

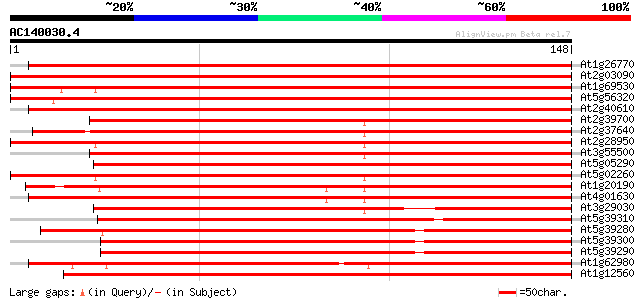
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140030.4 - phase: 0 /pseudo
(148 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g26770 expansin 10 264 1e-71
At2g03090 expansin like protein 264 1e-71
At1g69530 expansin-like protein 258 8e-70
At5g56320 expansin 232 5e-62
At2g40610 putative expansin 232 5e-62
At2g39700 putative expansin 231 1e-61
At2g37640 putative expansin 228 7e-61
At2g28950 putative expansin 227 2e-60
At3g55500 expansin-like protein 223 4e-59
At5g05290 expansin At-EXP2 (gb|AAB38073.1) 222 6e-59
At5g02260 expansin precursor - like protein 221 8e-59
At1g20190 expansin S2 precursor like protein 207 2e-54
At4g01630 putative expansin 201 2e-52
At3g29030 expansin At-EXP5 190 2e-49
At5g39310 expansin-like protein 182 6e-47
At5g39280 expansin-like protein 179 4e-46
At5g39300 expansin-like protein 176 4e-45
At5g39290 expansin-like protein 169 6e-43
At1g62980 expansin At-EXP6, putative 167 1e-42
At1g12560 hypothetical protein 165 9e-42
>At1g26770 expansin 10
Length = 249
Score = 264 bits (675), Expect = 1e-71
Identities = 120/143 (83%), Positives = 129/143 (89%)
Query: 6 LVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
LV++ M SSV YGGGW NAHATFYGG DASGTMGGACGYGNLYSQGYGT+TAALST
Sbjct: 7 LVMIMVGVMASSVSGYGGGWINAHATFYGGGDASGTMGGACGYGNLYSQGYGTSTAALST 66
Query: 66 ALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
ALFNNGLSCGSC+EIRC ND +WCLPGSIVVTATNFCPPNNAL NN+GGWCNPPL+HFDL
Sbjct: 67 ALFNNGLSCGSCFEIRCENDGKWCLPGSIVVTATNFCPPNNALANNNGGWCNPPLEHFDL 126
Query: 126 AQPVFLRIAQYKAGIVPVDFRRV 148
AQPVF RIAQY+AGIVPV +RRV
Sbjct: 127 AQPVFQRIAQYRAGIVPVSYRRV 149
>At2g03090 expansin like protein
Length = 253
Score = 264 bits (674), Expect = 1e-71
Identities = 116/148 (78%), Positives = 128/148 (86%)
Query: 1 MAFIGLVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 60
M +G+ L C M SV+ Y GW NAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT
Sbjct: 6 MGLLGIALFCFAAMVCSVHGYDAGWVNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNT 65
Query: 61 AALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPL 120
AALSTALFNNGLSCG+C+EI+C +D WCLPG+I+VTATNFCPPNNALPNN GGWCNPPL
Sbjct: 66 AALSTALFNNGLSCGACFEIKCQSDGAWCLPGAIIVTATNFCPPNNALPNNAGGWCNPPL 125
Query: 121 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
HFDL+QPVF RIAQYKAG+VPV +RRV
Sbjct: 126 HHFDLSQPVFQRIAQYKAGVVPVSYRRV 153
>At1g69530 expansin-like protein
Length = 250
Score = 258 bits (659), Expect = 8e-70
Identities = 120/150 (80%), Positives = 130/150 (86%), Gaps = 2/150 (1%)
Query: 1 MAFIGLVLVCSL-TMFSSVYAY-GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGT 58
MA + + + +L M S V Y GGGW NAHATFYGG DASGTMGGACGYGNLYSQGYGT
Sbjct: 1 MALVTFLFIATLGAMTSHVNGYAGGGWVNAHATFYGGGDASGTMGGACGYGNLYSQGYGT 60
Query: 59 NTAALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNP 118
NTAALSTALFNNGLSCG+C+EIRC ND +WCLPGSIVVTATNFCPPNNALPNN GGWCNP
Sbjct: 61 NTAALSTALFNNGLSCGACFEIRCQNDGKWCLPGSIVVTATNFCPPNNALPNNAGGWCNP 120
Query: 119 PLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
P QHFDL+QPVF RIAQY+AGIVPV +RRV
Sbjct: 121 PQQHFDLSQPVFQRIAQYRAGIVPVAYRRV 150
>At5g56320 expansin
Length = 255
Score = 232 bits (592), Expect = 5e-62
Identities = 104/152 (68%), Positives = 121/152 (79%), Gaps = 4/152 (2%)
Query: 1 MAFIGLVLVC----SLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGY 56
M F G +++ + M+ SV Y GW NA ATFYGG+DASGTMGGACGYGNLYSQGY
Sbjct: 1 MEFFGKMIISLSLMMMIMWKSVDGYSSGWVNARATFYGGADASGTMGGACGYGNLYSQGY 60
Query: 57 GTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWC 116
GTNTAALSTALFN G SCG+C++I+C +D +WC+ G+I VT TNFCPPN A NN GGWC
Sbjct: 61 GTNTAALSTALFNGGQSCGACFQIKCVDDPKWCIGGTITVTGTNFCPPNFAQANNAGGWC 120
Query: 117 NPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
NPP HFDLAQP+FLRIAQYKAG+VPV +RRV
Sbjct: 121 NPPQHHFDLAQPIFLRIAQYKAGVVPVQYRRV 152
>At2g40610 putative expansin
Length = 253
Score = 232 bits (592), Expect = 5e-62
Identities = 101/143 (70%), Positives = 117/143 (81%)
Query: 6 LVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
++ + S+ + GGW HATFYGG DASGTMGGACGYGNLY QGYGTNTAALST
Sbjct: 11 IISIISVLFLQGTHGDDGGWQGGHATFYGGEDASGTMGGACGYGNLYGQGYGTNTAALST 70
Query: 66 ALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
ALFNNGL+CG+CYE++C +D RWCL +I VTATNFCPPN L N++GGWCNPPLQHFDL
Sbjct: 71 ALFNNGLTCGACYEMKCNDDPRWCLGSTITVTATNFCPPNPGLSNDNGGWCNPPLQHFDL 130
Query: 126 AQPVFLRIAQYKAGIVPVDFRRV 148
A+P FL+IAQY+AGIVPV FRRV
Sbjct: 131 AEPAFLQIAQYRAGIVPVSFRRV 153
>At2g39700 putative expansin
Length = 257
Score = 231 bits (589), Expect = 1e-61
Identities = 102/129 (79%), Positives = 117/129 (90%), Gaps = 2/129 (1%)
Query: 22 GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIR 81
GG W NAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNG+SCG+C+E++
Sbjct: 28 GGAWQNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGMSCGACFELK 87
Query: 82 CANDHRWCLPG--SIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAG 139
CAND +WC G SI++TATNFCPPN A P+++GGWCNPP +HFDLA PVFL+IAQY+AG
Sbjct: 88 CANDPQWCHSGSPSILITATNFCPPNLAQPSDNGGWCNPPREHFDLAMPVFLKIAQYRAG 147
Query: 140 IVPVDFRRV 148
IVPV +RRV
Sbjct: 148 IVPVSYRRV 156
>At2g37640 putative expansin
Length = 262
Score = 228 bits (582), Expect = 7e-61
Identities = 104/144 (72%), Positives = 120/144 (83%), Gaps = 3/144 (2%)
Query: 7 VLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTA 66
+L + VY+ GG W NAHATFYGGSDASGTMGGACGYGNLYSQGYG NTAALSTA
Sbjct: 19 LLTATNAKIPGVYS-GGPWQNAHATFYGGSDASGTMGGACGYGNLYSQGYGVNTAALSTA 77
Query: 67 LFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNNDGGWCNPPLQHFD 124
LFNNG SCG+C+EI+C +D RWC+PG SI+VTATNFCPPN A P++DGGWCNPP +HFD
Sbjct: 78 LFNNGFSCGACFEIKCTDDPRWCVPGNPSILVTATNFCPPNFAQPSDDGGWCNPPREHFD 137
Query: 125 LAQPVFLRIAQYKAGIVPVDFRRV 148
LA P+FL+I Y+AGIVPV +RRV
Sbjct: 138 LAMPMFLKIGLYRAGIVPVSYRRV 161
>At2g28950 putative expansin
Length = 257
Score = 227 bits (579), Expect = 2e-60
Identities = 107/156 (68%), Positives = 124/156 (78%), Gaps = 8/156 (5%)
Query: 1 MAFIGLVLVCSLTMFSSVYAY------GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQ 54
MA +GLVL T+ + A GGGW AHATFYGGSDASGTMGGACGYGNLYSQ
Sbjct: 1 MAMLGLVLSVLTTILALSEARIPGVYNGGGWETAHATFYGGSDASGTMGGACGYGNLYSQ 60
Query: 55 GYGTNTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNND 112
GYG NTAALSTALFNNG SCG+C+E++CA+D +WC G SI +TATNFCPPN A P+++
Sbjct: 61 GYGVNTAALSTALFNNGFSCGACFELKCASDPKWCHSGSPSIFITATNFCPPNFAQPSDN 120
Query: 113 GGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
GGWCNPP HFDLA P+FL+IA+Y+AGIVPV FRRV
Sbjct: 121 GGWCNPPRPHFDLAMPMFLKIAEYRAGIVPVSFRRV 156
>At3g55500 expansin-like protein
Length = 260
Score = 223 bits (567), Expect = 4e-59
Identities = 97/129 (75%), Positives = 113/129 (87%), Gaps = 2/129 (1%)
Query: 22 GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIR 81
GG W AHATFYGG+DASGTMGGACGYGNLYSQGYGTNTAALST+LFN+G SCG+C+EI+
Sbjct: 31 GGSWQTAHATFYGGNDASGTMGGACGYGNLYSQGYGTNTAALSTSLFNSGQSCGACFEIK 90
Query: 82 CANDHRWCLPG--SIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAG 139
C ND +WC PG S+ VTATNFCPPN A P+++GGWCNPP HFDLA PVFL+IA+Y+AG
Sbjct: 91 CVNDPKWCHPGNPSVFVTATNFCPPNLAQPSDNGGWCNPPRSHFDLAMPVFLKIAEYRAG 150
Query: 140 IVPVDFRRV 148
IVP+ +RRV
Sbjct: 151 IVPISYRRV 159
>At5g05290 expansin At-EXP2 (gb|AAB38073.1)
Length = 255
Score = 222 bits (565), Expect = 6e-59
Identities = 95/126 (75%), Positives = 112/126 (88%)
Query: 23 GGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRC 82
GGW HATFYGG+DASGTMGGACGYGNL+SQGYG TAALSTALFN+G CG+C+E++C
Sbjct: 30 GGWERGHATFYGGADASGTMGGACGYGNLHSQGYGLQTAALSTALFNSGQKCGACFELQC 89
Query: 83 ANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVP 142
+D WC+PGSI+V+ATNFCPPN AL N++GGWCNPPL+HFDLA+P FL+IAQY+AGIVP
Sbjct: 90 EDDPEWCIPGSIIVSATNFCPPNFALANDNGGWCNPPLKHFDLAEPAFLQIAQYRAGIVP 149
Query: 143 VDFRRV 148
V FRRV
Sbjct: 150 VAFRRV 155
>At5g02260 expansin precursor - like protein
Length = 258
Score = 221 bits (564), Expect = 8e-59
Identities = 101/152 (66%), Positives = 123/152 (80%), Gaps = 4/152 (2%)
Query: 1 MAFIGLVLVCSLTMFSSVYAY--GGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGT 58
+ F+ +++V + T + + GG W NAHATFYG +DASGTMGGACGYGNLYSQGYG
Sbjct: 6 ITFMAVMVVTAFTANAKIPGVYTGGPWINAHATFYGEADASGTMGGACGYGNLYSQGYGV 65
Query: 59 NTAALSTALFNNGLSCGSCYEIRCANDHRWCLPG--SIVVTATNFCPPNNALPNNDGGWC 116
NTAALSTALFNNGLSCGSC+E++C ND WCLPG SI++TATNFCPPN +++GGWC
Sbjct: 66 NTAALSTALFNNGLSCGSCFELKCINDPGWCLPGNPSILITATNFCPPNFNQASDNGGWC 125
Query: 117 NPPLQHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
NPP +HFDLA P+FL IA+YKAGIVPV +RR+
Sbjct: 126 NPPREHFDLAMPMFLSIAKYKAGIVPVSYRRI 157
>At1g20190 expansin S2 precursor like protein
Length = 252
Score = 207 bits (527), Expect = 2e-54
Identities = 102/148 (68%), Positives = 118/148 (78%), Gaps = 6/148 (4%)
Query: 5 GLVLVCSLTMFSSVYAYG-GGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAAL 63
GL ++ +L F +V A+ G TN HATFYGGSDASGTMGGACGYG+LYS GYGT TAAL
Sbjct: 7 GLAVLAAL--FIAVDAFRPSGLTNGHATFYGGSDASGTMGGACGYGDLYSAGYGTMTAAL 64
Query: 64 STALFNNGLSCGSCYEIRC--ANDHRWCLPG-SIVVTATNFCPPNNALPNNDGGWCNPPL 120
STALFN+G SCG CY I C A D RWCL G S+V+TATNFCPPN ALPNN+GGWCNPPL
Sbjct: 65 STALFNDGASCGECYRITCDHAADSRWCLKGASVVITATNFCPPNFALPNNNGGWCNPPL 124
Query: 121 QHFDLAQPVFLRIAQYKAGIVPVDFRRV 148
+HFD+AQP + +I Y+ GIVPV F+RV
Sbjct: 125 KHFDMAQPAWEKIGIYRGGIVPVVFQRV 152
>At4g01630 putative expansin
Length = 255
Score = 201 bits (510), Expect = 2e-52
Identities = 92/146 (63%), Positives = 111/146 (76%), Gaps = 3/146 (2%)
Query: 6 LVLVCSLTMFSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
+ ++ S F + + GW AHATFYGGSDASGTMGGACGYGNLY+ GY TNTAALST
Sbjct: 9 VAMIFSTMFFMKISSVSAGWLQAHATFYGGSDASGTMGGACGYGNLYTDGYKTNTAALST 68
Query: 66 ALFNNGLSCGSCYEIRC--ANDHRWCLPG-SIVVTATNFCPPNNALPNNDGGWCNPPLQH 122
ALFN+G SCG CY+I C +WCL G SI +TATNFCPPN A +++GGWCNPP H
Sbjct: 69 ALFNDGKSCGGCYQILCDATKVPQWCLKGKSITITATNFCPPNFAQASDNGGWCNPPRPH 128
Query: 123 FDLAQPVFLRIAQYKAGIVPVDFRRV 148
FD+AQP FL IA+YKAGIVP+ +++V
Sbjct: 129 FDMAQPAFLTIAKYKAGIVPILYKKV 154
>At3g29030 expansin At-EXP5
Length = 255
Score = 190 bits (483), Expect = 2e-49
Identities = 88/127 (69%), Positives = 102/127 (80%), Gaps = 9/127 (7%)
Query: 23 GGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRC 82
G W NAHATFYGG DASGTMGGACGYGNLYSQGYG TAALSTALF+ GLSCG+C+E+ C
Sbjct: 36 GPWINAHATFYGGGDASGTMGGACGYGNLYSQGYGLETAALSTALFDQGLSCGACFELMC 95
Query: 83 ANDHRWCLPG-SIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIV 141
ND +WC+ G SIVVTATNFCPP GG C+PP HFDL+QP++ +IA YK+GI+
Sbjct: 96 VNDPQWCIKGRSIVVTATNFCPP--------GGACDPPNHHFDLSQPIYEKIALYKSGII 147
Query: 142 PVDFRRV 148
PV +RRV
Sbjct: 148 PVMYRRV 154
>At5g39310 expansin-like protein
Length = 296
Score = 182 bits (462), Expect = 6e-47
Identities = 81/125 (64%), Positives = 98/125 (77%), Gaps = 2/125 (1%)
Query: 24 GWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCA 83
GW + ATFYG + T GACGYG+L+ QGYG TAALSTALFNNG CG+CYEI C
Sbjct: 73 GWGHGRATFYGDINGGETQQGACGYGDLHKQGYGLETAALSTALFNNGSRCGACYEIMCE 132
Query: 84 NDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPV 143
+ +WCLPGSI +TATNFCPP+ PN++ WCNPP +HFDL+QP+FL+IA+YKAG+VPV
Sbjct: 133 HAPQWCLPGSIKITATNFCPPDFTKPNDN--WCNPPQKHFDLSQPMFLKIAKYKAGVVPV 190
Query: 144 DFRRV 148
FRRV
Sbjct: 191 KFRRV 195
>At5g39280 expansin-like protein
Length = 259
Score = 179 bits (455), Expect = 4e-46
Identities = 81/143 (56%), Positives = 102/143 (70%), Gaps = 5/143 (3%)
Query: 9 VCSLTMFSSVYAYGG---GWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALST 65
V +F V + G W +A ATFYG T GACGYG+L+ QGYG TAALST
Sbjct: 19 VAEAPVFDDVVSPNGLDSSWYDARATFYGDIHGGETQQGACGYGDLFKQGYGLETAALST 78
Query: 66 ALFNNGLSCGSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDL 125
ALFN G +CG+CY+I C ND +WCLPGS+ +TATNFCPP+ + +G WCNPP +HFDL
Sbjct: 79 ALFNEGYTCGACYQIMCVNDPQWCLPGSVKITATNFCPPDYS--KTEGVWCNPPQKHFDL 136
Query: 126 AQPVFLRIAQYKAGIVPVDFRRV 148
+ P+FL+IAQYKAG+VPV +RR+
Sbjct: 137 SLPMFLKIAQYKAGVVPVKYRRI 159
>At5g39300 expansin-like protein
Length = 260
Score = 176 bits (446), Expect = 4e-45
Identities = 76/124 (61%), Positives = 96/124 (77%), Gaps = 2/124 (1%)
Query: 25 WTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCAN 84
W +A ATFYG T GACGYG+L+ QGYG TAALSTALFN G +CG+CY+I C +
Sbjct: 39 WYDARATFYGDIHGGETQQGACGYGDLFKQGYGLETAALSTALFNEGYTCGACYQIMCVH 98
Query: 85 DHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVD 144
D +WCLPG+I +TATNFCPP+ + +G WCNPP +HFDL+ P+FL+IAQYKAG+VPV
Sbjct: 99 DPQWCLPGTIKITATNFCPPDYS--KTEGVWCNPPQKHFDLSLPMFLKIAQYKAGVVPVK 156
Query: 145 FRRV 148
+RR+
Sbjct: 157 YRRI 160
>At5g39290 expansin-like protein
Length = 263
Score = 169 bits (427), Expect = 6e-43
Identities = 75/124 (60%), Positives = 92/124 (73%), Gaps = 2/124 (1%)
Query: 25 WTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSCGSCYEIRCAN 84
W +A ATFYG T GACGYGNL+ QGYG TAALSTALFN+G +CG+CYEI C
Sbjct: 42 WYDARATFYGDIHGGDTQQGACGYGNLFRQGYGLATAALSTALFNDGYTCGACYEIMCTR 101
Query: 85 DHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIAQYKAGIVPVD 144
D +WCLPGS+ +TATNFCP N + WCNPP +HFDL+ +FL+IA+YKAG+VPV
Sbjct: 102 DPQWCLPGSVKITATNFCPANYS--KTTDLWCNPPQKHFDLSLAMFLKIAKYKAGVVPVR 159
Query: 145 FRRV 148
+RR+
Sbjct: 160 YRRI 163
>At1g62980 expansin At-EXP6, putative
Length = 257
Score = 167 bits (424), Expect = 1e-42
Identities = 80/147 (54%), Positives = 100/147 (67%), Gaps = 5/147 (3%)
Query: 6 LVLVCSLTMF-SSVYAYGGG-WTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAAL 63
LV++ + M +S+ Y G W A ATFYG S TMGGACGYGN+Y GYG T AL
Sbjct: 10 LVILSMMAMIGTSMATYAGTPWRTASATFYGDDTGSATMGGACGYGNMYDSGYGVATTAL 69
Query: 64 STALFNNGLSCGSCYEIRCANDHRWCLPGS--IVVTATNFCPPNNALPNNDGGWCNPPLQ 121
STALFN G +CG C++++C + C GS VVTATN CPPN +N+GGWCNPP
Sbjct: 70 STALFNEGYACGQCFQLKCVSSPN-CYYGSPATVVTATNICPPNYGQASNNGGWCNPPRV 128
Query: 122 HFDLAQPVFLRIAQYKAGIVPVDFRRV 148
HFDL +P F++IA +KAGI+PV +RRV
Sbjct: 129 HFDLTKPAFMKIANWKAGIIPVSYRRV 155
>At1g12560 hypothetical protein
Length = 262
Score = 165 bits (417), Expect = 9e-42
Identities = 74/134 (55%), Positives = 92/134 (68%)
Query: 15 FSSVYAYGGGWTNAHATFYGGSDASGTMGGACGYGNLYSQGYGTNTAALSTALFNNGLSC 74
F + Y G W AHATFYG TMGGACGYGNL++ GYG +TAALST LFN+G C
Sbjct: 27 FVAGYYRPGPWRYAHATFYGDETGGETMGGACGYGNLFNSGYGLSTAALSTTLFNDGYGC 86
Query: 75 GSCYEIRCANDHRWCLPGSIVVTATNFCPPNNALPNNDGGWCNPPLQHFDLAQPVFLRIA 134
G C++I C+ S VVTATN CPPN +N GGWCNPP HFD+A+P F+++A
Sbjct: 87 GQCFQITCSKSPHCYSGKSTVVTATNLCPPNWYQDSNAGGWCNPPRTHFDMAKPAFMKLA 146
Query: 135 QYKAGIVPVDFRRV 148
++AGI+PV +RRV
Sbjct: 147 YWRAGIIPVAYRRV 160
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.140 0.471
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,833,829
Number of Sequences: 26719
Number of extensions: 170484
Number of successful extensions: 778
Number of sequences better than 10.0: 69
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 535
Number of HSP's gapped (non-prelim): 199
length of query: 148
length of database: 11,318,596
effective HSP length: 90
effective length of query: 58
effective length of database: 8,913,886
effective search space: 517005388
effective search space used: 517005388
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 55 (25.8 bits)
Medicago: description of AC140030.4


