

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140030.7 - phase: 0
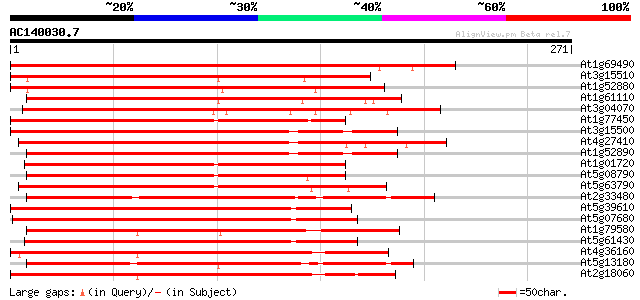
(271 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g69490 unknown protein 312 1e-85
At3g15510 jasmonic acid regulatory protein like 241 4e-64
At1g52880 NAM-like protein 239 9e-64
At1g61110 unknown protein 239 1e-63
At3g04070 NAM-like protein (no apical meristem) 231 3e-61
At1g77450 GRAB1-like protein 224 3e-59
At3g15500 putative jasmonic acid regulatory protein 224 4e-59
At4g27410 unknown protein 222 2e-58
At1g52890 NAM-like protein 221 3e-58
At1g01720 unknown protein with NAC domain protein 219 2e-57
At5g08790 ATAF2 protein 210 6e-55
At5g63790 unknown protein 208 2e-54
At2g33480 putative NAM (no apical meristem)-like protein 197 6e-51
At5g39610 NAM / CUC2 - like protein 191 3e-49
At5g07680 NAM (no apical meristem)-like protein 191 4e-49
At1g79580 unknown protein 190 6e-49
At5g61430 NAM, no apical meristem, - like protein 189 2e-48
At4g36160 NAM like protein 188 2e-48
At5g13180 NAM-like protein 187 4e-48
At2g18060 putative NAM (no apical meristem)-like protein 187 4e-48
>At1g69490 unknown protein
Length = 268
Score = 312 bits (799), Expect = 1e-85
Identities = 147/225 (65%), Positives = 174/225 (77%), Gaps = 10/225 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
ME ++ S LPPGFRFHPTDEELIV+YL NQ SKPCP SIIPEVDIYKFDPW+LP+K+EF
Sbjct: 1 MEVTSQSTLPPGFRFHPTDEELIVYYLRNQTMSKPCPVSIIPEVDIYKFDPWQLPEKTEF 60
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
ENEWYFFSPRERKYPNGVRPNRA +SGYWKATGTDKAI SGS +GVKK+LVFYKGRPP
Sbjct: 61 GENEWYFFSPRERKYPNGVRPNRAAVSGYWKATGTDKAIHSGSSNVGVKKALVFYKGRPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTH--- 177
KG+KTDWIMHEYRL S+K ++K GSMRLD+WVLCRIYKK+ K L ++E +
Sbjct: 121 KGIKTDWIMHEYRLHDSRKASTKRNGSMRLDEWVLCRIYKKRGASKLLNEQEGFMDEVLM 180
Query: 178 QFNDSIITNNDDGELE-------MMNLTRSCSLTYLLDMNYFGPI 215
+ ++ N + E M L R+CSL +LL+M+Y GP+
Sbjct: 181 EDETKVVVNEAERRTEEEIMMMTSMKLPRTCSLAHLLEMDYMGPV 225
>At3g15510 jasmonic acid regulatory protein like
Length = 364
Score = 241 bits (614), Expect = 4e-64
Identities = 115/191 (60%), Positives = 138/191 (72%), Gaps = 17/191 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL+VHYL +A S P P +II EVD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVVHYLKRKAASAPLPVAIIAEVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKK 110
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + G++++GVKK
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVLASDGNQKVGVKK 120
Query: 111 SLVFYKGRPPKGVKTDWIMHEYRLIGSQKQT-------SKHIGSMRLDDWVLCRIYKKKH 163
+LVFY G+PPKGVK+DWIMHEYRLI ++ S+RLDDWVLCRIYKK +
Sbjct: 121 ALVFYSGKPPKGVKSDWIMHEYRLIENKPNNRPPGCDFGNKKNSLRLDDWVLCRIYKKNN 180
Query: 164 MGKTLQQKEDY 174
+ + +D+
Sbjct: 181 ASRHVDNDKDH 191
>At1g52880 NAM-like protein
Length = 320
Score = 239 bits (611), Expect = 9e-64
Identities = 118/197 (59%), Positives = 139/197 (69%), Gaps = 16/197 (8%)
Query: 1 MESSASS--------ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPW 52
MES+ SS LPPGFRFHPTDEEL++HYL +A S P P +II +VD+YKFDPW
Sbjct: 1 MESTDSSGGPPPPQPNLPPGFRFHPTDEELVIHYLKRKADSVPLPVAIIADVDLYKFDPW 60
Query: 53 ELPDKSEFEENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKS----GSKQIGV 108
ELP K+ F E EWYFFSPR+RKYPNG RPNRA SGYWKATGTDK + S GSK++GV
Sbjct: 61 ELPAKASFGEQEWYFFSPRDRKYPNGARPNRAATSGYWKATGTDKPVISTGGGGSKKVGV 120
Query: 109 KKSLVFYKGRPPKGVKTDWIMHEYRLIGSQKQTSKHIG----SMRLDDWVLCRIYKKKHM 164
KK+LVFY G+PPKGVK+DWIMHEYRL ++ G S+RLDDWVLCRIYKK +
Sbjct: 121 KKALVFYSGKPPKGVKSDWIMHEYRLTDNKPTHICDFGNKKNSLRLDDWVLCRIYKKNNS 180
Query: 165 GKTLQQKEDYSTHQFND 181
+ + H ND
Sbjct: 181 TASRHHHHLHHIHLDND 197
>At1g61110 unknown protein
Length = 323
Score = 239 bits (610), Expect = 1e-63
Identities = 118/202 (58%), Positives = 140/202 (68%), Gaps = 21/202 (10%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+VHYL +A S P P SII E+D+YKFDPWELP K+ F E+EWYFF
Sbjct: 16 LPPGFRFHPTDEELVVHYLKKKADSVPLPVSIIAEIDLYKFDPWELPSKASFGEHEWYFF 75
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI-KSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
SPR+RKYPNGVRPNRA SGYWKATGTDK I S ++GVKK+LVFY G+PPKG+KTDW
Sbjct: 76 SPRDRKYPNGVRPNRAATSGYWKATGTDKPIFTCNSHKVGVKKALVFYGGKPPKGIKTDW 135
Query: 128 IMHEYRLIGSQKQ--------TSKHIGSMRLDDWVLCRIYKKKHMGKTLQQ----KEDY- 174
IMHEYRL T+ S+RLDDWVLCRIYKK + + +ED
Sbjct: 136 IMHEYRLTDGNLSTAAKPPDLTTTRKNSLRLDDWVLCRIYKKNSSQRPTMERVLLREDLM 195
Query: 175 -------STHQFNDSIITNNDD 189
S + + S++ NND+
Sbjct: 196 EGMLSKSSANSSSTSVLDNNDN 217
>At3g04070 NAM-like protein (no apical meristem)
Length = 359
Score = 231 bits (589), Expect = 3e-61
Identities = 123/236 (52%), Positives = 147/236 (62%), Gaps = 34/236 (14%)
Query: 7 SELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWY 66
S LPPGFRFHPTDEELI+HYL + +S P P SII +VDIYK DPW+LP K+ F E EWY
Sbjct: 8 SSLPPGFRFHPTDEELILHYLRKKVSSSPVPLSIIADVDIYKSDPWDLPAKAPFGEKEWY 67
Query: 67 FFSPRERKYPNGVRPNRATLSGYWKATGTDK--AIKSGS---KQIGVKKSLVFYKGRPPK 121
FFSPR+RKYPNG RPNRA SGYWKATGTDK A+ +G + IG+KK+LVFY+G+PPK
Sbjct: 68 FFSPRDRKYPNGARPNRAAASGYWKATGTDKLIAVPNGEGFHENIGIKKALVFYRGKPPK 127
Query: 122 GVKTDWIMHEYRL--------IGSQKQTSKHIG-----------SMRLDDWVLCRIYKKK 162
GVKT+WIMHEYRL I S + + SMRLDDWVLCRIYKK
Sbjct: 128 GVKTNWIMHEYRLADSLSPKRINSSRSGGSEVNNNFGDRNSKEYSMRLDDWVLCRIYKKS 187
Query: 163 H---------MGKTLQQKEDYSTHQFND-SIITNNDDGELEMMNLTRSCSLTYLLD 208
H + + Q+ E+ F D N + + SCS + LLD
Sbjct: 188 HASLSSPDVALVTSNQEHEENDNEPFVDRGTFLPNLQNDQPLKRQKSSCSFSNLLD 243
>At1g77450 GRAB1-like protein
Length = 253
Score = 224 bits (572), Expect = 3e-59
Identities = 101/162 (62%), Positives = 123/162 (75%), Gaps = 2/162 (1%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
M+S A + PPGFRFHPTDEEL++ YLC + S+P PA II E+D+Y++DPW+LPD + +
Sbjct: 2 MKSGADLQFPPGFRFHPTDEELVLMYLCRKCASQPIPAPIITELDLYRYDPWDLPDMALY 61
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
E EWYFFSPR+RKYPNG RPNRA +GYWKATG DK I K +G+KK+LVFY G+PP
Sbjct: 62 GEKEWYFFSPRDRKYPNGSRPNRAAGTGYWKATGADKPI-GRPKPVGIKKALVFYSGKPP 120
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK 162
G KT+WIMHEYRL + K S+RLDDWVLCRIY KK
Sbjct: 121 NGEKTNWIMHEYRLADVDRSVRKK-NSLRLDDWVLCRIYNKK 161
>At3g15500 putative jasmonic acid regulatory protein
Length = 317
Score = 224 bits (571), Expect = 4e-59
Identities = 106/187 (56%), Positives = 132/187 (69%), Gaps = 8/187 (4%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
++ A LPPGFRF+PTDEEL+V YLC +A +I E+D+YKFDPW LP K+ F
Sbjct: 6 LDPLAQLSLPPGFRFYPTDEELMVEYLCRKAAGHDFSLQLIAEIDLYKFDPWVLPSKALF 65
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
E EWYFFSPR+RKYPNG RPNR SGYWKATGTDK I + +++G+KK+LVFY G+ P
Sbjct: 66 GEKEWYFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKVISTEGRRVGIKKALVFYIGKAP 125
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFN 180
KG KT+WIMHEYRLI + S+ GS +LDDWVLCRIYKK +T QK+ Y+ +
Sbjct: 126 KGTKTNWIMHEYRLI----EPSRRNGSTKLDDWVLCRIYKK----QTSAQKQAYNNLMTS 177
Query: 181 DSIITNN 187
+NN
Sbjct: 178 GREYSNN 184
>At4g27410 unknown protein
Length = 297
Score = 222 bits (565), Expect = 2e-58
Identities = 113/220 (51%), Positives = 146/220 (66%), Gaps = 17/220 (7%)
Query: 5 ASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENE 64
A LPPGFRF+PTDEEL+V YLC + +I ++D+YKFDPW+LP K+ F E E
Sbjct: 10 AQLSLPPGFRFYPTDEELLVQYLCRKVAGYHFSLQVIGDIDLYKFDPWDLPSKALFGEKE 69
Query: 65 WYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVK 124
WYFFSPR+RKYPNG RPNR SGYWKATGTDK I + +++G+KK+LVFY G+ PKG K
Sbjct: 70 WYFFSPRDRKYPNGSRPNRVAGSGYWKATGTDKIITADGRRVGIKKALVFYAGKAPKGTK 129
Query: 125 TDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKK----KHMGKTLQQ--KEDYSTHQ 178
T+WIMHEYRLI + S+ GS +LDDWVLCRIYKK + T Q +E++ST+
Sbjct: 130 TNWIMHEYRLI----EHSRSHGSSKLDDWVLCRIYKKTSGSQRQAVTPVQACREEHSTNG 185
Query: 179 FNDSIITNNDDG-------ELEMMNLTRSCSLTYLLDMNY 211
+ S + DD + + NL R SL +L+ N+
Sbjct: 186 SSSSSSSQLDDVLDSFPEIKDQSFNLPRMNSLRTILNGNF 225
>At1g52890 NAM-like protein
Length = 317
Score = 221 bits (563), Expect = 3e-58
Identities = 105/179 (58%), Positives = 129/179 (71%), Gaps = 8/179 (4%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRF+PTDEEL+V YLC +A +I E+D+YKFDPW LP+K+ F E EWYFF
Sbjct: 14 LPPGFRFYPTDEELMVQYLCRKAAGYDFSLQLIAEIDLYKFDPWVLPNKALFGEKEWYFF 73
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDWI 128
SPR+RKYPNG RPNR SGYWKATGTDK I + +++G+KK+LVFY G+ PKG KT+WI
Sbjct: 74 SPRDRKYPNGSRPNRVAGSGYWKATGTDKIISTEGQRVGIKKALVFYIGKAPKGTKTNWI 133
Query: 129 MHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFNDSIITNN 187
MHEYRLI + S+ GS +LDDWVLCRIYKK ++ QK+ Y N +NN
Sbjct: 134 MHEYRLI----EPSRRNGSTKLDDWVLCRIYKK----QSSAQKQVYDNGIANAREFSNN 184
>At1g01720 unknown protein with NAC domain protein
Length = 289
Score = 219 bits (557), Expect = 2e-57
Identities = 98/155 (63%), Positives = 117/155 (75%), Gaps = 1/155 (0%)
Query: 8 ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYF 67
+LPPGFRFHPTDEEL++HYLC + S+ II E+D+YK+DPWELP + + E EWYF
Sbjct: 6 QLPPGFRFHPTDEELVMHYLCRKCASQSIAVPIIAEIDLYKYDPWELPGLALYGEKEWYF 65
Query: 68 FSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
FSPR+RKYPNG RPNR+ SGYWKATG DK I K +G+KK+LVFY G+ PKG KT+W
Sbjct: 66 FSPRDRKYPNGSRPNRSAGSGYWKATGADKPI-GLPKPVGIKKALVFYAGKAPKGEKTNW 124
Query: 128 IMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKK 162
IMHEYRL + K S+RLDDWVLCRIY KK
Sbjct: 125 IMHEYRLADVDRSVRKKKNSLRLDDWVLCRIYNKK 159
>At5g08790 ATAF2 protein
Length = 283
Score = 210 bits (535), Expect = 6e-55
Identities = 96/155 (61%), Positives = 116/155 (73%), Gaps = 2/155 (1%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LP GFRFHPTDEEL+ YLC + S+ A +I E+D+YKF+PWELP+ S + E EWYFF
Sbjct: 7 LPAGFRFHPTDEELVKFYLCRKCASEQISAPVIAEIDLYKFNPWELPEMSLYGEKEWYFF 66
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDWI 128
SPR+RKYPNG RPNRA +GYWKATG DK I K +G+KK+LVFY G+ PKG+KT+WI
Sbjct: 67 SPRDRKYPNGSRPNRAAGTGYWKATGADKPI-GKPKTLGIKKALVFYAGKAPKGIKTNWI 125
Query: 129 MHEYRLIGSQKQTS-KHIGSMRLDDWVLCRIYKKK 162
MHEYRL + S ++RLDDWVLCRIY KK
Sbjct: 126 MHEYRLANVDRSASVNKKNNLRLDDWVLCRIYNKK 160
>At5g63790 unknown protein
Length = 312
Score = 208 bits (530), Expect = 2e-54
Identities = 100/180 (55%), Positives = 125/180 (68%), Gaps = 3/180 (1%)
Query: 5 ASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENE 64
A LP GFRFHPTDEEL+ YLC + S+P +I E+D+YKF+PWELP+ + + E E
Sbjct: 46 AELNLPAGFRFHPTDEELVKFYLCRRCASEPINVPVIAEIDLYKFNPWELPEMALYGEKE 105
Query: 65 WYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVK 124
WYFFS R+RKYPNG RPNRA +GYWKATG DK I K +G+KK+LVFY G+ PKG+K
Sbjct: 106 WYFFSHRDRKYPNGSRPNRAAGTGYWKATGADKPI-GKPKTLGIKKALVFYAGKAPKGIK 164
Query: 125 TDWIMHEYRLIGSQKQTSKH-IGSMRLDDWVLCRIYKKK-HMGKTLQQKEDYSTHQFNDS 182
T+WIMHEYRL + S + ++RLDDWVLCRIY KK M K L + T + + S
Sbjct: 165 TNWIMHEYRLANVDRSASTNKKNNLRLDDWVLCRIYNKKGTMEKYLPAAAEKPTEKMSTS 224
>At2g33480 putative NAM (no apical meristem)-like protein
Length = 268
Score = 197 bits (500), Expect = 6e-51
Identities = 100/197 (50%), Positives = 127/197 (63%), Gaps = 9/197 (4%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+V YL + T P PAS+IPE D+ K DPW+LP E +E YFF
Sbjct: 15 LPPGFRFHPTDEELVVQYLRRKVTGLPLPASVIPETDVCKSDPWDLPGDCE---SEMYFF 71
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDWI 128
S RE KYPNG R NR+T SGYWKATG DK I +G+KK+LVFYKG+PP G +T+W+
Sbjct: 72 STREAKYPNGNRSNRSTGSGYWKATGLDKQIGKKKLVVGMKKTLVFYKGKPPNGTRTNWV 131
Query: 129 MHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFNDSIITNND 188
+HEYRL+ SQ Q S + +M +WVLCR++ KK ++KED N+
Sbjct: 132 LHEYRLVDSQ-QDSLYGQNM---NWVLCRVFLKKRSNSNSKRKEDEKEEVENEK--ETET 185
Query: 189 DGELEMMNLTRSCSLTY 205
+ E E N +C + Y
Sbjct: 186 EREREEENKKSTCPIFY 202
>At5g39610 NAM / CUC2 - like protein
Length = 285
Score = 191 bits (486), Expect = 3e-49
Identities = 89/165 (53%), Positives = 117/165 (69%), Gaps = 2/165 (1%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
+E +LPPGFRFHPTDEELI HYL + + A+ I EVD+ K +PW+LP K++
Sbjct: 12 VEDEEHIDLPPGFRFHPTDEELITHYLKPKVFNTFFSATAIGEVDLNKIEPWDLPWKAKM 71
Query: 61 EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPP 120
E EWYFF R+RKYP G+R NRAT +GYWKATG DK I G +G+KK+LVFYKGR P
Sbjct: 72 GEKEWYFFCVRDRKYPTGLRTNRATEAGYWKATGKDKEIFKGKSLVGMKKTLVFYKGRAP 131
Query: 121 KGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMG 165
KGVKT+W+MHEYRL G K +++ ++WV+CR+++K+ G
Sbjct: 132 KGVKTNWVMHEYRLEG--KYCIENLPQTAKNEWVICRVFQKRADG 174
>At5g07680 NAM (no apical meristem)-like protein
Length = 329
Score = 191 bits (485), Expect = 4e-49
Identities = 89/167 (53%), Positives = 114/167 (67%), Gaps = 2/167 (1%)
Query: 2 ESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFE 61
E +LPPGFRFHPTDEELI HYL + A I EVD+ K +PWELP K++
Sbjct: 10 EDDEQMDLPPGFRFHPTDEELITHYLHKKVLDLGFSAKAIGEVDLNKAEPWELPYKAKIG 69
Query: 62 ENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPK 121
E EWYFF R+RKYP G+R NRAT +GYWKATG DK I G +G+KK+LVFY+GR PK
Sbjct: 70 EKEWYFFCVRDRKYPTGLRTNRATQAGYWKATGKDKEIFRGKSLVGMKKTLVFYRGRAPK 129
Query: 122 GVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTL 168
G KT+W+MHEYRL G K ++ ++ ++WV+CR++ K GK +
Sbjct: 130 GQKTNWVMHEYRLDG--KLSAHNLPKTAKNEWVICRVFHKTAGGKKI 174
>At1g79580 unknown protein
Length = 371
Score = 190 bits (483), Expect = 6e-49
Identities = 89/184 (48%), Positives = 125/184 (67%), Gaps = 11/184 (5%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF---EENEW 65
+PPGFRFHPT+EEL+ +YL + + +P +I EVD+ K +PWEL +K +NEW
Sbjct: 17 VPPGFRFHPTEEELLYYYLKKKVSYEPIDLDVIREVDLNKLEPWELKEKCRIGSGPQNEW 76
Query: 66 YFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIK-SGSKQIGVKKSLVFYKGRPPKGVK 124
YFFS +++KYP G R NRAT +G+WKATG DK+I + SK+IG++K+LVFY GR P G K
Sbjct: 77 YFFSHKDKKYPTGTRTNRATAAGFWKATGRDKSIHLNSSKKIGLRKTLVFYTGRAPHGQK 136
Query: 125 TDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFNDSII 184
T+WIMHEYRL S+ + + D WV+CR++KKK+ + Q+++ H + I
Sbjct: 137 TEWIMHEYRLDDSENEIQE-------DGWVVCRVFKKKNHFRGFHQEQEQDHHHHHQYIS 189
Query: 185 TNND 188
TNND
Sbjct: 190 TNND 193
>At5g61430 NAM, no apical meristem, - like protein
Length = 336
Score = 189 bits (479), Expect = 2e-48
Identities = 87/161 (54%), Positives = 113/161 (70%), Gaps = 2/161 (1%)
Query: 8 ELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYF 67
+LPPGFRFHPTDEELI HYL + A I EVD+ K +PWELP ++ E EWYF
Sbjct: 15 DLPPGFRFHPTDEELITHYLHKKVLDTSFSAKAIGEVDLNKSEPWELPWMAKMGEKEWYF 74
Query: 68 FSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKGRPPKGVKTDW 127
F R+RKYP G+R NRAT +GYWKATG DK I G +G+KK+LVFY+GR PKG KT+W
Sbjct: 75 FCVRDRKYPTGLRTNRATEAGYWKATGKDKEIYRGKSLVGMKKTLVFYRGRAPKGQKTNW 134
Query: 128 IMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTL 168
+MHEYRL G K ++ ++ ++WV+CR+++K GK +
Sbjct: 135 VMHEYRLEG--KFSAHNLPKTAKNEWVICRVFQKSAGGKKI 173
>At4g36160 NAM like protein
Length = 377
Score = 188 bits (478), Expect = 2e-48
Identities = 90/187 (48%), Positives = 121/187 (64%), Gaps = 10/187 (5%)
Query: 1 MES-SASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSE 59
MES S +PPGFRFHPTDEEL+ +YL + S+ +I ++D+Y+ +PW+L +
Sbjct: 1 MESVDQSCSVPPGFRFHPTDEELVGYYLRKKVASQKIDLDVIRDIDLYRIEPWDLQESCR 60
Query: 60 F---EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYK 116
E NEWYFFS +++KYP G R NRAT++G+WKATG DKA+ SK IG++K+LVFYK
Sbjct: 61 IGYEERNEWYFFSHKDKKYPTGTRTNRATMAGFWKATGRDKAVYDKSKLIGMRKTLVFYK 120
Query: 117 GRPPKGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYST 176
GR P G KTDWIMHEYRL + + G WV+CR +KKK M + E +S+
Sbjct: 121 GRAPNGQKTDWIMHEYRLESDENAPPQEEG------WVVCRAFKKKPMTGQAKNTETWSS 174
Query: 177 HQFNDSI 183
F D +
Sbjct: 175 SYFYDEL 181
>At5g13180 NAM-like protein
Length = 252
Score = 187 bits (476), Expect = 4e-48
Identities = 98/189 (51%), Positives = 125/189 (65%), Gaps = 12/189 (6%)
Query: 9 LPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEFEENEWYFF 68
LPPGFRFHPTDEEL+V YL + S P PASIIPE D+ + DPW+LP E E YFF
Sbjct: 14 LPPGFRFHPTDEELVVQYLKRKVCSSPLPASIIPEFDVCRADPWDLPGNL---EKERYFF 70
Query: 69 SPRERKYPNGVRPNRATLSGYWKATGTDKAI--KSGSKQIGVKKSLVFYKGRPPKGVKTD 126
S RE KYPNG R NRAT SGYWKATG DK + G++ +G+KK+LVFYKG+PP G +TD
Sbjct: 71 STREAKYPNGNRSNRATGSGYWKATGIDKRVVTSRGNQIVGLKKTLVFYKGKPPHGSRTD 130
Query: 127 WIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTHQFNDSIITN 186
WIMHEYRL S + +G + +WVLCRI+ KK G + S + +++ N
Sbjct: 131 WIMHEYRLSSSPPSS---MGPTQ--NWVLCRIFLKKRAGNKNDDDDGDSRNLRHNN--NN 183
Query: 187 NDDGELEMM 195
N ++E++
Sbjct: 184 NSSDQIEII 192
>At2g18060 putative NAM (no apical meristem)-like protein
Length = 365
Score = 187 bits (476), Expect = 4e-48
Identities = 87/189 (46%), Positives = 124/189 (65%), Gaps = 10/189 (5%)
Query: 1 MESSASSELPPGFRFHPTDEELIVHYLCNQATSKPCPASIIPEVDIYKFDPWELPDKSEF 60
ME S +PPGFRFHPTDEEL+ +YL + S+ +I ++D+Y+ +PW+L ++
Sbjct: 1 MEPMESCSVPPGFRFHPTDEELVGYYLRKKIASQKIDLDVIRDIDLYRIEPWDLQEQCRI 60
Query: 61 ---EENEWYFFSPRERKYPNGVRPNRATLSGYWKATGTDKAIKSGSKQIGVKKSLVFYKG 117
E+NEWYFFS +++KYP G R NRAT++G+WKATG DKA+ +K IG++K+LVFYKG
Sbjct: 61 GYEEQNEWYFFSHKDKKYPTGTRTNRATMAGFWKATGRDKAVYDKTKLIGMRKTLVFYKG 120
Query: 118 RPPKGVKTDWIMHEYRLIGSQKQTSKHIGSMRLDDWVLCRIYKKKHMGKTLQQKEDYSTH 177
R P G K+DWIMHEYRL + + G WV+CR +KK+ G+ + E +S+
Sbjct: 121 RAPNGKKSDWIMHEYRLESDENAPPQEEG------WVVCRAFKKRATGQA-KNTETWSSS 173
Query: 178 QFNDSIITN 186
F D + N
Sbjct: 174 YFYDEVAPN 182
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.134 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,930,986
Number of Sequences: 26719
Number of extensions: 321534
Number of successful extensions: 949
Number of sequences better than 10.0: 118
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 15
Number of HSP's that attempted gapping in prelim test: 658
Number of HSP's gapped (non-prelim): 127
length of query: 271
length of database: 11,318,596
effective HSP length: 98
effective length of query: 173
effective length of database: 8,700,134
effective search space: 1505123182
effective search space used: 1505123182
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140030.7


