E V F CNSWV + K + RI FTN
Sbjct: 173 SAFGSPGAITVTNKHQKEFFLESITIEGFACGP-VHFPCNSWVQSQKDHPSKRILFTNQP 231
Query: 176 YLPSQTPAPLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGG 235
YLPS+TP+ L RE+EL+ LRG+G GERK +DR+YDYD+YND+GNPD L RP LGG
Sbjct: 232 YLPSETPSGLRTLREKELENLRGNGKGERKLSDRIYDYDVYNDIGNPDISRELARPTLGG 291
Query: 236 SSTYPYPRRVRSGRKPTRKDPKSE----KPGVIYVPRDENFGHLKSSDFLMYGIKSLSQN 291
+PYPRR R+GR T D SE KP +YVPRDE F K + F +K++ N
Sbjct: 292 RE-FPYPRRCRTGRSSTDTDMMSERRVEKPLPMYVPRDEQFEESKQNTFAACRLKAVLHN 350
Query: 292 VIPLFKSVIFDLNFTPNEFDSFDEVRGLFEGGIKLPT---DILSQISPLPALKEIFRTDG 348
+IP K+ I +F +F E+ L++ G+ L D + + PLP + +
Sbjct: 351 LIPSLKASIL-----AEDFANFGEIDSLYKEGLLLKLGFQDDMFKKFPLPKIVTTLQKSS 405
Query: 349 EQVLKFPPPHVIKVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQ-N 407
E +L++ P ++ K AW+ D+EF R+ +AG+NP I + +PP S LD +YG +
Sbjct: 406 EGLLRYDTPKIVSKDKYAWLRDDEFARQAIAGINPVNIERVTSYPPVSNLDPEIYGPGLH 465
Query: 408 STITKEHLATNLGDITVEEALNGKKLFLLDYHDAFMPYLERINI--NAKAYATRTILFLK 465
S +T++H+ L +TV++AL +LF++DYHD ++P+L+RIN KAYATRTILFL
Sbjct: 466 SALTEDHIIGQLDGLTVQQALETNRLFMVDYHDIYLPFLDRINALDGRKAYATRTILFLT 525
Query: 466 DDGTLKPIAIELSLPHSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLIS 525
GTLKPIAIELSLP + S+ V P D + +W LAKAHV ND+ HQL++
Sbjct: 526 RLGTLKPIAIELSLPSQSSSNQKSKRVVTPPVD-ATSNWMWQLAKAHVGSNDAGVHQLVN 584
Query: 526 HWLNTHAVVEPFIIATNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFL 585
HWL THA +EPFI+A +R LS +HPI KLL PH R T+ IN +ARQ+LI+A G+IE F
Sbjct: 585 HWLRTHACLEPFILAAHRQLSAMHPIFKLLDPHMRYTLEINAVARQTLISADGVIESCFT 644
Query: 586 PGPNSVEISSIVYRD-WVFTDQALPADLIKR 615
G +EISS Y++ W F + LPADLI+R
Sbjct: 645 AGQYGLEISSAAYKNKWRFDMEGLPADLIRR 675
>At1g17420 lipoxygenase
Length = 919
Score = 439 bits (1130), Expect = e-123
Identities = 248/564 (43%), Positives = 344/564 (60%), Gaps = 21/564 (3%)
Query: 63 LGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPG 122
+GRN+ ++LIS T+ D K + K + A + ++ F DA FG PG
Sbjct: 115 IGRNIVLELIS-TQLDPKTKLPKKSNAAVLKDWSKKSKTKAERVHYTAEFTVDAAFGSPG 173
Query: 123 AFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTP 182
A + N Q EFFL S+T+E V F CNSWV + K + RIFFTN YLP++TP
Sbjct: 174 AITVMNKHQKEFFLESITIEGFA-LGPVHFPCNSWVQSQKDHPDKRIFFTNQPYLPNETP 232
Query: 183 APLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYP 242
+ L RE+EL+ LRGDG+G RK +DR+YD+D+YNDLGNPD L RP LGG PYP
Sbjct: 233 SGLRVLREKELKNLRGDGSGVRKLSDRIYDFDVYNDLGNPDKSSELSRPKLGGKEV-PYP 291
Query: 243 RRVRSGRKPTRKDPKSE----KPGVIYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKS 298
RR R+GR+ T D +E KP +YVPRDE F K F +K++ ++IP K+
Sbjct: 292 RRCRTGRQSTVSDKDAESRVEKPLPMYVPRDEQFEESKQDTFAAGRLKAVLHHLIPSLKA 351
Query: 299 VIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQIS---PLP-ALKEIFRTDGEQVLKF 354
I +F F E+ L++ G+ L I PLP + + + + +LK+
Sbjct: 352 SI-----VAEDFADFGEIDRLYKEGLLLKLGFQDDIFKKFPLPKVVVDTLQESTKGLLKY 406
Query: 355 PPPHVIKVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEH 414
P ++ K+AW+ D+EF R+ +AG+NP I ++ FPP S LD +YG Q+S +T +H
Sbjct: 407 DTPKILSKDKNAWLRDDEFARQAIAGINPVNIERVKTFPPVSNLDPKIYGPQHSALTDDH 466
Query: 415 LATNLGDITVEEALNGKKLFLLDYHDAFMPYLERINI--NAKAYATRTILFLKDDGTLKP 472
+ +L +V++AL +L++LDYHD F+P+L+RIN KAYATRTI FL GTLKP
Sbjct: 467 IIGHLDGFSVQQALEENRLYMLDYHDIFLPFLDRINALDGRKAYATRTIFFLTRLGTLKP 526
Query: 473 IAIELSLPHSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHA 532
+AIELSLP +G ++ S+ +V P + + +W LAKAHV ND+ HQL++HWL THA
Sbjct: 527 VAIELSLP-PHGPKHRSK-RVLTPPVDATSNWMWQLAKAHVSSNDAGVHQLVNHWLRTHA 584
Query: 533 VVEPFIIATNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFLPGPNSVE 592
+EPFI+A +R LS +HPI KLL PH R T+ IN LARQSLI+A G+IE F G +E
Sbjct: 585 CLEPFILAAHRQLSAMHPIFKLLDPHMRYTLEINALARQSLISADGVIEGGFTAGAYGME 644
Query: 593 ISSIVYR-DWVFTDQALPADLIKR 615
+S+ Y+ W F + LPADLI+R
Sbjct: 645 MSAAAYKSSWRFDMEGLPADLIRR 668
>At3g45140 lipoxygenase (AtLox2)
Length = 896
Score = 429 bits (1104), Expect = e-120
Identities = 255/600 (42%), Positives = 353/600 (58%), Gaps = 38/600 (6%)
Query: 29 ITSVGKGGVINTAGNIIGGVTGIVGGVVDTATAFLGRNVSMQLISATKTDANGKGLVGKE 88
I ++ G I + G+T G +D GR++ ++LISA KTD + E
Sbjct: 70 IQNIKVKGYITAQEEFLEGITWSRG--LDDIADIRGRSLLVELISA-KTDQR----ITVE 122
Query: 89 TFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPGAFYIRNYTQAEFFLVSVTLEDIPNRE 148
+ + + P + + FE +FG GA I+N + FL V L+ +P
Sbjct: 123 DYAQRVWAEAP-----DEKYECEFEMPEDFGPVGAIKIQNQYHRQLFLKGVELK-LPGG- 175
Query: 149 SVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTPAPLNHYREEELQTLRG---DGTGERK 205
S+ F C SWV RIFF++ +YLPSQTP PL YR+EEL+TL+G + GE
Sbjct: 176 SITFTCESWVAPKSVDPTKRIFFSDKSYLPSQTPEPLKKYRKEELETLQGKNREEVGEFT 235
Query: 206 EADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYPRRVRSGRKPTRKDPKSEKP--GV 263
+ +R+YDYD+YND+G+PD L RPV+GG T+PYPRR ++GRKP DP SE+ G
Sbjct: 236 KFERIYDYDVYNDVGDPDNDPELARPVIGGL-THPYPRRCKTGRKPCETDPSSEQRYGGE 294
Query: 264 IYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKSVIFDLNFTPNE-FDSFDEVRGLFEG 322
YVPRDE F K + F + + ++ P +SV+ +P E F F ++ LFE
Sbjct: 295 FYVPRDEEFSTAKGTSFTGKAVLAALPSIFPQIESVLL----SPQEPFPHFKAIQNLFEE 350
Query: 323 GIKLPTDILSQISPLPALKEIFRTDGE---QVLKFPPPHVIKVSKSAWMTDEEFGREMVA 379
GI+LP D LP L I + GE +L+F P +I + +W+ D+EF R+ +A
Sbjct: 351 GIQLPKDA----GLLPLLPRIIKALGEAQDDILQFDAPVLINRDRFSWLRDDEFARQTLA 406
Query: 380 GVNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNL-GDITVEEALNGKKLFLLDY 438
G+NP I+L++E+P S LD VYGD S IT E + + G++TV+EAL K+LF+LDY
Sbjct: 407 GLNPYSIQLVEEWPLISKLDPAVYGDPTSLITWEIVEREVKGNMTVDEALKNKRLFVLDY 466
Query: 439 HDAFMPYLERINI--NAKAYATRTILFLKDDGTLKPIAIELSLPHSNGVQYGSESKVFLP 496
HD +PY+ ++ N YA+RT+ FL DD TL+P+AIEL+ P + + +VF P
Sbjct: 467 HDLLLPYVNKVRELNNTTLYASRTLFFLSDDSTLRPVAIELTCPPN--INKPQWKQVFTP 524
Query: 497 ADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLHPINKLLF 556
+ +W LAK H I +D+ YHQLISHWL THA EP+IIA NR LS +HPI +LL
Sbjct: 525 GYDATSCWLWNLAKTHAISHDAGYHQLISHWLRTHACTEPYIIAANRQLSAMHPIYRLLH 584
Query: 557 PHFRDTININGLARQSLINAGGIIEQTFLPGPNSVEISSIVY-RDWVFTDQALPADLIKR 615
PHFR T+ IN ARQSL+N GGIIE F PG ++E+SS VY + W F + LPADLIKR
Sbjct: 585 PHFRYTMEINARARQSLVNGGGIIETCFWPGKYALELSSAVYGKLWRFDQEGLPADLIKR 644
>At1g67560 putative lipoxygenase
Length = 917
Score = 391 bits (1005), Expect = e-109
Identities = 234/573 (40%), Positives = 327/573 (56%), Gaps = 38/573 (6%)
Query: 63 LGRNVSMQLISATKTDANGKGLVGKETFLSKHLPQLPTLGARQDAFSVFFEYDANFGIPG 122
+G+ + +QL+S GKG E+ + LP+ R F+ F NFG PG
Sbjct: 111 IGQGMLIQLVSEEIDPETGKGRKSLESPVMG-LPKA-VKDPRYLVFTADFTVPINFGKPG 168
Query: 123 AFYIRNYTQAEFFLVSVTLEDIPNRESVQFICNSWVYNFKSYKKDRIFFTNDTYLPSQTP 182
A + N E L + +ED + +++ F N+W+++ + RI F + LPS+TP
Sbjct: 169 AILVTNLLSTEICLSEIIIED--STDTILFPANTWIHSKNDNPQARIIFRSQPCLPSETP 226
Query: 183 APLNHYREEELQTLRGDGTGERKEADRVYDYDIYNDLGNPDGGDALVRPVLGGSSTYPYP 242
+ RE++L ++RGDG GERK +R+YDYD+YNDLG+P + VRPVLG T PYP
Sbjct: 227 DGIKELREKDLVSVRGDGKGERKPHERIYDYDVYNDLGDPRKTER-VRPVLGVPET-PYP 284
Query: 243 RRVRSGRKPTRKDPKSEKPGV----IYVPRDENFGHLKSSDFLMYGIKSLSQNVIPLFKS 298
RR R+GR KDP E G YVPRDE F +K F K+L N++P +
Sbjct: 285 RRCRTGRPLVSKDPPCESRGKEKEEFYVPRDEVFEEIKRDTFRAGRFKALFHNLVPSIAA 344
Query: 299 VIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISP--------LPALKEIFRTDGEQ 350
+ +L+ F F ++ L++ I +L P + E
Sbjct: 345 ALSNLDIP---FTCFSDIDNLYKSNI-----VLGHTEPKDTGLGGFIGGFMNGILNVTET 396
Query: 351 VLKFPPPHVIKVSKSAWMTDEEFGREMVAGVNPNVIRLLQEFPPKSTLDTTVYGDQNSTI 410
+LK+ P VIK + AW+ D EFGR+ +AGVNP I LL+E P +S LD +YG Q S +
Sbjct: 397 LLKYDTPAVIKWDRFAWLRDNEFGRQALAGVNPVNIELLKELPIRSNLDPALYGPQESVL 456
Query: 411 TKEHLATNLGDI--TVEEALNGKKLFLLDYHDAFMPYLERINI----NAKAYATRTILFL 464
T+E +A + T+E+AL K+LFL+DYHD +P++E+IN K YA+RTI F
Sbjct: 457 TEEIIAREVEHYGTTIEKALEEKRLFLVDYHDILLPFVEKINSIKEDPRKTYASRTIFFY 516
Query: 465 KDDGTLKPIAIELSLPHSNGVQYGSESK-VFLPADEGVESTIWLLAKAHVIVNDSCYHQL 523
+G L+P+AIELSLP + SE+K V+ + IW LAKAHV ND+ HQL
Sbjct: 517 SKNGALRPLAIELSLPPTAE----SENKFVYTHGHDATTHWIWKLAKAHVCSNDAGVHQL 572
Query: 524 ISHWLNTHAVVEPFIIATNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQT 583
++HWL THA +EP+IIATNR LS +HP+ KLL PH R T+ IN AR+SLIN GGIIE
Sbjct: 573 VNHWLRTHASMEPYIIATNRQLSTMHPVYKLLHPHMRYTLEINARARKSLINGGGIIESC 632
Query: 584 FLPGPNSVEISSIVYRD-WVFTDQALPADLIKR 615
F PG ++E+SS Y+ W F + LPADL++R
Sbjct: 633 FTPGKYAMELSSAAYKSMWRFDMEGLPADLVRR 665
>At3g17990 unknown protein
Length = 165
Score = 34.7 bits (78), Expect = 0.16
Identities = 39/144 (27%), Positives = 59/144 (40%), Gaps = 27/144 (18%)
Query: 383 PNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEEALNGKKLFLLDYHDAF 442
P V+ LL + KS L+ L +G T E A +L LD+ D
Sbjct: 16 PEVLSLLPPYEGKSVLE---------------LGAGIGRFTGELAQKAGELIALDFIDNV 60
Query: 443 MPYLERININAKAYATRTILFLKDDGTLKPIAI---ELSLPHSNGV-QYGSESKVFLPAD 498
+ E IN + K + F+ D T + I L L SN + Y S+ +V L A+
Sbjct: 61 IKKNESINGHYK-----NVKFMCADVTSPDLKITDGSLDLIFSNWLLMYLSDKEVELLAE 115
Query: 499 EGVESTIWLLAKAHVIVNDSCYHQ 522
V W+ ++ +SC+HQ
Sbjct: 116 RMVG---WIKVGGYIFFRESCFHQ 136
>At5g04590 sulphite reductase
Length = 642
Score = 33.1 bits (74), Expect = 0.47
Identities = 49/226 (21%), Positives = 87/226 (37%), Gaps = 34/226 (15%)
Query: 222 PDGGDALVRPVLGGSSTYPYPRRVRSGRKPT----RKDPKSEKPGVIYVPRDE-----NF 272
P+ ++ V+ + S Y R R GR + K+P + P +Y+ D+
Sbjct: 100 PNVNESAVQLIKFHGSYQQYNREERGGRSYSFMLRTKNPSGKVPNQLYLTMDDLADEFGI 159
Query: 273 GHLKSSD---FLMYGIKSLSQNVIPLFKSVIFDLNFTPNEFDSFDE---------VRGLF 320
G L+ + F ++G+ L QN+ + S+I ++ T + V+ +
Sbjct: 160 GTLRLTTRQTFQLHGV--LKQNLKTVMSSIIKNMGSTLGACGDLNRNVLAPAAPYVKKDY 217
Query: 321 EGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKSAWMTDEEFGREMVAG 380
+ +I + +SP DGEQ + PP V+K D G V
Sbjct: 218 LFAQETADNIAALLSPQSGFYYDMWVDGEQFMTAEPPEVVKA-----RNDNSHGTNFVD- 271
Query: 381 VNPNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEE 426
+P I Q P K + TV D + + L ++G + V +
Sbjct: 272 -SPEPIYGTQFLPRKFKVAVTVPTDNSVDL----LTNDIGVVVVSD 312
>At1g48600 phosphoethanolamine N-methyltransferase, putative
Length = 475
Score = 30.8 bits (68), Expect = 2.3
Identities = 34/144 (23%), Positives = 61/144 (41%), Gaps = 27/144 (18%)
Query: 383 PNVIRLLQEFPPKSTLDTTVYGDQNSTITKEHLATNLGDITVEEALNGKKLFLLDYHDAF 442
P V+ L+ + KS L+ L +G T E A ++ LD+ ++
Sbjct: 27 PEVLSLIPPYEGKSVLE---------------LGAGIGRFTGELAQKAGEVIALDFIESA 71
Query: 443 MPYLERININAKAYATRTILFLKDDGTLKPIAIE---LSLPHSNGV-QYGSESKVFLPAD 498
+ E +N + K I F+ D T + I+ + L SN + Y S+ +V L A+
Sbjct: 72 IQKNESVNGHYK-----NIKFMCADVTSPDLKIKDGSIDLIFSNWLLMYLSDKEVELMAE 126
Query: 499 EGVESTIWLLAKAHVIVNDSCYHQ 522
+ W+ ++ +SC+HQ
Sbjct: 127 RMIG---WVKPGGYIFFRESCFHQ 147
>At5g27880 putative protein
Length = 278
Score = 30.4 bits (67), Expect = 3.0
Identities = 11/30 (36%), Positives = 18/30 (59%)
Query: 336 PLPALKEIFRTDGEQVLKFPPPHVIKVSKS 365
PLP+ ++ ++ VL FPPP K+S +
Sbjct: 199 PLPSYNNLYPSNSRNVLPFPPPQTTKLSSN 228
>At5g49310 importin alpha
Length = 519
Score = 29.6 bits (65), Expect = 5.2
Identities = 23/75 (30%), Positives = 35/75 (46%), Gaps = 5/75 (6%)
Query: 320 FEGGIKLPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKSAWMTDEEFGREMVA 379
F G P D++ + LP LK + +D EQVL +S + ++E + A
Sbjct: 227 FRGKPSPPFDLVKHV--LPVLKRLVYSDDEQVL---IDACWALSNLSDASNENIQSVIEA 281
Query: 380 GVNPNVIRLLQEFPP 394
GV P ++ LLQ P
Sbjct: 282 GVVPRLVELLQHASP 296
>At5g27730 unknown protein
Length = 472
Score = 29.6 bits (65), Expect = 5.2
Identities = 27/101 (26%), Positives = 45/101 (43%), Gaps = 6/101 (5%)
Query: 490 ESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNTHAVVEPFIIATNRHLSVLH 549
E + L + + STI + H+I++ + + HW++T V+ + H + L
Sbjct: 290 EPEGILSSISAILSTIIGVHFGHIILHLKGHSARLKHWISTGLVL--LALGLTLHFTHLM 347
Query: 550 PINKLL--FPHFRDTININGLARQSLINAGGIIE--QTFLP 586
P+NK L F + T L SL + I+E FLP
Sbjct: 348 PLNKQLYSFSYICVTSGAAALVFSSLYSLVDILEWKHMFLP 388
>At4g11990 unknown protein
Length = 501
Score = 29.6 bits (65), Expect = 5.2
Identities = 23/85 (27%), Positives = 36/85 (42%), Gaps = 4/85 (4%)
Query: 326 LPTDILSQISPLPALKEIFRTDGEQVLKFPPPHVIKVSKSAWMTDEEFGREMVAGVNPNV 385
LPTD+ S++S L R + L+FP P +K SK + + G E + N
Sbjct: 418 LPTDLFSKLS----LSSELRPNNGSRLRFPQPEQVKGSKENRLNSFQAGNERTSKFMGNP 473
Query: 386 IRLLQEFPPKSTLDTTVYGDQNSTI 410
+ P S T+ G +T+
Sbjct: 474 MVQPSVVPETSRQWTSRSGKLEATV 498
>At4g14605 unknown protein
Length = 444
Score = 29.3 bits (64), Expect = 6.8
Identities = 12/38 (31%), Positives = 23/38 (59%)
Query: 413 EHLATNLGDITVEEALNGKKLFLLDYHDAFMPYLERIN 450
+HL + L + L G++L L+ D+ +PYLE+++
Sbjct: 97 DHLVSRLHSVHKARYLVGRELTTLEIRDSLIPYLEQLH 134
>At3g16660 unknown protein
Length = 180
Score = 29.3 bits (64), Expect = 6.8
Identities = 11/24 (45%), Positives = 17/24 (70%)
Query: 33 GKGGVINTAGNIIGGVTGIVGGVV 56
G GG+ G ++GG+ G+VGG+V
Sbjct: 33 GSGGLGGLIGGLVGGLGGLVGGLV 56
>At3g48190 ataxia-telangiectasia mutated protein AtATM
Length = 3856
Score = 28.9 bits (63), Expect = 8.8
Identities = 24/115 (20%), Positives = 46/115 (39%), Gaps = 11/115 (9%)
Query: 471 KPIAIELSLPHSNGVQYGSESKVFLPADEGVESTIWLLAKAHVIVNDSCYHQLISHWLNT 530
K IA E+S +NG ++ F +D+ + + +W L + L+S +L+
Sbjct: 2384 KLIASEVSQEDTNG----ETAETFWQSDDEIVNAVWTLVRVSASDEADSMRLLVSDFLSR 2439
Query: 531 HAVVEPFIIATNRHLSVLHPINKLLFPHFRDTININGLARQSLINAGGIIEQTFL 585
+ +P H V H L+ H N ++ + GI ++T +
Sbjct: 2440 IGIRDP-------HTVVFHLPGNLVSMHGLQGFGHNTGSKVRSLTENGISDETLI 2487
>At2g42910 putative ribose phosphate pyrophosphokinase
Length = 337
Score = 28.9 bits (63), Expect = 8.8
Identities = 18/50 (36%), Positives = 24/50 (48%)
Query: 299 VIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPALKEIFRTDG 348
VI+D++ F D+V LFE GI L T L Q+ + F DG
Sbjct: 155 VIYDIHALQERFYFADQVLPLFETGIPLLTKRLQQLPETEKVIVAFPDDG 204
>At1g04660 unknown protein
Length = 212
Score = 28.9 bits (63), Expect = 8.8
Identities = 14/29 (48%), Positives = 18/29 (61%), Gaps = 1/29 (3%)
Query: 29 ITSVGKGGVINTAGNIIGGVTGIVGGVVD 57
+ +GK G I G I GG G+VGGV+D
Sbjct: 182 VGGLGKAGGIGVGGGI-GGGHGVVGGVID 209
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.139 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,718,026
Number of Sequences: 26719
Number of extensions: 693358
Number of successful extensions: 1626
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 1561
Number of HSP's gapped (non-prelim): 23
length of query: 617
length of database: 11,318,596
effective HSP length: 105
effective length of query: 512
effective length of database: 8,513,101
effective search space: 4358707712
effective search space used: 4358707712
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140032.3


