

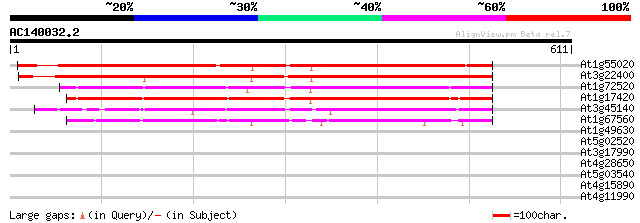
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140032.2 - phase: 0 /pseudo
(611 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g55020 unknown protein 524 e-149
At3g22400 putative lipoxygenase 491 e-139
At1g72520 putative lipoxygenase 348 4e-96
At1g17420 lipoxygenase 347 1e-95
At3g45140 lipoxygenase (AtLox2) 327 9e-90
At1g67560 putative lipoxygenase 286 2e-77
At1g49630 putative hydrogenase (At1g49630) 33 0.46
At5g02520 unknown protein 32 1.0
At3g17990 unknown protein 32 1.3
At4g28650 receptor protein kinase-like protein 30 3.0
At5g03540 unknown protein (At5g03540) 30 3.9
At4g15890 hypothetical protein 29 6.7
At4g11990 unknown protein 29 8.7
>At1g55020 unknown protein
Length = 859
Score = 524 bits (1349), Expect = e-149
Identities = 278/530 (52%), Positives = 356/530 (66%), Gaps = 42/530 (7%)
Query: 9 QKIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATAFLGRNVSM 68
+K+KGTVVLM KNVLDFN + +D FLG +++
Sbjct: 19 KKVKGTVVLMKKNVLDFNDFNA----------------------SFLDRLHEFLGNKITL 56
Query: 69 QLISATKTDASG--KGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPGAFYIR 126
+L+S+ TD+ KG +GK L + + +L A + AF + F+Y+ +FG PGAF IR
Sbjct: 57 RLVSSDVTDSENGSKGKLGKAAHLEDWITTITSLTAGESAFKVTFDYETDFGYPGAFLIR 116
Query: 127 NYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPAPLNH 186
N +EF L +TLED+P G V + CNSW+Y K Y +R+FF+N YLP +TPA L
Sbjct: 117 NSHFSEFLLKSLTLEDVPGHGRVHYICNSWIYPAKHYTTDRVFFSNKTYLPHETPATLLK 176
Query: 187 FREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVRT 246
+REEEL +LRG G GE KEWDR+YDY YNDLG P RP+LGG+ +PYPRR RT
Sbjct: 177 YREEELVSLRGTGEGELKEWDRVYDYAYYNDLGVPPKNP---RPVLGGTQEYPYPRRGRT 233
Query: 247 GRKPTRKDLKSEKPGAI------YVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIF 300
GRKPT++D ++E I YVPRDE FGHLK SDFL Y +K+++Q + P L++V
Sbjct: 234 GRKPTKEDPQTESRLPITSSLDIYVPRDERFGHLKMSDFLAYALKAIAQFIQPALEAVFD 293
Query: 301 DLNFTPNEFDSFDEVRGLFEGGIKLPT----DILSQISPLPALKEILRTDGEQALKFPPP 356
D TP EFDSF++V ++E GI LP D + + PL LKEI RTDG++ LKFP P
Sbjct: 294 D---TPKEFDSFEDVLKIYEEGIDLPNQALIDSIVKNIPLEMLKEIFRTDGQKFLKFPVP 350
Query: 357 QVIRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLET 416
QVI+ K+AW TDEEF REMLAG+NP VI+LL+EFPPKS LD+ YG+QNSTITK H+E
Sbjct: 351 QVIKEDKTAWRTDEEFAREMLAGLNPVVIQLLKEFPPKSKLDSESYGNQNSTITKSHIEH 410
Query: 417 NLGDITVEEALDGKRLFLLDYHDAFMPYLERIN-LNAKAYATRTILFLQDDGTLKPLAIE 475
NL +TVEEAL+ +RLF+LD+HD MPYL R+N K YA+RT+LFL+DDGTLKPL IE
Sbjct: 411 NLDGLTVEEALEKERLFILDHHDTLMPYLGRVNTTTTKTYASRTLLFLKDDGTLKPLVIE 470
Query: 476 LSLPHSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
LSLPH NG ++GA S+V+ P EGV ++W LAKA V VNDS HQL+SH
Sbjct: 471 LSLPHPNGDKFGAVSEVYTPG-EGVYDSLWQLAKAFVGVNDSGNHQLISH 519
>At3g22400 putative lipoxygenase
Length = 864
Score = 491 bits (1263), Expect = e-139
Identities = 262/534 (49%), Positives = 352/534 (65%), Gaps = 43/534 (8%)
Query: 10 KIKGTVVLMPKNVLDFNAITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATAFLGRNVSMQ 69
KI+G VV+M KN+LDF ++ ++D LGR VS+
Sbjct: 12 KIEGEVVVMKKNLLDFK----------------------DVMASLLDRVNELLGRRVSLH 49
Query: 70 LISATKTDASG--KGLVGKETFLSKHLPQLPT-LGARQDAFSIFFEYDANFGIPGAFYIR 126
LIS+ + D + +G +GK L K + ++ T + A + AF + F++D + G P AF I+
Sbjct: 50 LISSHQPDPANEKRGRLGKAAHLEKWVTKIKTSVTAEETAFGVTFDWDESMGPPAAFVIK 109
Query: 127 NYTQAEFFLVRVTLEDIPN----RGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPA 182
N+ ++F+L +TL P+ ++ F CNSW+Y Y+++R+FF+N AYLPS+TP
Sbjct: 110 NHHHSQFYLKSLTLRGFPDGEGGATAIHFICNSWIYPNHRYRSDRVFFSNKAYLPSETPE 169
Query: 183 PLNHFREEELQNLRGDGTG-ERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYP 241
+ REEEL+NLRG+ G E KEWDR+YDY YNDLG PD G VRP+LGGS PYP
Sbjct: 170 LIKELREEELKNLRGNEKGGEFKEWDRVYDYAYYNDLGAPDKGPDSVRPVLGGSPELPYP 229
Query: 242 RRVRTGRKPTRKDLKSEKPGA-----IYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQ 296
RR +TGRK T+ D KSE A IYVPRDE F H+K SDFL Y +KS++Q ++P +
Sbjct: 230 RRGKTGRKSTKSDPKSESRLALLNLNIYVPRDERFSHVKFSDFLAYALKSVTQVLVPEIA 289
Query: 297 SVIFDLNFTPNEFDSFDEVRGLFEGGIKLPT----DILSQISPLPALKEILRTDGEQALK 352
SV + T NEFDSF++V L++G IKL L + P +E++R DGE+ LK
Sbjct: 290 SVC---DKTINEFDSFEDVFHLYDGSIKLANGHTISKLRDVIPWEMFRELVRNDGERFLK 346
Query: 353 FPPPQVIRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKE 412
+P P +++ S+SAW TDEEF REMLAG+NP VI LQEFPPKS LD+ YG+Q+S+I E
Sbjct: 347 YPLPDILKESRSAWRTDEEFAREMLAGLNPVVISRLQEFPPKSCLDSAKYGNQHSSIRTE 406
Query: 413 HLETNLGDITVEEALDGKRLFLLDYHDAFMPYLERIN-LNAKAYATRTILFLQDDGTLKP 471
H+E+N+ + V+EAL+ +L++LD+HDA MPYL RIN N K YATRT+L LQ DGTLKP
Sbjct: 407 HIESNMNGLNVQEALEQNKLYILDHHDALMPYLTRINSTNTKTYATRTLLLLQADGTLKP 466
Query: 472 LAIELSLPHSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
LAIELSLPH+ G YG+ SKVF PA +GVE ++W LAKA+ VNDS YHQL+SH
Sbjct: 467 LAIELSLPHAQGESYGSVSKVFTPAEKGVEGSVWQLAKAYAAVNDSGYHQLISH 520
>At1g72520 putative lipoxygenase
Length = 926
Score = 348 bits (894), Expect = 4e-96
Identities = 202/481 (41%), Positives = 283/481 (57%), Gaps = 19/481 (3%)
Query: 55 VDTATAFLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYD 114
+D T +GRNV ++L+S T+ D + + K + A + ++ F D
Sbjct: 114 LDAFTDKIGRNVVLELMS-TQVDPKTNEPKKSKAAVLKDWSKKSNSKAERVHYTAEFTVD 172
Query: 115 ANFGIPGAFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDA 174
+ FG PGA + N Q EFFL +T+E G V F CNSWV + K + + RI FTN
Sbjct: 173 SAFGSPGAITVTNKHQKEFFLESITIEGFAC-GPVHFPCNSWVQSQKDHPSKRILFTNQP 231
Query: 175 YLPSQTPAPLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGG 234
YLPS+TP+ L RE+EL+NLRG+G GERK DRIYDYDVYND+GNPD L RP LGG
Sbjct: 232 YLPSETPSGLRTLREKELENLRGNGKGERKLSDRIYDYDVYNDIGNPDISRELARPTLGG 291
Query: 235 SSTHPYPRRVRTGRKPTRKDLKS----EKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQD 290
PYPRR RTGR T D+ S EKP +YVPRDE F K + F +K++ +
Sbjct: 292 RE-FPYPRRCRTGRSSTDTDMMSERRVEKPLPMYVPRDEQFEESKQNTFAACRLKAVLHN 350
Query: 291 VLPLLQSVIFDLNFTPNEFDSFDEVRGLFEGGIKLP---TDILSQISPLPALKEILRTDG 347
++P L++ I +F +F E+ L++ G+ L D + + PLP + L+
Sbjct: 351 LIPSLKASIL-----AEDFANFGEIDSLYKEGLLLKLGFQDDMFKKFPLPKIVTTLQKSS 405
Query: 348 EQALKFPPPQVIRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYG-DQN 406
E L++ P+++ K AW D+EF R+ +AG+NP I + +PP S LD +YG +
Sbjct: 406 EGLLRYDTPKIVSKDKYAWLRDDEFARQAIAGINPVNIERVTSYPPVSNLDPEIYGPGLH 465
Query: 407 STITKEHLETNLGDITVEEALDGKRLFLLDYHDAFMPYLERINL--NAKAYATRTILFLQ 464
S +T++H+ L +TV++AL+ RLF++DYHD ++P+L+RIN KAYATRTILFL
Sbjct: 466 SALTEDHIIGQLDGLTVQQALETNRLFMVDYHDIYLPFLDRINALDGRKAYATRTILFLT 525
Query: 465 DDGTLKPLAIELSLPHSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMS 524
GTLKP+AIELSLP S +V P + + +W LAKAHV ND+ HQL++
Sbjct: 526 RLGTLKPIAIELSLP-SQSSSNQKSKRVVTPPVDATSNWMWQLAKAHVGSNDAGVHQLVN 584
Query: 525 H 525
H
Sbjct: 585 H 585
>At1g17420 lipoxygenase
Length = 919
Score = 347 bits (890), Expect = 1e-95
Identities = 204/474 (43%), Positives = 287/474 (60%), Gaps = 20/474 (4%)
Query: 62 LGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPG 121
+GRN+ ++LIS T+ D K + K + A + ++ F DA FG PG
Sbjct: 115 IGRNIVLELIS-TQLDPKTKLPKKSNAAVLKDWSKKSKTKAERVHYTAEFTVDAAFGSPG 173
Query: 122 AFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTP 181
A + N Q EFFL +T+E G V F CNSWV + K + + RIFFTN YLP++TP
Sbjct: 174 AITVMNKHQKEFFLESITIEGFA-LGPVHFPCNSWVQSQKDHPDKRIFFTNQPYLPNETP 232
Query: 182 APLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYP 241
+ L RE+EL+NLRGDG+G RK DRIYD+DVYNDLGNPD L RP LGG PYP
Sbjct: 233 SGLRVLREKELKNLRGDGSGVRKLSDRIYDFDVYNDLGNPDKSSELSRPKLGGKEV-PYP 291
Query: 242 RRVRTGRKPT--RKDLKS--EKPGAIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQS 297
RR RTGR+ T KD +S EKP +YVPRDE F K F +K++ ++P L++
Sbjct: 292 RRCRTGRQSTVSDKDAESRVEKPLPMYVPRDEQFEESKQDTFAAGRLKAVLHHLIPSLKA 351
Query: 298 VIFDLNFTPNEFDSFDEVRGLFEGGIKLP---TDILSQISPLP-ALKEILRTDGEQALKF 353
I +F F E+ L++ G+ L D + + PLP + + L+ + LK+
Sbjct: 352 SI-----VAEDFADFGEIDRLYKEGLLLKLGFQDDIFKKFPLPKVVVDTLQESTKGLLKY 406
Query: 354 PPPQVIRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEH 413
P+++ K+AW D+EF R+ +AG+NP I ++ FPP S LD +YG Q+S +T +H
Sbjct: 407 DTPKILSKDKNAWLRDDEFARQAIAGINPVNIERVKTFPPVSNLDPKIYGPQHSALTDDH 466
Query: 414 LETNLGDITVEEALDGKRLFLLDYHDAFMPYLERINL--NAKAYATRTILFLQDDGTLKP 471
+ +L +V++AL+ RL++LDYHD F+P+L+RIN KAYATRTI FL GTLKP
Sbjct: 467 IIGHLDGFSVQQALEENRLYMLDYHDIFLPFLDRINALDGRKAYATRTIFFLTRLGTLKP 526
Query: 472 LAIELSLPHSNGVQYGAESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
+AIELSLP +G ++ ++ +V P + + +W LAKAHV ND+ HQL++H
Sbjct: 527 VAIELSLP-PHGPKHRSK-RVLTPPVDATSNWMWQLAKAHVSSNDAGVHQLVNH 578
>At3g45140 lipoxygenase (AtLox2)
Length = 896
Score = 327 bits (839), Expect = 9e-90
Identities = 206/511 (40%), Positives = 292/511 (56%), Gaps = 39/511 (7%)
Query: 28 ITSIGKGGVLDAAGNLIGGVTSIVGGVVDTATAFLGRNVSMQLISATKTDASGKGLVGKE 87
I +I G + A + G+T G +D GR++ ++LISA KTD + E
Sbjct: 70 IQNIKVKGYITAQEEFLEGITWSRG--LDDIADIRGRSLLVELISA-KTDQR----ITVE 122
Query: 88 TFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPGAFYIRNYTQAEFFLVRVTLEDIPNRG 147
+ + + P + + FE +FG GA I+N + FL V L+ +P G
Sbjct: 123 DYAQRVWAEAP-----DEKYECEFEMPEDFGPVGAIKIQNQYHRQLFLKGVELK-LPG-G 175
Query: 148 SVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTPAPLNHFREEELQNLRG---DGTGERK 204
S+ F C SWV RIFF++ +YLPSQTP PL +R+EEL+ L+G + GE
Sbjct: 176 SITFTCESWVAPKSVDPTKRIFFSDKSYLPSQTPEPLKKYRKEELETLQGKNREEVGEFT 235
Query: 205 EWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVRTGRKPTRKDLKSEKP--GA 262
+++RIYDYDVYND+G+PD L RP++GG THPYPRR +TGRKP D SE+ G
Sbjct: 236 KFERIYDYDVYNDVGDPDNDPELARPVIGGL-THPYPRRCKTGRKPCETDPSSEQRYGGE 294
Query: 263 IYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQSVIFDLNFTPNE-FDSFDEVRGLFEG 321
YVPRDE F K + F + + + P ++SV+ +P E F F ++ LFE
Sbjct: 295 FYVPRDEEFSTAKGTSFTGKAVLAALPSIFPQIESVLL----SPQEPFPHFKAIQNLFEE 350
Query: 322 GIKLPTDILSQISPLPALKEILRTDGE---QALKFPPPQVIRVSKSAWNTDEEFGREMLA 378
GI+LP D LP L I++ GE L+F P +I + +W D+EF R+ LA
Sbjct: 351 GIQLPKDA----GLLPLLPRIIKALGEAQDDILQFDAPVLINRDRFSWLRDDEFARQTLA 406
Query: 379 GVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNL-GDITVEEALDGKRLFLLDY 437
G+NP I+L++E+P S LD VYGD S IT E +E + G++TV+EAL KRLF+LDY
Sbjct: 407 GLNPYSIQLVEEWPLISKLDPAVYGDPTSLITWEIVEREVKGNMTVDEALKNKRLFVLDY 466
Query: 438 HDAFMPYLERIN--LNAKAYATRTILFLQDDGTLKPLAIELSL-PHSNGVQYGAESKVFL 494
HD +PY+ ++ N YA+RT+ FL DD TL+P+AIEL+ P+ N Q+ +VF
Sbjct: 467 HDLLLPYVNKVRELNNTTLYASRTLFFLSDDSTLRPVAIELTCPPNINKPQW---KQVFT 523
Query: 495 PAHEGVESTIWMLAKAHVIVNDSCYHQLMSH 525
P ++ +W LAK H I +D+ YHQL+SH
Sbjct: 524 PGYDATSCWLWNLAKTHAISHDAGYHQLISH 554
>At1g67560 putative lipoxygenase
Length = 917
Score = 286 bits (732), Expect = 2e-77
Identities = 187/486 (38%), Positives = 262/486 (53%), Gaps = 43/486 (8%)
Query: 62 LGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPTLGARQDAFSIFFEYDANFGIPG 121
+G+ + +QL+S +GKG E+ + LP+ R F+ F NFG PG
Sbjct: 111 IGQGMLIQLVSEEIDPETGKGRKSLESPVMG-LPKA-VKDPRYLVFTADFTVPINFGKPG 168
Query: 122 AFYIRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSWVYNFKSYKNNRIFFTNDAYLPSQTP 181
A + N E L + +ED + ++ F N+W+++ RI F + LPS+TP
Sbjct: 169 AILVTNLLSTEICLSEIIIED--STDTILFPANTWIHSKNDNPQARIIFRSQPCLPSETP 226
Query: 182 APLNHFREEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYP 241
+ RE++L ++RGDG GERK +RIYDYDVYNDLG+P + VRP+LG T PYP
Sbjct: 227 DGIKELREKDLVSVRGDGKGERKPHERIYDYDVYNDLGDPRKTER-VRPVLGVPET-PYP 284
Query: 242 RRVRTGRKPTRKDLKSEKPG----AIYVPRDENFGHLKSSDFLMYGIKSLSQDVLPLLQS 297
RR RTGR KD E G YVPRDE F +K F K+L +++P + +
Sbjct: 285 RRCRTGRPLVSKDPPCESRGKEKEEFYVPRDEVFEEIKRDTFRAGRFKALFHNLVPSIAA 344
Query: 298 VIFDLNFTPNEFDSFDEVRGLFEGGIKLPTDILSQISPLPA---------LKEILRTDGE 348
+ +L+ F F ++ L++ I +L P + IL E
Sbjct: 345 ALSNLDI---PFTCFSDIDNLYKSNI-----VLGHTEPKDTGLGGFIGGFMNGILNVT-E 395
Query: 349 QALKFPPPQVIRVSKSAWNTDEEFGREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNST 408
LK+ P VI+ + AW D EFGR+ LAGVNP I LL+E P +S LD +YG Q S
Sbjct: 396 TLLKYDTPAVIKWDRFAWLRDNEFGRQALAGVNPVNIELLKELPIRSNLDPALYGPQESV 455
Query: 409 ITKEHL--ETNLGDITVEEALDGKRLFLLDYHDAFMPYLERINL----NAKAYATRTILF 462
+T+E + E T+E+AL+ KRLFL+DYHD +P++E+IN K YA+RTI F
Sbjct: 456 LTEEIIAREVEHYGTTIEKALEEKRLFLVDYHDILLPFVEKINSIKEDPRKTYASRTIFF 515
Query: 463 LQDDGTLKPLAIELSLPHSNGVQYGAESK---VFLPAHEGVESTIWMLAKAHVIVNDSCY 519
+G L+PLAIELSLP + AES+ V+ H+ IW LAKAHV ND+
Sbjct: 516 YSKNGALRPLAIELSLPPT------AESENKFVYTHGHDATTHWIWKLAKAHVCSNDAGV 569
Query: 520 HQLMSH 525
HQL++H
Sbjct: 570 HQLVNH 575
>At1g49630 putative hydrogenase (At1g49630)
Length = 1080
Score = 33.1 bits (74), Expect = 0.46
Identities = 35/139 (25%), Positives = 54/139 (38%), Gaps = 28/139 (20%)
Query: 60 AFLGRNVSMQLISATKTDASGKGLVGKETFLSKHLPQLPT------------LGARQDAF 107
+FL RN + A GK L E ++ K L LP L R +A
Sbjct: 835 SFLSRNGCI-----VNMTADGKSLTNTEKYVGKFLDLLPENPSGELVTWDARLPLRNEAI 889
Query: 108 SIFFE----------YDANFGIPGAFY-IRNYTQAEFFLVRVTLEDIPNRGSVQFDCNSW 156
I + Y + + + G+ Y I + + RV + GS FD +S
Sbjct: 890 VIPTQVNYVGKAGNIYSSGYKLDGSSYVISKHISNTWLWDRVRVSGGAYGGSCDFDSHSG 949
Query: 157 VYNFKSYKNNRIFFTNDAY 175
V++F SY++ + T D Y
Sbjct: 950 VFSFLSYRDPNLLKTLDIY 968
>At5g02520 unknown protein
Length = 584
Score = 32.0 bits (71), Expect = 1.0
Identities = 36/152 (23%), Positives = 60/152 (38%), Gaps = 7/152 (4%)
Query: 189 EEELQNLRGDGTGERKEWDRIYDYDVYNDLGNPDGGDALVRPILGGSSTHPYPRRVR--T 246
E E+QNL DG + + + DV D D ++ P +G H V T
Sbjct: 245 ESEVQNLCNDGDNGSEGFIKAKSSDVEKDKSEAIDND-VISPAVGSGIKHTGADNVDKVT 303
Query: 247 GRKPTRKDLKSEKPGAIYVPRDENFGHL----KSSDFLMYGIKSLSQDVLPLLQSVIFDL 302
T + L SE+ + V L KSS GI S +L +V+ +
Sbjct: 304 SASATGESLTSEQQNGLLVTTASPHSLLKDLAKSSKPEKKGISKKSGKILRSDDNVVDPM 363
Query: 303 NFTPNEFDSFDEVRGLFEGGIKLPTDILSQIS 334
N++ + S + R + ++ PT +++ S
Sbjct: 364 NYSGTKVKSAENKRKIDASKLQSPTSNVAEHS 395
>At3g17990 unknown protein
Length = 165
Score = 31.6 bits (70), Expect = 1.3
Identities = 39/144 (27%), Positives = 57/144 (39%), Gaps = 27/144 (18%)
Query: 382 PNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDITVEEALDGKRLFLLDYHDAF 441
P V+ LL + KS L+ L +G T E A L LD+ D
Sbjct: 16 PEVLSLLPPYEGKSVLE---------------LGAGIGRFTGELAQKAGELIALDFIDNV 60
Query: 442 MPYLERINLNAKAYATRTILFLQDDGTLKPLAI---ELSLPHSNGV-QYGAESKVFLPAH 497
+ E IN + K + F+ D T L I L L SN + Y ++ +V L A
Sbjct: 61 IKKNESINGHYK-----NVKFMCADVTSPDLKITDGSLDLIFSNWLLMYLSDKEVELLAE 115
Query: 498 EGVESTIWMLAKAHVIVNDSCYHQ 521
V W+ ++ +SC+HQ
Sbjct: 116 RMVG---WIKVGGYIFFRESCFHQ 136
>At4g28650 receptor protein kinase-like protein
Length = 1013
Score = 30.4 bits (67), Expect = 3.0
Identities = 44/176 (25%), Positives = 80/176 (45%), Gaps = 23/176 (13%)
Query: 261 GAIYVPRDENFG--HLKSSDFLMYGIKSLSQDVLPLLQSVIFDL--NF----TPNEFDSF 312
G++++ +E+ G HL +S + G +L++D+ L+ + DL NF P+ F +
Sbjct: 130 GSLFLFSNESLGLVHLNASGNNLSG--NLTEDLGNLVSLEVLDLRGNFFQGSLPSSFKNL 187
Query: 313 DEVRGLFEGGIKLPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVSKSAWNTDEEF 372
++R L G L ++ S + LP+L+ + G K P P S D
Sbjct: 188 QKLRFLGLSGNNLTGELPSVLGQLPSLETAIL--GYNEFKGPIPPEFGNINSLKYLDLAI 245
Query: 373 GREMLAGVNPNVIRLLQEFPPKSTLDATVYGDQNSTITKEHLETNLGDITVEEALD 428
G+ L+G P+ + L+ +L+ + + N T T + +G IT + LD
Sbjct: 246 GK--LSGEIPSELGKLK------SLETLLLYENNFTGT---IPREIGSITTLKVLD 290
>At5g03540 unknown protein (At5g03540)
Length = 638
Score = 30.0 bits (66), Expect = 3.9
Identities = 16/55 (29%), Positives = 28/55 (50%)
Query: 488 AESKVFLPAHEGVESTIWMLAKAHVIVNDSCYHQLMSHCDGTIHHSNK*TSKCTS 542
AE+KV HE +ES + +A+ I+ ++ DG ++H+N +K S
Sbjct: 86 AETKVLKGPHEDLESYLDAIAQLRKIIRYFMSNKSFKSSDGVLNHANSLLAKAQS 140
>At4g15890 hypothetical protein
Length = 1314
Score = 29.3 bits (64), Expect = 6.7
Identities = 26/76 (34%), Positives = 33/76 (43%), Gaps = 16/76 (21%)
Query: 20 KNVLDFNAITSIGKGGVLD---------------AAGNLIGGVTSIVGGVVD-TATAFLG 63
K L FN TS GKG V D AA L+ +TS++GG D + +G
Sbjct: 407 KQALGFNGETSEGKGAVTDLLKKRCVDEKAAVRRAALLLVTKLTSLMGGCFDGSILKTMG 466
Query: 64 RNVSMQLISATKTDAS 79
+ S LIS K S
Sbjct: 467 TSCSDPLISIRKAAVS 482
>At4g11990 unknown protein
Length = 501
Score = 28.9 bits (63), Expect = 8.7
Identities = 15/39 (38%), Positives = 23/39 (58%), Gaps = 4/39 (10%)
Query: 325 LPTDILSQISPLPALKEILRTDGEQALKFPPPQVIRVSK 363
LPTD+ S++S L LR + L+FP P+ ++ SK
Sbjct: 418 LPTDLFSKLS----LSSELRPNNGSRLRFPQPEQVKGSK 452
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.325 0.142 0.438
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,280,342
Number of Sequences: 26719
Number of extensions: 665427
Number of successful extensions: 1840
Number of sequences better than 10.0: 13
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1787
Number of HSP's gapped (non-prelim): 15
length of query: 611
length of database: 11,318,596
effective HSP length: 105
effective length of query: 506
effective length of database: 8,513,101
effective search space: 4307629106
effective search space used: 4307629106
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140032.2


