

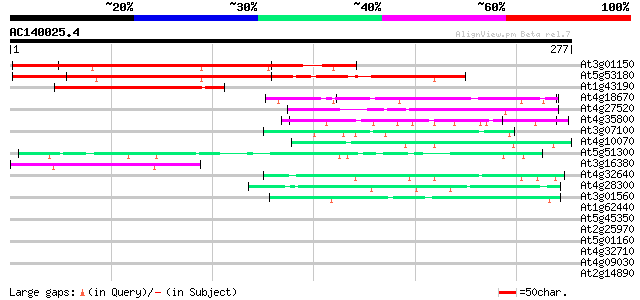
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140025.4 + phase: 0
(277 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g01150 polypyrimidine tract-binding protein like 220 6e-58
At5g53180 polypyrimidine tract-binding RNA transport protein-like 203 9e-53
At1g43190 hypothetical protein 73 2e-13
At4g18670 extensin-like protein 49 2e-06
At4g27520 early nodulin-like 2 predicted GPI-anchored protein 48 7e-06
At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain 47 1e-05
At3g07100 putative Sec24-like COPII protein 45 6e-05
At4g10070 putative DNA-directed RNA polymerase 44 7e-05
At5g51300 unknown protein 44 1e-04
At3g16380 putative poly(A) binding protein 43 2e-04
At4g32640 putative protein 42 5e-04
At4g28300 predicted proline-rich protein 42 5e-04
At3g01560 unknown protein 41 6e-04
At1g62440 putative extensin-like protein (gnl|PID|e1310400 39 0.004
At5g45350 unknown protein 38 0.005
At2g25970 pseudogene 38 0.007
At5g01160 unknown protein 37 0.009
At4g32710 putative protein kinase 37 0.009
At4g09030 arabinogalactan-protein AGP10 37 0.012
At2g14890 arabinogalactan-protein AGP9 37 0.012
>At3g01150 polypyrimidine tract-binding protein like
Length = 399
Score = 220 bits (561), Expect = 6e-58
Identities = 116/186 (62%), Positives = 133/186 (71%), Gaps = 25/186 (13%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPVN +A++G+ QPA+G DGK++E++SNVLL IENMQYAVTVDV++TVFSA+GTVQKIA
Sbjct: 216 LPVNQTAMDGSMQPALGADGKKVESQSNVLLGLIENMQYAVTVDVLHTVFSAYGTVQKIA 275
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
+FEKNG TQALIQY D+ TAA A+EALEGHCIYDGGYCKL LSYSRHTDLNVKAFSDKSR
Sbjct: 276 IFEKNGSTQALIQYSDIPTAAMAKEALEGHCIYDGGYCKLRLSYSRHTDLNVKAFSDKSR 335
Query: 122 DYTVP---------------LVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSP-AYQTQVP 165
DYT+P P WQNPQA Y GGSP Y + P
Sbjct: 336 DYTLPDLSLLVAQKGPAVSGSAPPAGWQNPQAQSQYSGY---------GGSPYMYPSSDP 386
Query: 166 GGQVPS 171
G PS
Sbjct: 387 NGASPS 392
Score = 101 bits (252), Expect = 4e-22
Identities = 57/112 (50%), Positives = 73/112 (64%), Gaps = 7/112 (6%)
Query: 25 ETESNVLLASIENMQ-YAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAA 83
+ NVLL + E ++ + V++DVI+ VFSAFG V KIA FEK QAL+Q+ DV TA+A
Sbjct: 106 DVPGNVLLVTFEGVESHEVSIDVIHLVFSAFGFVHKIATFEKAAGFQALVQFTDVETASA 165
Query: 84 AREALEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
AR AL+G I G C L +SYS HTDLN+K S +SRDYT P +P
Sbjct: 166 ARSALDGRSIPRYLLSAHVGSCSLRMSYSAHTDLNIKFQSHRSRDYTNPYLP 217
Score = 27.3 bits (59), Expect = 9.2
Identities = 19/50 (38%), Positives = 23/50 (46%), Gaps = 8/50 (16%)
Query: 232 SGGSTSGPGSSPHMQQNLGAQGMVRPGAP---PNVRPGGASPSGQ-HYYG 277
SG + +P Q G G+P P+ P GASPSGQ YYG
Sbjct: 354 SGSAPPAGWQNPQAQSQYSGYG----GSPYMYPSSDPNGASPSGQPPYYG 399
>At5g53180 polypyrimidine tract-binding RNA transport protein-like
Length = 429
Score = 203 bits (516), Expect = 9e-53
Identities = 118/225 (52%), Positives = 139/225 (61%), Gaps = 13/225 (5%)
Query: 2 LPVNHSAIEGAAQPAIGPDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIA 61
LPV SAI+ Q A+G DGK++E ESNVLLASIENMQYAVT+DV++ VF+AFG VQKIA
Sbjct: 217 LPVAPSAIDSTGQVAVGVDGKKMEPESNVLLASIENMQYAVTLDVLHMVFAAFGEVQKIA 276
Query: 62 MFEKNGQTQALIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKAFSDKSR 121
MF+KNG QALIQY DV TA A+EALEGHCIYDGG+CKLH++YSRHTDL++K +D+SR
Sbjct: 277 MFDKNGGVQALIQYSDVQTAVVAKEALEGHCIYDGGFCKLHITYSRHTDLSIKVNNDRSR 336
Query: 122 DYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRP 181
DYT+P P P+ Q P P Y N Y GGS Q Q P G W V+P
Sbjct: 337 DYTMPNPPVPMPQQPVQNP-YAGNPQQY--HAAGGSHHQQQQQPQG---GW------VQP 384
Query: 182 GYVPVPGAYPGQTGAFPTMPSYGSAAM-PTASSPLAQSSHPGAPH 225
G G G + PS S P P PG H
Sbjct: 385 GGQGSMGMGGGGHNHYMAPPSSSSMHQGPGGHMPPQHYGGPGPMH 429
Score = 90.1 bits (222), Expect = 1e-18
Identities = 53/108 (49%), Positives = 66/108 (61%), Gaps = 7/108 (6%)
Query: 29 NVLLASIENMQYA-VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREA 87
NVLL +IE V++DV++ VFSAFG V KI FEK QAL+Q+ D TA AA+ A
Sbjct: 111 NVLLVTIEGDDARMVSIDVLHLVFSAFGFVHKITTFEKTAGYQALVQFTDAETATAAKLA 170
Query: 88 LEGHCI------YDGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVP 129
L+G I G C L ++YS HTDL VK S +SRDYT P +P
Sbjct: 171 LDGRSIPRYLLAETVGQCSLKITYSAHTDLTVKFQSHRSRDYTNPYLP 218
>At1g43190 hypothetical protein
Length = 396
Score = 72.8 bits (177), Expect = 2e-13
Identities = 38/84 (45%), Positives = 56/84 (66%), Gaps = 1/84 (1%)
Query: 23 RIETESNVLLASIENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAA 82
R + + +LL +I +M Y +TVDV++ VFS +G V+K+ F+K+ QALIQY AA
Sbjct: 93 REDEPNRILLVTIHHMLYPITVDVLHQVFSPYGFVEKLVTFQKSAGFQALIQYQVQQCAA 152
Query: 83 AAREALEGHCIYDGGYCKLHLSYS 106
+AR AL+G IYD G C+L + +S
Sbjct: 153 SARTALQGRNIYD-GCCQLDIQFS 175
Score = 28.5 bits (62), Expect = 4.1
Identities = 15/43 (34%), Positives = 22/43 (50%)
Query: 42 VTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAA 84
VT + + G V +FE NG+ QAL+Q+ + AA A
Sbjct: 330 VTEEEVMNHVQEHGAVVNTKVFEMNGKKQALVQFENEEEAAEA 372
>At4g18670 extensin-like protein
Length = 839
Score = 49.3 bits (116), Expect = 2e-06
Identities = 47/147 (31%), Positives = 58/147 (38%), Gaps = 25/147 (17%)
Query: 127 LVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSP--AYQTQVPGGQVPSWDLTQHAVRPGYV 184
+VP+P P +P P SP T PGGSP + T PGG PS T PG
Sbjct: 454 IVPSPPSTTP--SPGSPPTSPT--TPTPGGSPPSSPTTPTPGGSPPSSPTTP---TPGGS 506
Query: 185 PVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPH 244
P G P PS + T SP + + PG+P + PS + S P SP
Sbjct: 507 PPSSPTTPSPGGSPPSPSISPSPPITVPSPPSTPTSPGSPPS----PSSPTPSSPIPSP- 561
Query: 245 MQQNLGAQGMVRPGAPPN-VRPGGASP 270
P PP + PG SP
Sbjct: 562 ----------PTPSTPPTPISPGQNSP 578
Score = 48.1 bits (113), Expect = 5e-06
Identities = 35/110 (31%), Positives = 44/110 (39%), Gaps = 3/110 (2%)
Query: 162 TQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHP 221
T PGG PS ++ P VP P P G+ P PS + T SP + + P
Sbjct: 416 TPSPGGSPPSPSISPSP--PITVPSPPTTPSPGGS-PPSPSIVPSPPSTTPSPGSPPTSP 472
Query: 222 GAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPS 271
P PS +T PG SP G P +P PGG+ PS
Sbjct: 473 TTPTPGGSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPSPGGSPPS 522
Score = 45.1 bits (105), Expect = 4e-05
Identities = 45/157 (28%), Positives = 66/157 (41%), Gaps = 22/157 (14%)
Query: 127 LVPAP-VWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVP 185
+VP+P +P +P P+ SP+ VP SP T PGG PS + P P
Sbjct: 409 VVPSPPTTPSPGGSPPSPSISPSPPITVP--SPP-TTPSPGGSPPSPSIVPSP--PSTTP 463
Query: 186 VPGAYP---------GQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGST 236
PG+ P G + PT P+ G + + ++P S P +P + GGS
Sbjct: 464 SPGSPPTSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPS---PGGSP 520
Query: 237 SGPGSSPHMQQNLGA--QGMVRPGAPPNVRPGGASPS 271
P SP + + PG+PP+ P +PS
Sbjct: 521 PSPSISPSPPITVPSPPSTPTSPGSPPS--PSSPTPS 555
>At4g27520 early nodulin-like 2 predicted GPI-anchored protein
Length = 349
Score = 47.8 bits (112), Expect = 7e-06
Identities = 38/140 (27%), Positives = 61/140 (43%), Gaps = 21/140 (15%)
Query: 138 AAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAF 197
++P+ PT SP T PGG+ + ++ AV P P PG+ ++G+
Sbjct: 165 SSPVSPTTSPPGSTTPPGGAHSPKSS-------------SAVSPATSP-PGSMAPKSGS- 209
Query: 198 PTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP------HMQQNLGA 251
P P+ A P ++SP++ SS P + P ST P S+P M +
Sbjct: 210 PVSPTTSPPAPPKSTSPVSPSSAPMTSPPAPMAPKSSSTIPPSSAPMTSPPGSMAPKSSS 269
Query: 252 QGMVRPGAPPNVRPGGASPS 271
P P++ PGG++ S
Sbjct: 270 PVSNSPTVSPSLAPGGSTSS 289
>At4g35800 DNA-directed RNA polymerase (EC 2.7.7.6) II largest chain
Length = 1840
Score = 47.0 bits (110), Expect = 1e-05
Identities = 39/113 (34%), Positives = 47/113 (41%), Gaps = 9/113 (7%)
Query: 135 NPQAAPMYPTNSPAYQTQVPG---GSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAY- 190
+P ++P Y +SP Y PG SP Y PG S T PGY P AY
Sbjct: 1538 SPSSSPGYSPSSPGYSPTSPGYSPTSPGYSPTSPGYSPTS--PTYSPSSPGYSPTSPAYS 1595
Query: 191 PGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSP 243
P PT PSY + + SP + S P +P PS TS P SP
Sbjct: 1596 PTSPSYSPTSPSYSPTS--PSYSPTSPSYSPTSPSYSPTSPSYSPTS-PAYSP 1645
Score = 42.4 bits (98), Expect = 3e-04
Identities = 41/133 (30%), Positives = 51/133 (37%), Gaps = 14/133 (10%)
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAY-PGQTGAF 197
+P Y SPAY P SP P PS+ T P Y P +Y P
Sbjct: 1633 SPSYSPTSPAYSPTSPAYSPTSPAYSPTS--PSYSPTS----PSYSPTSPSYSPTSPSYS 1686
Query: 198 PTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRP 257
PT PSY + A SP + P +P PS G TS P +P + +
Sbjct: 1687 PTSPSYSPTS--PAYSPTSPGYSPTSPSYSPTSPSYGPTS-PSYNPQSAKYSPSIAY--- 1740
Query: 258 GAPPNVRPGGASP 270
+P N R ASP
Sbjct: 1741 -SPSNARLSPASP 1752
Score = 41.6 bits (96), Expect = 5e-04
Identities = 46/149 (30%), Positives = 56/149 (36%), Gaps = 19/149 (12%)
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHA---VRPGYVPVPGAY-PGQT 194
+P Y SPAY P SP + P PS+ T + P Y P AY P
Sbjct: 1647 SPAYSPTSPAYSPTSPSYSPTSPSYSPTS--PSYSPTSPSYSPTSPSYSPTSPAYSPTSP 1704
Query: 195 GAFPTMPSYGSAAM---PTASSPLAQS-----SHPGAPHNVNLQP-SGGSTSGPGSSPHM 245
G PT PSY + PT+ S QS S +P N L P S S + P SP
Sbjct: 1705 GYSPTSPSYSPTSPSYGPTSPSYNPQSAKYSPSIAYSPSNARLSPASPYSPTSPNYSPTS 1764
Query: 246 QQNLGAQGMVRPG----APPNVRPGGASP 270
P +P + GASP
Sbjct: 1765 PSYSPTSPSYSPSSPTYSPSSPYSSGASP 1793
Score = 41.2 bits (95), Expect = 6e-04
Identities = 40/141 (28%), Positives = 57/141 (40%), Gaps = 16/141 (11%)
Query: 139 APMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAF- 197
+P Y SPAY PG SP PS+ T P Y P +Y Q+ +
Sbjct: 1689 SPSYSPTSPAYSPTSPGYSPT---------SPSYSPTS----PSYGPTSPSYNPQSAKYS 1735
Query: 198 PTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGG--STSGPGSSPHMQQNLGAQGMV 255
P++ S A + +SP + +S +P + + P+ S S P SP + GA
Sbjct: 1736 PSIAYSPSNARLSPASPYSPTSPNYSPTSPSYSPTSPSYSPSSPTYSPSSPYSSGASPDY 1795
Query: 256 RPGAPPNVRPGGASPSGQHYY 276
P A + G SPS Y
Sbjct: 1796 SPSAGYSPTLPGYSPSSTGQY 1816
>At3g07100 putative Sec24-like COPII protein
Length = 1054
Score = 44.7 bits (104), Expect = 6e-05
Identities = 45/146 (30%), Positives = 56/146 (37%), Gaps = 25/146 (17%)
Query: 126 PLVPAPVWQNPQAAPMYPTNSPAY------------QTQVPGGSPAYQTQ--VPGGQV-- 169
P P P QNP P P + V G P T +PG
Sbjct: 114 PSPPFPTTQNPPQGPPPPQTLAGHLSPPMSLRPQQPMAPVAMGPPPQSTTSGLPGANAYP 173
Query: 170 PSWDLTQHAVRPGYV----PVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPH 225
P+ D A RPG+ PV +YPG G+ P+ P Y S + A +P S P P
Sbjct: 174 PATDYHMPA-RPGFQQSMPPVTPSYPGVGGSQPSFPGYPSKQVLQAPTPFQTSQGPPGPP 232
Query: 226 NVNLQPSGGSTSGPGSSPHM--QQNL 249
V+ P T G P+M QQNL
Sbjct: 233 PVSSYPP--HTGGFAQRPNMAAQQNL 256
Score = 36.6 bits (83), Expect = 0.015
Identities = 41/153 (26%), Positives = 57/153 (36%), Gaps = 25/153 (16%)
Query: 143 PTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPS 202
P +P T PG + +Q+PG + P + P Y P PG P Q FP+ P
Sbjct: 63 PPPAPPVGTMRPGQPSPFVSQIPGSRPPP-PSSNSFPSPAYGP-PGGAPFQR--FPS-PP 117
Query: 203 YGSAAMPTASSPLAQS--SHPGAPHNVNLQ----------PSGGSTSG--------PGSS 242
+ + P P Q+ H P ++ Q P +TSG P +
Sbjct: 118 FPTTQNPPQGPPPPQTLAGHLSPPMSLRPQQPMAPVAMGPPPQSTTSGLPGANAYPPATD 177
Query: 243 PHMQQNLGAQGMVRPGAPPNVRPGGASPSGQHY 275
HM G Q + P P GG+ PS Y
Sbjct: 178 YHMPARPGFQQSMPPVTPSYPGVGGSQPSFPGY 210
Score = 29.3 bits (64), Expect = 2.4
Identities = 22/64 (34%), Positives = 26/64 (40%), Gaps = 4/64 (6%)
Query: 211 ASSPLAQSSHPGAPHNVNLQPSGGSTSG--PGSSPHMQQNLGAQGMVRPGAPP--NVRPG 266
ASSP A + PG P P+G G P + Q P APP +RPG
Sbjct: 16 ASSPFASAPPPGIPPQSGGPPTGSEAVGFRPFTPSASQPTRPFTASGPPPAPPVGTMRPG 75
Query: 267 GASP 270
SP
Sbjct: 76 QPSP 79
>At4g10070 putative DNA-directed RNA polymerase
Length = 748
Score = 44.3 bits (103), Expect = 7e-05
Identities = 46/166 (27%), Positives = 58/166 (34%), Gaps = 30/166 (18%)
Query: 140 PMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQT----- 194
PM P +Y G S YQ Q PG V + + Q V+ GY P A +
Sbjct: 528 PMQPPYGGSYPPAGGGQSGYYQMQQPG--VRPYGMQQGPVQQGYGPPQPAAAASSGDVPY 585
Query: 195 -GAFPTMPSYGSAAM---------PTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPH 244
GA P PSYGS M ++ P+ Q ++P T P + P
Sbjct: 586 QGATPAAPSYGSTNMAPQQQQYGYTSSDGPVQQQTYPSYSSAPPSDAYNNGTQTPATGPA 645
Query: 245 MQQ-----------NLGAQGMVRPGAPPNVRPGG--ASPSGQHYYG 277
QQ GAQ G V P G P+ Q YG
Sbjct: 646 YQQQSVQPASSTYDQTGAQQAAAAGYGGQVAPTGGYTYPTSQPAYG 691
Score = 34.3 bits (77), Expect = 0.075
Identities = 40/127 (31%), Positives = 55/127 (42%), Gaps = 20/127 (15%)
Query: 131 PVWQNPQAAPMYPTNSP--AYQ--TQVPGGSPAYQTQVPGGQVPSWDLT--QHAVRPGYV 184
PV Q Q P Y + P AY TQ P PAYQ Q ++D T Q A GY
Sbjct: 615 PVQQ--QTYPSYSSAPPSDAYNNGTQTPATGPAYQQQSVQPASSTYDQTGAQQAAAAGY- 671
Query: 185 PVPGAYPGQTG--AFPT-MPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGS 241
G TG +PT P+YGS A + ++P Q+ + P + + PG+
Sbjct: 672 ---GGQVAPTGGYTYPTSQPAYGSQAAYSQAAP-TQTGYEQQP----ATQAAVYATAPGT 723
Query: 242 SPHMQQN 248
+P Q+
Sbjct: 724 APVKTQS 730
Score = 32.7 bits (73), Expect = 0.22
Identities = 40/143 (27%), Positives = 53/143 (36%), Gaps = 15/143 (10%)
Query: 145 NSPAYQTQVPGGSPAYQTQVPGGQVPSWDL---TQHAVRPGYVPVP--GAYPGQTGAFPT 199
N PAY+ Q PGG P + ++ P P +D ++ + Y P G YP Q P
Sbjct: 379 NQPAYRPQGPGGPPQWGSRGPHAPHP-YDYHPRGPYSSQGSYYNSPGFGGYPPQ--HMPP 435
Query: 200 MPSYGS--AAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGP--GSSPHMQQNLG---AQ 252
YG+ P S P GA + P G P G P Q + G +
Sbjct: 436 RGGYGTDWDQRPPYSGPYNYYGRQGAQSAGPVPPPSGPVPSPAFGGPPLSQVSYGYGQSH 495
Query: 253 GMVRPGAPPNVRPGGASPSGQHY 275
G A P + G GQ Y
Sbjct: 496 GPEYGHAAPYSQTGYQQTYGQTY 518
>At5g51300 unknown protein
Length = 804
Score = 43.5 bits (101), Expect = 1e-04
Identities = 71/281 (25%), Positives = 108/281 (38%), Gaps = 68/281 (24%)
Query: 5 NHSAIEGAAQPAIG-----PDGKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTV-- 57
N++ +A P +G P K + E+N+ + + M + D + +FS+FG +
Sbjct: 453 NNAGNGASAHPGLGSTPTKPPSKEYD-ETNLYIGFLPPM---LEDDGLINLFSSFGEIVM 508
Query: 58 QKIAMFEKNGQTQA--LIQYPDVTTAAAAREALEGHCIYDGGYCKLHLSYSRHTDLNVKA 115
K+ G ++ ++Y DV A A +A+ G+ ++G + ++
Sbjct: 509 AKVIKDRVTGLSKGYGFVKYADVQMANTAVQAMNGYR-FEGRTLAVRIA----------- 556
Query: 116 FSDKSRDYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQ--------TQVP-G 166
KS P P+ AP PT Q PG P+ Q VP G
Sbjct: 557 --GKS--------PPPIAPPGPPAPQPPTQGYPPSNQPPGAYPSQQYATGGYSTAPVPWG 606
Query: 167 GQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHN 226
VPS+ + +A+ P P PG+Y G MP YG +P P +
Sbjct: 607 PPVPSY--SPYALPP---PPPGSYHPVHGQH--MPPYG-------------MQYPPPPPH 646
Query: 227 VNLQPSGGSTSGPGSS-PHMQQNLGAQ---GMVRPGAPPNV 263
V P G+T P SS P G Q G PPNV
Sbjct: 647 VTQAPPPGTTQNPSSSEPQQSFPPGVQADSGAATSSIPPNV 687
Score = 35.0 bits (79), Expect = 0.044
Identities = 37/130 (28%), Positives = 47/130 (35%), Gaps = 4/130 (3%)
Query: 134 QNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQ 193
Q P P P TQ P S Q+ PG Q S T Y A PGQ
Sbjct: 639 QYPPPPPHVTQAPPPGTTQNPSSSEPQQSFPPGVQADSGAATSSIPPNVYGSSVTAMPGQ 698
Query: 194 TGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQG 253
+ + PSY +A P +P A +S N+ P + S + H Q + A
Sbjct: 699 P-PYMSYPSYYNAVPP--PTPPAPASSTDHSQNMGNMPWANNPS-VSTPDHSQGLVNAPW 754
Query: 254 MVRPGAPPNV 263
P PP V
Sbjct: 755 APNPPMPPTV 764
>At3g16380 putative poly(A) binding protein
Length = 537
Score = 42.7 bits (99), Expect = 2e-04
Identities = 25/99 (25%), Positives = 48/99 (48%), Gaps = 5/99 (5%)
Query: 1 MLPVNHSAIEGAAQPAIGPD---GKRIETESNVLLASIENMQYAVTVDVINTVFSAFGTV 57
M +NHS ++G A + R T + ++N+ ++T + +F FG++
Sbjct: 79 MTRLNHSDLKGKAMRIMWSQRDLAYRRRTRTGFANLYVKNLDSSITSSCLERMFCPFGSI 138
Query: 58 QKIAMFEKNGQTQ--ALIQYPDVTTAAAAREALEGHCIY 94
+ E+NGQ++ +Q+ +A +AR AL G +Y
Sbjct: 139 LSCKVVEENGQSKGFGFVQFDTEQSAVSARSALHGSMVY 177
Score = 31.2 bits (69), Expect = 0.64
Identities = 23/101 (22%), Positives = 41/101 (39%), Gaps = 1/101 (0%)
Query: 35 IENMQYAVTVDVINTVFSAFGTVQKIAMFEKNGQTQALIQYPDVTTAAAAREALEGHCIY 94
++N+ VT D ++T+FS +GTV + + + + A++A+E C
Sbjct: 206 VKNLIETVTDDCLHTLFSQYGTVSSVVVMRDGMGRSRGFGFVNFCNPENAKKAMESLCGL 265
Query: 95 DGGYCKLHLSYSRHTDLNVKAFSDKSRDYTVPLVPAPVWQN 135
G KL + + D + K D + P W N
Sbjct: 266 QLGSKKLFVGKALKKDERREMLKQKFSDNFI-AKPNMRWSN 305
>At4g32640 putative protein
Length = 1069
Score = 41.6 bits (96), Expect = 5e-04
Identities = 52/192 (27%), Positives = 65/192 (33%), Gaps = 47/192 (24%)
Query: 126 PLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQV---------------P 170
P P P Q+PQ +P SP+Y G SP + P G
Sbjct: 55 PRPPPPFGQSPQP---FPQQSPSYGAPQRGPSPMSRPGPPAGMARPGGPPPVSQPAGFQS 111
Query: 171 SWDLTQHAVRPGYVPVPGAYPGQTGA-----------FPTMPSYGSAAM--PTASSPLAQ 217
+ L + P P G+ P G FP GS A P S P+A
Sbjct: 112 NVPLNRPTGPPSRQPSFGSRPSMPGGPVAQPAASSSGFPAFGPSGSVAAGPPPGSRPMAF 171
Query: 218 SSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGA--QGMVRPGA------PPNVRPGGA- 268
S P +++ PS G GP S+ H G +G PGA P VRP A
Sbjct: 172 GSPPPVGSGMSMPPS-GMIGGPVSNGHQMVGSGGFPRGTQFPGAAVTTPQAPYVRPPSAP 230
Query: 269 ------SPSGQH 274
P G H
Sbjct: 231 YARTPPQPLGSH 242
>At4g28300 predicted proline-rich protein
Length = 496
Score = 41.6 bits (96), Expect = 5e-04
Identities = 42/160 (26%), Positives = 62/160 (38%), Gaps = 13/160 (8%)
Query: 119 KSRDYTVPLVPAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQH- 177
+ Y +P P + P AP+ P ++P Q Q P + P PS TQ
Sbjct: 234 QQHQYYMPPPPTQLQNTP--APV-PVSTPPSQLQAPPAQSQFMPPPPAPSHPSSAQTQSF 290
Query: 178 -AVRPGYVPVPGAYPGQTGAFPT-MPSYGSAAMPTASSPLA---QSSHPGAPHNVNLQPS 232
+ + P P A P +G +PT P+ P S P + QS + G P ++Q
Sbjct: 291 PQYQQNWPPQPQARPQSSGGYPTYSPAPPGNQPPVESLPSSMQMQSPYSGPPQQ-SMQAY 349
Query: 233 GGSTSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSG 272
G + P +P Q + G P+ G PSG
Sbjct: 350 GYGAAPPPQAPPQQTKMSYSPQTGDGYLPS---GPPPPSG 386
Score = 36.2 bits (82), Expect = 0.020
Identities = 42/169 (24%), Positives = 52/169 (29%), Gaps = 33/169 (19%)
Query: 129 PAPVWQNPQAA-----PMYPTN---SPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVR 180
P P +P +A P Y N P + Q GG P Y PG Q P L
Sbjct: 275 PPPAPSHPSSAQTQSFPQYQQNWPPQPQARPQSSGGYPTYSPAPPGNQPPVESL------ 328
Query: 181 PGYVPVPGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHN-------------- 226
P + + Y G +M +YG A P +P Q+ +P
Sbjct: 329 PSSMQMQSPYSGPPQ--QSMQAYGYGAAPPPQAPPQQTKMSYSPQTGDGYLPSGPPPPSG 386
Query: 227 -VNLQPSGGSTSGPGSSPHMQQNLG--AQGMVRPGAPPNVRPGGASPSG 272
N GG P P QQ QG G P G G
Sbjct: 387 YANAMYEGGRMQYPPPQPQQQQQQAHYLQGPQGGGYSPQPHQAGGGNIG 435
>At3g01560 unknown protein
Length = 511
Score = 41.2 bits (95), Expect = 6e-04
Identities = 43/152 (28%), Positives = 53/152 (34%), Gaps = 13/152 (8%)
Query: 129 PAPVWQNPQAAPMYPTNSPAYQTQVPGGS--PAYQTQVPGGQVPSWDLTQHAVRPGYVPV 186
P P+ Q P + P +S A +Q P P TQ Q P H P P
Sbjct: 242 PVPMQQFPLTSFPQPPSSTAAPSQPPSSQLPPQLPTQFSSQQEPYCPPPSHPQPPPSNPP 301
Query: 187 PGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQ 246
P Y P PSY S P Q P + +N QP S P + P Q
Sbjct: 302 P--YQAPQTQTPHQPSYQS---PPQQPQYPQQPPPSSGYNPEEQPPYQMQSYPPNPPRQQ 356
Query: 247 QNLGAQGMVRPGAPPNVRP------GGASPSG 272
G+ + PP +P GG S SG
Sbjct: 357 PPAGSTPSQQFYNPPQPQPSMYDGAGGRSNSG 388
>At1g62440 putative extensin-like protein (gnl|PID|e1310400
Length = 786
Score = 38.5 bits (88), Expect = 0.004
Identities = 33/104 (31%), Positives = 40/104 (37%), Gaps = 11/104 (10%)
Query: 129 PAPVWQNPQAAPMYPTNSPAYQ--TQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPV 186
P Q P YP+ SP Y T P Y TQ P P T +AV+ P
Sbjct: 540 PPKYEQTPSPREYYPSPSPPYYQYTSSPPPPTYYATQSPPPPPPP---TYYAVQSPPPPP 596
Query: 187 PGAYPGQTGAFPTMPSYGSAAM------PTASSPLAQSSHPGAP 224
P YP T + P P Y + + P SP+ QS P P
Sbjct: 597 PVYYPPVTASPPPPPVYYTPVIQSPPPPPVYYSPVTQSPPPPPP 640
Score = 33.9 bits (76), Expect = 0.098
Identities = 43/163 (26%), Positives = 55/163 (33%), Gaps = 17/163 (10%)
Query: 126 PLVPAPVWQNPQAAPMYPTNSPAYQTQV---PGGSPAYQTQVPGGQVPSWDLTQHAVRPG 182
P P PV+ P A P P Y T V P P Y + V P + V
Sbjct: 592 PPPPPPVYYPPVTAS--PPPPPVYYTPVIQSPPPPPVYYSPVTQSPPPPPPVYYPPVTQS 649
Query: 183 YVPVPGAYPGQTGAFPTMPSY-------GSAAMPTASSPLAQSSHPGAPHNVNLQPSGGS 235
P P YP T + P P Y P P+A+S P +P P
Sbjct: 650 PPPSPVYYPPVTQSPPPPPVYYLPVTQSPPPPSPVYYPPVAKSPPPPSP---VYYPPVTQ 706
Query: 236 TSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGA--SPSGQHYY 276
+ P S+P + P + P G SPS H+Y
Sbjct: 707 SPPPPSTPVEYHPPASPNQSPPPEYQSPPPKGCNDSPSNDHHY 749
>At5g45350 unknown protein
Length = 177
Score = 38.1 bits (87), Expect = 0.005
Identities = 30/106 (28%), Positives = 38/106 (35%), Gaps = 7/106 (6%)
Query: 177 HAVRPGYVPVPGAYP----GQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPS 232
H P P PGAYP Q G P +Y A P + P A +P AP P+
Sbjct: 14 HGYPPAGYPPPGAYPPAGYPQQGYPPPPGAYPPAGYPPGAYPPAPGGYPPAPGYGGYPPA 73
Query: 233 ---GGSTSGPGSSPHMQQNLGAQGMVRPGAPPNVRPGGASPSGQHY 275
GG PG + A G + G A+ G H+
Sbjct: 74 PGYGGYPPAPGHGGYPPAGYPAHHSGHAGGIGGMIAGAAAAYGAHH 119
Score = 32.7 bits (73), Expect = 0.22
Identities = 26/85 (30%), Positives = 27/85 (31%), Gaps = 5/85 (5%)
Query: 143 PTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRP---GYVPVPG--AYPGQTGAF 197
P P P G P P G P A P GY P PG YP G
Sbjct: 18 PAGYPPPGAYPPAGYPQQGYPPPPGAYPPAGYPPGAYPPAPGGYPPAPGYGGYPPAPGYG 77
Query: 198 PTMPSYGSAAMPTASSPLAQSSHPG 222
P+ G P A P S H G
Sbjct: 78 GYPPAPGHGGYPPAGYPAHHSGHAG 102
>At2g25970 pseudogene
Length = 632
Score = 37.7 bits (86), Expect = 0.007
Identities = 31/131 (23%), Positives = 48/131 (35%), Gaps = 18/131 (13%)
Query: 143 PTNSPAYQTQVPGG-SPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTG------ 195
PT+ Y +Q + Y P + P++ +Q + PG PG+Y Q+G
Sbjct: 510 PTSQAGYSSQPAAAYNSGYGAPPPASKPPTYGQSQQS--PG---APGSYGSQSGYAQPAA 564
Query: 196 -AFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGM 254
+ P+YG P + P A + S G+T+G G +Q
Sbjct: 565 SGYGQPPAYGYGQAPQGYGSYGGYTQPAAGGGYSSDGSAGATAGGGGG-----TPASQSA 619
Query: 255 VRPGAPPNVRP 265
P PP P
Sbjct: 620 APPAGPPKASP 630
Score = 30.8 bits (68), Expect = 0.83
Identities = 35/141 (24%), Positives = 51/141 (35%), Gaps = 28/141 (19%)
Query: 150 QTQVP--GGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVP------GAYP-------GQT 194
Q+Q P GGS A T G ++ QHA G GAY GQ
Sbjct: 397 QSQQPSSGGSSAPPTDTTG-----YNYYQHASGYGQAGQGYQQDGYGAYNASQQSGYGQA 451
Query: 195 GAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGM 254
+ YGS P+ +Q++ P + + +G G++ G+ G
Sbjct: 452 AGYDQQGGYGSTTNPSQEEDASQAAPPSSAQS--------GQAGYGTTGQQPPAQGSTGQ 503
Query: 255 VRPGAPPNVRPGGASPSGQHY 275
GAPP + G +S Y
Sbjct: 504 AGYGAPPTSQAGYSSQPAAAY 524
>At5g01160 unknown protein
Length = 360
Score = 37.4 bits (85), Expect = 0.009
Identities = 39/135 (28%), Positives = 56/135 (40%), Gaps = 21/135 (15%)
Query: 142 YPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMP 201
YP +S ++ PG A + PG + P + + ++P +PVP P +P
Sbjct: 213 YPPDSD--NSRPPGFETA--SPKPGIRFPDYPQPMNLMQPPSLPVP------MNQNPGLP 262
Query: 202 S-YGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSG-----PGSSPHMQQNLGAQGMV 255
+G + PT S +Q GA + + SGGS P SP NL QG
Sbjct: 263 QQFGFPSYPTTESGSSQQFFNGAQYEMTRTESGGSEQSSLLGYPPPSP--MTNLNFQGSY 320
Query: 256 RPGAPPNVRPGGASP 270
PP+ PG A P
Sbjct: 321 ---PPPSWNPGMAPP 332
>At4g32710 putative protein kinase
Length = 731
Score = 37.4 bits (85), Expect = 0.009
Identities = 38/157 (24%), Positives = 56/157 (35%), Gaps = 17/157 (10%)
Query: 129 PAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYV--PV 186
P P+ ++P P+ + P+ P GSP P V PG P
Sbjct: 80 PPPLLESPPPPPLESPSPPSPHVSAPSGSPPLPFLPAKPSPPPSSPPSETVPPGNTISPP 139
Query: 187 PGAYPGQTGAFPTMPSYGSAAMPTASSPLAQSS-HPGAPHNVNL---QPSGGSTSGPGSS 242
P + P + + P +A+ P S P +S P P +N PS + S P +S
Sbjct: 140 PRSLPSE-----STPPVNTASPPPPSPPRRRSGPKPSFPPPINSSPPNPSPNTPSLPETS 194
Query: 243 PHMQQNLGAQGMVRPGAPPNVRPGGA------SPSGQ 273
P + L PP + A P+GQ
Sbjct: 195 PPPKPPLSTTPFPSSSTPPPKKSPAAVTLPFFGPAGQ 231
Score = 32.3 bits (72), Expect = 0.29
Identities = 35/130 (26%), Positives = 46/130 (34%), Gaps = 7/130 (5%)
Query: 143 PTNSPAYQTQVPGGSPAYQTQVPGGQVPSWD----LTQHAVRPGYVPVPGAYPGQTGAFP 198
P++SPA T P S VP P L P +P P T + P
Sbjct: 5 PSSSPAPATSPPAMSLPPADSVPDTSSPPAPPLSPLPPPLSSPPPLPSPPPLSAPTASPP 64
Query: 199 TMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGAQGMVRPG 258
+P + P S P P P + P S P SP + L A+ P
Sbjct: 65 PLPVESPPSPPIESPPPPLLESPPPPPLESPSPPSPHVSAPSGSPPL-PFLPAKPSPPPS 123
Query: 259 APPN--VRPG 266
+PP+ V PG
Sbjct: 124 SPPSETVPPG 133
>At4g09030 arabinogalactan-protein AGP10
Length = 127
Score = 37.0 bits (84), Expect = 0.012
Identities = 28/100 (28%), Positives = 36/100 (36%), Gaps = 12/100 (12%)
Query: 152 QVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPGAYPGQTGAFPTMPSYGSAAMPTA 211
Q PG +P Q P ++ P P P A P + P A
Sbjct: 22 QAPGPAPTRSPLPSPAQPPRTAAPTPSITPTPTPTPSATP-----------TAAPVSPPA 70
Query: 212 SSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQNLGA 251
SPL S+ P AP +L P G +GP S + N A
Sbjct: 71 GSPLPSSASPPAP-PTSLTPDGAPVAGPTGSTPVDNNNAA 109
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 37.0 bits (84), Expect = 0.012
Identities = 38/127 (29%), Positives = 43/127 (32%), Gaps = 20/127 (15%)
Query: 129 PAPVWQNPQAAPMYPTNSPAYQTQVPGGSPAYQTQVPGGQVPSWDLTQHAVRPGYVPVPG 188
P PV P A+P T P P SP T P P L A P VP P
Sbjct: 71 PPPVSSPPPASPPPATPPPVASPPPPVASPPPATPPPVATPPPAPL---ASPPAQVPAPA 127
Query: 189 AYPGQTGAFPTMPSYGSAAMPTASSPLAQSSHPGAPHNVNLQPSGGSTSGPGSSPHMQQN 248
PT + P++S PL S PG PS S S S +
Sbjct: 128 ---------PTTKPDSPSPSPSSSPPLPSSDAPG--------PSTDSISPAPSPTDVNDQ 170
Query: 249 LGAQGMV 255
GA MV
Sbjct: 171 NGASKMV 177
Score = 33.9 bits (76), Expect = 0.098
Identities = 26/84 (30%), Positives = 36/84 (41%), Gaps = 5/84 (5%)
Query: 192 GQTGAFPTMPSYGSAAMPTASSPLAQSSHP--GAPHNVNLQPSGGSTSGPGSSPHMQQNL 249
G TG PT P + A PT ++P ++ P AP V P +T+ P ++P +
Sbjct: 17 GVTGQAPTSPPTATPAPPTPTTPPPAATPPPVSAPPPVTTSPPPVTTAPPPANPPPPVSS 76
Query: 250 GAQGMVRPGAPPNVR---PGGASP 270
P PP V P ASP
Sbjct: 77 PPPASPPPATPPPVASPPPPVASP 100
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.312 0.130 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,878,522
Number of Sequences: 26719
Number of extensions: 320108
Number of successful extensions: 1057
Number of sequences better than 10.0: 122
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 98
Number of HSP's that attempted gapping in prelim test: 814
Number of HSP's gapped (non-prelim): 242
length of query: 277
length of database: 11,318,596
effective HSP length: 98
effective length of query: 179
effective length of database: 8,700,134
effective search space: 1557323986
effective search space used: 1557323986
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 59 (27.3 bits)
Medicago: description of AC140025.4


