

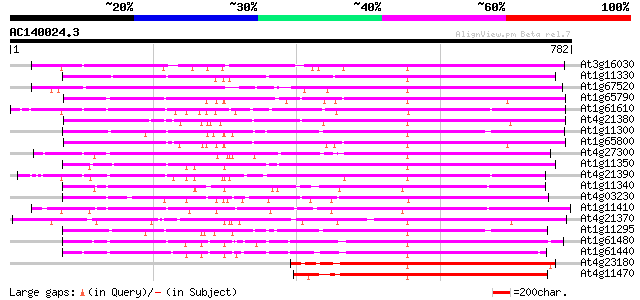
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140024.3 + phase: 0 /pseudo
(782 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g16030 putative receptor kinase 439 e-123
At1g11330 receptor-like protein kinase, putative 430 e-120
At1g67520 putative receptor protein kinase 417 e-116
At1g65790 receptor kinase, putative 416 e-116
At1g61610 413 e-115
At4g21380 receptor-like serine/threonine protein kinase ARK3 408 e-114
At1g11300 putative s-locus protein kinase (PPC:1.7.2) 401 e-112
At1g65800 receptor kinase, putative 398 e-111
At4g27300 putative receptor protein kinase 394 e-109
At1g11350 putative receptor-like protein kinase gi|4008010; simi... 393 e-109
At4g21390 serine/threonine kinase - like protein 392 e-109
At1g11340 receptor kinase, putative 389 e-108
At4g03230 putative receptor kinase 385 e-107
At1g11410 putative brassinosteroid insensitive protein; similar ... 384 e-106
At4g21370 receptor kinase - like protein 380 e-105
At1g11295 putative s-locus protein kinase (PPC:1.7.2) 377 e-104
At1g61480 S-like receptor protein kinase; member of gene cluster... 360 2e-99
At1g61440 S-like receptor protein kinase; member of gene cluster... 330 1e-90
At4g23180 receptor-like protein kinase 4 (RLK4) 330 2e-90
At4g11470 serine/threonine kinase-like protein 323 2e-88
>At3g16030 putative receptor kinase
Length = 850
Score = 439 bits (1130), Expect = e-123
Identities = 308/847 (36%), Positives = 430/847 (50%), Gaps = 131/847 (15%)
Query: 31 SDSLKPGDTLNSKSKLCSEQGKFCLYFDSEEAHLVVSSGV-------DGAVVWMYDRNQP 83
+D+L G L +L S F L F + E G+ GAV W+ +RN P
Sbjct: 24 TDTLLQGQYLKDGQELVSAFNIFKLKFFNFENSSNWYLGIWYNNFYLSGAV-WIANRNNP 82
Query: 84 IAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTNDTVATMLDTGNFVLQQLHPNGT- 142
+ S L++D G L+I ++ S + T +T +LD+GN LQ++ +G+
Sbjct: 83 VLGRSGSLTVDSLGRLRI--LRGASSLLELSSTETTGNTTLKLLDSGNLQLQEMDSDGSM 140
Query: 143 KSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEWEPK-EGELN 201
K LWQSFD P DTLLP MKLG N KTG W L S L +LP G + L
Sbjct: 141 KRTLWQSFDYPTDTLLPGMKLGFNVKTGKRWELTSWLGDTLPASGSFVFGMDDNITNRLT 200
Query: 202 IRKSGKVHWKSG----------KLKSNGMFENIPAKVQRIYQYIIVSNKDEDSFAFEVKD 251
I G V+W SG KL +NG + VS + E F + +
Sbjct: 201 ILWLGNVYWASGLWFKGGFSLEKLNTNGFI------------FSFVSTESEHYFMYSGDE 248
Query: 252 GK----FIRWFISPKGRL--ISDAGSTSN---ADMCYGYKSDEGCQVANADMC------- 295
F R I +G L I+ G + + +G + + GC N C
Sbjct: 249 NYGGPLFPRIRIDQQGSLQKINLDGVKKHVHCSPSVFGEELEYGCYQQNFRNCVPARYKE 308
Query: 296 -----------YGYNSDGGCQKWEEIPNCREPGEVFRKMVGRPNKDNATTDEPANGYD-- 342
+GY +K ++ C G FR+ V ++ +E
Sbjct: 309 VTGSWDCSPFGFGYTYT---RKTYDLSYCSRFGYTFRETVSPSAENGFVFNEIGRRLSSY 365
Query: 343 DCKMRCWRNCNCYGFEELYSNFTGCIYYSWNSTQDVDLDKK-NNFYALVKPTKSPPNSHG 401
DC ++C +NC+C + + TGC ++ + T + Y +K +K
Sbjct: 366 DCYVKCLQNCSCVAYASTNGDGTGCEIWNTDPTNENSASHHPRTIYIRIKGSKLAAT--- 422
Query: 402 KRRIWIGAAIATALLILCP---LILFLAKKK------------------QKYALQGKK-S 439
W+ + +L ++ P LI++L +K Q +L K+ S
Sbjct: 423 ----WL--VVVASLFLIIPVTWLIIYLVLRKFKIKGTNFVSESLKMISSQSCSLTNKRLS 476
Query: 440 KRKEGKMKDLAESYDIKDLENDFKG--------HDIKVFNFTSILEATMDFSSENKLGQG 491
+ G D +E +G +++++F+F S+ AT FS NKLG+G
Sbjct: 477 TLRVGSTIDQEMLLLELGIERRRRGKRSARNNNNELQIFSFESVAFATDYFSDANKLGEG 536
Query: 492 GYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEER 551
G+GPVYKG L G+EVA+KRLS SGQG+VEF+NE LI +LQHTNLV+LLGCC+ ++E+
Sbjct: 537 GFGPVYKGRLIDGEEVAIKRLSLASGQGLVEFKNEAMLIAKLQHTNLVKLLGCCVEKDEK 596
Query: 552 IL----------------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKAS 589
+L LDWK R I+EGI QGLLYLHKYSRLK+IHRD+KA
Sbjct: 597 MLIYEYMPNKSLDYFLFDPLRKIVLDWKLRFRIMEGIIQGLLYLHKYSRLKVIHRDIKAG 656
Query: 590 NILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFG 649
NILLDE++NPKISDFGMAR+F QES NT R+ GT+GYMSPEY EG+ S KSDV+SFG
Sbjct: 657 NILLDEDMNPKISDFGMARIFGAQESKANTKRVAGTFGYMSPEYFREGLFSAKSDVFSFG 716
Query: 650 VLLLEIICGRKNNSF-HDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPD-EVQRCI 707
VL+LEIICGRKNNSF HD + PLNLI H W L+ + +++DPSL D+ V + +V RC+
Sbjct: 717 VLMLEIICGRKNNSFHHDSEGPLNLIVHVWNLFKENRVREVIDPSLGDSAVENPQVLRCV 776
Query: 708 HVGLLCVQQYANDRPTMSDVISML-TNKYKLTTLPRRPAFYIRREIYDGETTSKGPDTDT 766
V LLCVQQ A+DRP+M DV+SM+ + +LP+ PAFY E + P+ +
Sbjct: 777 QVALLCVQQNADDRPSMLDVVSMIYGDGNNALSLPKEPAFYDGPPRSSPEMEVEPPEMEN 836
Query: 767 YSTTAIS 773
S ++
Sbjct: 837 VSANRVT 843
>At1g11330 receptor-like protein kinase, putative
Length = 840
Score = 430 bits (1106), Expect = e-120
Identities = 274/748 (36%), Positives = 386/748 (50%), Gaps = 69/748 (9%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKI-EFQNRNVPIIIYYSPQPTNDTVATMLDTGNF 132
VVW+ +++ PI S V+S+ G L + + +NR V P N T ++D+GN
Sbjct: 83 VVWVANKDSPINDTSGVISIYQDGNLAVTDGRNRLVWSTNVSVPVAPNATWVQLMDSGNL 142
Query: 133 VLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLE 192
+LQ NG ILW+SF P D+ +P M LG + +TG N L S +H P+ G +
Sbjct: 143 MLQDNRNNG--EILWESFKHPYDSFMPRMTLGTDGRTGGNLKLTSWTSHDDPSTGNYTAG 200
Query: 193 WEPKE-GELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYIIVSNKD-EDSFAFEVK 250
P EL I K+ W+SG +F +P ++ N D + + +
Sbjct: 201 IAPFTFPELLIWKNNVPTWRSGPWNGQ-VFIGLPNMDSLLFLDGFNLNSDNQGTISMSYA 259
Query: 251 DGKFIRWF-ISPKGRLISDAGSTSNADMCYGYKSDE-------------GCQVANADMCY 296
+ F+ F + P+G + STS G K C C
Sbjct: 260 NDSFMYHFNLDPEGIIYQKDWSTSMRTWRIGVKFPYTDCDAYGRCGRFGSCHAGENPPCK 319
Query: 297 --------------GYNSDGGC--------QKWEEIPNCREPGEVFRKMVGRPNKDNATT 334
G N GC ++ + N G+ + + K +
Sbjct: 320 CVKGFVPKNNTEWNGGNWSNGCMRKAPLQCERQRNVSNGGGGGKADGFLKLQKMKVPISA 379
Query: 335 DEPANGYDDCKMRCWRNCNCYGFEELYSNFTGCIYYSWNSTQDVDLDKKNNFYALVKPTK 394
+ C C NC+C + Y GC+ +S + D+ + ++
Sbjct: 380 ERSEASEQVCPKVCLDNCSCTAYA--YDRGIGCMLWSGDLV-DMQSFLGSGIDLFIRVAH 436
Query: 395 SPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAESYD 454
S +H + I A + +LI +L +K +K + + ++ +M+ L D
Sbjct: 437 SELKTHSNLAVMIAAPVIGVMLIAAVCVLLACRKYKKRPAKDRSAELMFKRMEALTS--D 494
Query: 455 IKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSK 514
+ N K ++ +F F + +T FS NKLGQGG+GPVYKG L GQE+AVKRLS+
Sbjct: 495 NESASNQIKLKELPLFEFQVLATSTDSFSLRNKLGQGGFGPVYKGKLPEGQEIAVKRLSR 554
Query: 515 TSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL--------------------- 553
SGQG+ E NE+ +I +LQH NLV+LLGCCI EER+L
Sbjct: 555 KSGQGLEELMNEVVVISKLQHRNLVKLLGCCIEGEERMLVYEYMPKKSLDAYLFDPMKQK 614
Query: 554 -LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ 612
LDWK R NI+EGI +GLLYLH+ SRLKIIHRDLKASNILLDENLNPKISDFG+AR+F
Sbjct: 615 ILDWKTRFNIMEGICRGLLYLHRDSRLKIIHRDLKASNILLDENLNPKISDFGLARIFRA 674
Query: 613 QESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLN 672
E NT R+VGTYGYMSPEYAMEG S KSDV+S GV+ LEII GR+N+S H + LN
Sbjct: 675 NEDEANTRRVVGTYGYMSPEYAMEGFFSEKSDVFSLGVIFLEIISGRRNSSSHKEENNLN 734
Query: 673 LIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLT 732
L+ +AW+LWNDGE L DP++ D E+++C+H+GLLCVQ+ ANDRP +S+VI MLT
Sbjct: 735 LLAYAWKLWNDGEAASLADPAVFDKCFEKEIEKCVHIGLLCVQEVANDRPNVSNVIWMLT 794
Query: 733 NKYKLTTLPRRPAFYIRREIYDGETTSK 760
+ P++PAF +RR + E++ +
Sbjct: 795 TENMSLADPKQPAFIVRRGASEAESSDQ 822
>At1g67520 putative receptor protein kinase
Length = 833
Score = 417 bits (1072), Expect = e-116
Identities = 291/844 (34%), Positives = 416/844 (48%), Gaps = 145/844 (17%)
Query: 31 SDSLKPGDTLNSKSKLCSEQGKFCLYF----DSEEAHLVV-------SSGVDGAVVWMYD 79
+D+L G L +L S F L F +SE +L + ++ VW+ +
Sbjct: 24 TDTLHQGQFLKDGQELVSAFKIFKLKFFNFKNSENLYLGIWFNNLYLNTDSQDRPVWIAN 83
Query: 80 RNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSP-QPTNDTVATMLDTGNFVLQQLH 138
RN PI+ S L++D G LKI R ++ S + T +T +LD+GN LQ++
Sbjct: 84 RNNPISDRSGSLTVDSLGRLKIL---RGASTMLELSSIETTRNTTLQLLDSGNLQLQEMD 140
Query: 139 PNGT-KSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEWEPK- 196
+G+ K +LWQSFD P DTLLP MKLG + KT W L S L +LP G +
Sbjct: 141 ADGSMKRVLWQSFDYPTDTLLPGMKLGFDGKTRKRWELTSWLGDTLPASGSFVFGMDTNI 200
Query: 197 EGELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYIIVSNKDEDSFAFEVKDGK--F 254
L I G ++W SG E + ++ ++ + ++ + D + F
Sbjct: 201 TNVLTILWRGNMYWSSGLWNKGRFSEEELNECGFLFSFVSTKSGQYFMYSGDQDDARTFF 260
Query: 255 IRWFISPKGRLISDAGSTSNADMCYGYKSDEGCQVANADMCYGYNSDGGCQKWEEIPNCR 314
I +G L + Y ++ D YG+ S
Sbjct: 261 PTIMIDEQGILRREQMHRQRNRQNYRNRNCLAAGYVVRDEPYGFTS-------------- 306
Query: 315 EPGEVFRKMVGRPNKDNATTDEPANGYDDCKMRCWRNCNCYGFEELYSNFTGCIYYSWNS 374
FR V + + D C C +N +C + + TGC WN+
Sbjct: 307 -----FRVTVSSSASNGFVLSGTFSSVD-CSAICLQNSSCLAYASTEPDGTGCEI--WNT 358
Query: 375 TQDVDLDKKNNFYALVKPTKSPPNSHGKRRIWIGA------------AIATALLILCPLI 422
PT SH R I+I + L ++ P+I
Sbjct: 359 Y----------------PTNKGSASHSPRTIYIRGNGQENKKVAAWHIVVATLFLMTPII 402
Query: 423 LFLAKKK-QKYALQGKKSKR---------------------------------------- 441
F+ +K+ ++G+ R
Sbjct: 403 WFIIYLVLRKFNVKGRNCIRITHKTVLVSMVFLLTSSPSFFLFMIQDVFYFVEYTTFYGE 462
Query: 442 -------KEGKMKDLAESYDIKDLENDFKGH-DIKVFNFTSILEATMDFSSENKLGQGGY 493
+E +++L N+ K + ++++F+F S++ AT DFS ENKLG+GG+
Sbjct: 463 SSLLKVHQEMLLRELGIDRSCIHKRNERKSNNELQIFSFESVVSATDDFSDENKLGEGGF 522
Query: 494 GPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL 553
GPVYKG L G+EVA+KRLS SGQG+VEF+NE LI +LQHTNLVQ+LGCCI ++E++L
Sbjct: 523 GPVYKGKLLNGEEVAIKRLSLASGQGLVEFKNEAILIAKLQHTNLVQVLGCCIEKDEKML 582
Query: 554 ----------------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNI 591
LDW R I+EGI QGLLYLHKYSRLK+IHRD+KASNI
Sbjct: 583 IYEYMQNKSLDYFLFDPLRKNVLDWTLRFRIMEGIIQGLLYLHKYSRLKVIHRDIKASNI 642
Query: 592 LLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVL 651
LLDE++NPKISDFG+AR+F +E+ NT R+ GT+GYMSPEY EG+ S KSDV+SFGVL
Sbjct: 643 LLDEDMNPKISDFGLARIFGAEETRANTKRVAGTFGYMSPEYFREGLFSAKSDVFSFGVL 702
Query: 652 LLEIICGRKNNSF-HDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVP-DEVQRCIHV 709
+LEIICGRKNNSF HD++ PLNLI H W L+ + + +++D SL D+ + +V RC+ V
Sbjct: 703 MLEIICGRKNNSFHHDLEGPLNLIVHVWNLFKENKIREVIDLSLRDSALDYPQVLRCVQV 762
Query: 710 GLLCVQQYANDRPTMSDVISMLTNK-YKLTTLPRRPAFY--IRREIYDGETTSKGPDTDT 766
LLCVQ+ A DRP+M DV+SM+ + +LP+ PAFY RR + + + P+ +
Sbjct: 763 ALLCVQENAEDRPSMLDVVSMIYGEGNNALSLPKEPAFYDGPRRSFPEMKVEPQEPENVS 822
Query: 767 YSTT 770
S T
Sbjct: 823 ASIT 826
>At1g65790 receptor kinase, putative
Length = 843
Score = 416 bits (1068), Expect = e-116
Identities = 276/775 (35%), Positives = 423/775 (53%), Gaps = 89/775 (11%)
Query: 75 VWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTNDTVATMLDTGNFVL 134
VW+ +R+ P++ + L + + ++ + +R V + A +LD GNF+L
Sbjct: 79 VWVANRDNPLSSSNGTLKISGNNLVIFDQSDRPVWSTNITGGDVRSPVAAELLDNGNFLL 138
Query: 135 QQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEWE 194
+ + +LWQSFD P DTLL MKLG ++KTG N L S P+ GE S + E
Sbjct: 139 R----DSNNRLLWQSFDFPTDTLLAEMKLGWDQKTGFNRILRSWKTTDDPSSGEFSTKLE 194
Query: 195 PKE-GELNIRKSGKVHWKSGKLKSNGM-FENIPAKVQRIYQ-YIIVSNKDEDSFAFEV-K 250
E E I + ++SG NGM F ++P +Q Y Y ++K+E ++++ + K
Sbjct: 195 TSEFPEFYICSKESILYRSGPW--NGMRFSSVPGTIQVDYMVYNFTASKEEVTYSYRINK 252
Query: 251 DGKFIRWFISPKGRL--ISDAGST--------SNADMCYGYKSDEG---CQVANADMCY- 296
+ R +++ G L ++ +T S D+C YK C + CY
Sbjct: 253 TNLYSRLYLNSAGLLQRLTWFETTQSWKQLWYSPKDLCDNYKVCGNFGYCDSNSLPNCYC 312
Query: 297 --GY-----------NSDGGCQKWEEIPNCREPGEVFRKMVGRPNKDNATTDEPANGYDD 343
G+ + GC + + G K + P+ AT + G
Sbjct: 313 IKGFKPVNEQAWDLRDGSAGCMRKTRLSCDGRDGFTRLKRMKLPDT-TATIVDREIGLKV 371
Query: 344 CKMRCWRNCNCYGFE--ELYSNFTGCIYYSWNSTQDVDLDKKN------NFYALVKPTKS 395
CK RC +CNC F ++ + +GC+ + T+++ LD +N + Y + +
Sbjct: 372 CKERCLEDCNCTAFANADIRNGGSGCVIW----TREI-LDMRNYAKGGQDLYVRLAAAEL 426
Query: 396 PPNSHGKRRIWIGAAIATALLILCPLILF-LAKKKQKYALQGK-----KSKRKEGKMKDL 449
+I IG++I ++L+L ++F K+KQK ++ + + + ++ + D+
Sbjct: 427 EDKRIKNEKI-IGSSIGVSILLLLSFVIFHFWKRKQKRSITIQTPNVDQVRSQDSLINDV 485
Query: 450 AES---YDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQE 506
S Y K+ ++++ ++ + ++ AT +FS++NKLGQGG+G VYKG L G+E
Sbjct: 486 VVSRRGYTSKEKKSEYL--ELPLLELEALATATNNFSNDNKLGQGGFGIVYKGRLLDGKE 543
Query: 507 VAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------------- 553
+AVKRLSK S QG EF NE+ LI +LQH NLV+LLGCC+ + E++L
Sbjct: 544 IAVKRLSKMSSQGTDEFMNEVRLIAKLQHINLVRLLGCCVDKGEKMLIYEYLENLSLDSH 603
Query: 554 ---------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDF 604
L+W+KR +II GI++GLLYLH+ SR +IIHRDLKASN+LLD+N+ PKISDF
Sbjct: 604 LFDQTRSSNLNWQKRFDIINGIARGLLYLHQDSRCRIIHRDLKASNVLLDKNMTPKISDF 663
Query: 605 GMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSF 664
GMAR+F ++E+ NT R+VGTYGYMSPEYAM+GI S KSDV+SFGVLLLEII G++N F
Sbjct: 664 GMARIFGREETEANTRRVVGTYGYMSPEYAMDGIFSMKSDVFSFGVLLLEIISGKRNKGF 723
Query: 665 HDVDRPLNLIGHAWELWNDGEYLQLLDP----SLCDTFVPDEVQRCIHVGLLCVQQYAND 720
++ +R LNL+G W W +G L+++DP SL F E+ RCI +GLLCVQ+ A D
Sbjct: 724 YNSNRDLNLLGFVWRHWKEGNELEIVDPINIDSLSSKFPTHEILRCIQIGLLCVQERAED 783
Query: 721 RPTMSDVISMLTNKYKLTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAISTS 775
RP MS V+ ML ++ P+RP F I R + +++S D + I+ S
Sbjct: 784 RPVMSSVMVMLGSETTAIPQPKRPGFCIGRSPLEADSSSSTQRDDECTVNQITLS 838
>At1g61610
Length = 842
Score = 413 bits (1062), Expect = e-115
Identities = 301/856 (35%), Positives = 428/856 (49%), Gaps = 100/856 (11%)
Query: 1 MISFKRNKQIVWIYLWLWWNTTANICVKATSDSLKPGDTLNSKSKLCSEQGKFCLYFDSE 60
M F RN +V L ++ +N+ +TS+S T+ L SE F L F +
Sbjct: 1 MAGFNRNLTLVTTLL-IFHQLCSNVSC-STSNSFTRNHTIREGDSLISEDESFELGFFTP 58
Query: 61 EAHLVVSSGV------DGAVVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYY 114
+ + G+ VVW+ +R +P+ L + G L I +N I
Sbjct: 59 KNSTLRYVGIWYKNIEPQTVVWVANREKPLLDHKGALKIADDGNLVI-VNGQNETIWSTN 117
Query: 115 SPQPTNDTVATMLDTGNFVLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWS 174
+N+TVA + TG+ VL + + W+SF++P DT LP M++ VN G N +
Sbjct: 118 VEPESNNTVAVLFKTGDLVL--CSDSDRRKWYWESFNNPTDTFLPGMRVRVNPSLGENRA 175
Query: 175 LVSRLAHSLPTPGELSLEWEPKEG-ELNIRKSGKVHWKSGKLKSNGMFENIPAKVQ---R 230
+ + S P+PG+ S+ +P E+ I + K W+SG S +F IP ++
Sbjct: 176 FIPWKSESDPSPGKYSMGIDPVGALEIVIWEGEKRKWRSGPWNS-AIFTGIPDMLRFTNY 234
Query: 231 IYQYIIVSNKDEDS---FAFEVKDGK-FIRWFISPKG--------------RLISDAGST 272
IY + + S D D F + D F+R++I P G L+ ST
Sbjct: 235 IYGFKLSSPPDRDGSVYFTYVASDSSDFLRFWIRPDGVEEQFRWNKDIRNWNLLQWKPST 294
Query: 273 S--------NADMCYGYKSDE--------GCQVANADMCYGYNSDGGCQKWEEIP-NC-- 313
N +C K + G + + D + GGCQ+ +P NC
Sbjct: 295 ECEKYNRCGNYSVCDDSKEFDSGKCSCIDGFEPVHQDQWNNRDFSGGCQR--RVPLNCNQ 352
Query: 314 -----REPGEVFRKMVGRPNKDNATTDEPANGYDDCKMRCWRNCNCYGFEELYSNFTGCI 368
+E G K + P+ + N + CK C R+C+C + + GC+
Sbjct: 353 SLVAGQEDGFTVLKGIKVPDFGSVVLH---NNSETCKDVCARDCSCKAYALVVG--IGCM 407
Query: 369 YYSWNSTQDVDLDKKNNFYALVKPTKSPPNSHGKRRIWI--GAAIATALLILCPLILFLA 426
++ + ++ N + +WI + I LL LC IL+
Sbjct: 408 IWTRDLIDMEHFERGGNSINIRLAGSKLGGGKENSTLWIIVFSVIGAFLLGLCIWILWKF 467
Query: 427 KKKQKYALQGKKSKRKEGKMKDLAESYD-----IKDLEND-FKGHDIKVFNFTSILEATM 480
KK K L K+K+ + D+ E+ D IK L D D+ +F+F S+ AT
Sbjct: 468 KKSLKAFLW----KKKDITVSDIIENRDYSSSPIKVLVGDQVDTPDLPIFSFDSVASATG 523
Query: 481 DFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQ 540
DF+ ENKLGQGG+G VYKG + G+E+AVKRLS S QG+ EF+NE+ LI +LQH NLV+
Sbjct: 524 DFAEENKLGQGGFGTVYKGNFSEGREIAVKRLSGKSKQGLEEFKNEILLIAKLQHRNLVR 583
Query: 541 LLGCCIHEEERILL----------------------DWKKRLNIIEGISQGLLYLHKYSR 578
LLGCCI + E++LL DW+KR +I GI++GLLYLH+ SR
Sbjct: 584 LLGCCIEDNEKMLLYEYMPNKSLDRFLFDESKQGSLDWRKRWEVIGGIARGLLYLHRDSR 643
Query: 579 LKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGI 638
LKIIHRDLKASNILLD +NPKISDFGMAR+F ++ NT R+VGTYGYM+PEYAMEGI
Sbjct: 644 LKIIHRDLKASNILLDTEMNPKISDFGMARIFNYRQDHANTIRVVGTYGYMAPEYAMEGI 703
Query: 639 CSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTF 698
S KSDVYSFGVL+LEI+ GRKN SF D +LIG+AW LW+ G+ +++DP + DT
Sbjct: 704 FSEKSDVYSFGVLILEIVSGRKNVSFRGTDHG-SLIGYAWHLWSQGKTKEMIDPIVKDTR 762
Query: 699 VPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAFYIRREIYDGETT 758
E RCIHVG+LC Q RP M V+ ML ++ PR+P F+ D E
Sbjct: 763 DVTEAMRCIHVGMLCTQDSVIHRPNMGSVLLMLESQTSQLPPPRQPTFHSFLNSGDIELN 822
Query: 759 SKGPDTDTYSTTAIST 774
G D + + +T
Sbjct: 823 FDGHDVASVNDVTFTT 838
>At4g21380 receptor-like serine/threonine protein kinase ARK3
Length = 850
Score = 408 bits (1048), Expect = e-114
Identities = 278/772 (36%), Positives = 418/772 (54%), Gaps = 78/772 (10%)
Query: 75 VWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTNDTVATMLDTGNFVL 134
VW+ +R+ P++ L + S ++ ++ + V + VA +LD GNFVL
Sbjct: 81 VWVANRDTPLSSSIGTLKISDSNLVVLDQSDTPVWSTNLTGGDVRSPLVAELLDNGNFVL 140
Query: 135 QQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEWE 194
+ + +LWQSFD P DTLLP MKLG + KTG N + S + P+ G+ S + E
Sbjct: 141 RDSKNSAPDGVLWQSFDFPTDTLLPEMKLGWDAKTGFNRFIRSWKSPDDPSSGDFSFKLE 200
Query: 195 PKEG--ELNIRKSGKVHWKSGKLKSNGM-FENIPAKVQRIYQYII---VSNKDEDSFAFE 248
EG E+ + ++SG NG+ F +P + ++Y++ ++K+E +++F
Sbjct: 201 T-EGFPEIFLWNRESRMYRSGPW--NGIRFSGVPE--MQPFEYMVFNFTTSKEEVTYSFR 255
Query: 249 V-KDGKFIRWFISPKGRL-----ISDAGSTSN-----ADMC--------YGYKSDEGCQV 289
+ K + R IS G L I A + + D C YGY V
Sbjct: 256 ITKSDVYSRLSISSSGLLQRFTWIETAQNWNQFWYAPKDQCDEYKECGVYGYCDSNTSPV 315
Query: 290 ANA--------DMCYGY-NSDGGCQKWEEIPNCREPGEVFRKMVGRPNKDNATTDEPANG 340
N +G + GC + + G V K + P+ A+ D G
Sbjct: 316 CNCIKGFKPRNPQVWGLRDGSDGCVRKTLLSCGGGDGFVRLKKMKLPDTTTASVDRGI-G 374
Query: 341 YDDCKMRCWRNCNCYGFE--ELYSNFTGCIYYSWNSTQDVDLDKKN-NFYALVKPTKSPP 397
+C+ +C R+CNC F ++ + +GC+ ++ + K + Y + T
Sbjct: 375 VKECEQKCLRDCNCTAFANTDIRGSGSGCVTWTGELFDIRNYAKGGQDLYVRLAATDLED 434
Query: 398 NSHGKRRIWIGAAIATALLILCPLILF-LAKKKQKYALQ------GKKSKRKEGKMKDLA 450
+ +I IG++I ++L+L I+F L K+KQK ++ + + ++ M ++
Sbjct: 435 KRNRSAKI-IGSSIGVSVLLLLSFIIFFLWKRKQKRSILIETPIVDHQLRSRDLLMNEVV 493
Query: 451 -ESYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAV 509
S EN+ ++ + F + AT +FS+ NKLGQGG+G VYKG L GQE+AV
Sbjct: 494 ISSRRHISRENNTDDLELPLMEFEEVAMATNNFSNANKLGQGGFGIVYKGKLLDGQEMAV 553
Query: 510 KRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---------------- 553
KRLSKTS QG EF+NE+ LI LQH NLV+LL CC+ E++L
Sbjct: 554 KRLSKTSVQGTDEFKNEVKLIARLQHINLVRLLACCVDAGEKMLIYEYLENLSLDSHLFD 613
Query: 554 ------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMA 607
L+W+ R +II GI++GLLYLH+ SR +IIHRDLKASNILLD+ + PKISDFGMA
Sbjct: 614 KSRNSKLNWQMRFDIINGIARGLLYLHQDSRFRIIHRDLKASNILLDKYMTPKISDFGMA 673
Query: 608 RMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDV 667
R+F + E+ NT ++VGTYGYMSPEYAM+GI S KSDV+SFGVLLLEII ++N F++
Sbjct: 674 RIFGRDETEANTRKVVGTYGYMSPEYAMDGIFSMKSDVFSFGVLLLEIISSKRNKGFYNS 733
Query: 668 DRPLNLIGHAWELWNDGEYLQLLDPSLCD---TFVPDEVQRCIHVGLLCVQQYANDRPTM 724
DR LNL+G W W +G+ L+++DP + D TF E+ RCI +GLLCVQ+ A DRPTM
Sbjct: 734 DRDLNLLGCVWRNWKEGKGLEIIDPIITDSSSTFRQHEILRCIQIGLLCVQERAEDRPTM 793
Query: 725 SDVISMLTNKYKLTTLPRRPAFYIRREIYDGE-TTSKGPDTDTYSTTAISTS 775
S VI ML ++ P+ P + + R + D + ++SK D ++++ I+ S
Sbjct: 794 SLVILMLGSESTTIPQPKAPGYCLERSLLDTDSSSSKQRDDESWTVNQITVS 845
>At1g11300 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 401 bits (1030), Expect = e-112
Identities = 268/761 (35%), Positives = 392/761 (51%), Gaps = 82/761 (10%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPT-NDTVATMLDTGNF 132
V+W+ ++++PI S V+S+ G L + R V S Q + N TVA +LD+GN
Sbjct: 76 VIWVANKDKPINDSSGVISVSQDGNLVVTDGQRRVLWSTNVSTQASANSTVAELLDSGNL 135
Query: 133 VLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGH-NWSLVSRLAHSLPTPGE--- 188
VL++ + + LW+SF P D+ LP M +G N + G N ++ S + S P+PG
Sbjct: 136 VLKEA---SSDAYLWESFKYPTDSWLPNMLVGTNARIGGGNVTITSWKSPSDPSPGSYTA 192
Query: 189 -LSLEWEPKEGELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYIIVSNKDED-SFA 246
L L P+ +N + W+SG MF +P ++ Y + N D + S
Sbjct: 193 ALVLAAYPELFIMNNNNNNSTVWRSGPWNGQ-MFNGLPDVYAGVFLYRFIVNDDTNGSVT 251
Query: 247 FEVKDGKFIRWF-ISPKGRLISDAGSTSN----------ADMCYGYK---SDEGCQVANA 292
+ +R+F + +G +I S + A C Y+ C
Sbjct: 252 MSYANDSTLRYFYMDYRGSVIRRDWSETRRNWTVGLQVPATECDNYRRCGEFATCNPRKN 311
Query: 293 DMCY---GY-----------NSDGGCQKWE----EIPNCREPGEVFRKMVGRPNKDNATT 334
+C G+ N GGC + E N + F ++ D A
Sbjct: 312 PLCSCIRGFRPRNLIEWNNGNWSGGCTRRVPLQCERQNNNGSADGFLRLRRMKLPDFARR 371
Query: 335 DEPANGYDDCKMRCWRNCNCYGFEELYSNFTGCIYYSWNSTQDVDLDKKNNFYALVKPTK 394
E + +C C + C+C + GC+ ++ + +L + ++
Sbjct: 372 SEASE--PECLRTCLQTCSCIA--AAHGLGYGCMIWNGSLVDSQELSA-SGLDLYIRLAH 426
Query: 395 SPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAESYD 454
S + KR I IG +A + ++ +L + K K++K+K + + E +
Sbjct: 427 SEIKTKDKRPILIGTILAGGIFVVAACVLLARRIVMK-----KRAKKKGRDAEQIFERVE 481
Query: 455 IKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSK 514
N K ++ +F F + AT +FS NKLGQGG+GPVYKG L GQE+AVKRLS+
Sbjct: 482 ALAGGNKGKLKELPLFEFQVLAAATNNFSLRNKLGQGGFGPVYKGKLQEGQEIAVKRLSR 541
Query: 515 TSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL--------------------- 553
SGQG+ E NE+ +I +LQH NLV+LLGCCI EER+L
Sbjct: 542 ASGQGLEELVNEVVVISKLQHRNLVKLLGCCIAGEERMLVYEFMPKKSLDYYLFDSRRAK 601
Query: 554 -LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ 612
LDWK R NII GI +GLLYLH+ SRL+IIHRDLKASNILLDENL PKISDFG+AR+F
Sbjct: 602 LLDWKTRFNIINGICRGLLYLHRDSRLRIIHRDLKASNILLDENLIPKISDFGLARIFPG 661
Query: 613 QESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLN 672
E NT R+VGTYGYM+PEYAM G+ S KSDV+S GV+LLEII GR+N++
Sbjct: 662 NEDEANTRRVVGTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIISGRRNSN-------ST 714
Query: 673 LIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLT 732
L+ + W +WN+GE L+DP + D E+ +CIH+GLLCVQ+ ANDRP++S V SML+
Sbjct: 715 LLAYVWSIWNEGEINSLVDPEIFDLLFEKEIHKCIHIGLLCVQEAANDRPSVSTVCSMLS 774
Query: 733 NKYKLTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAIS 773
++ P++PAF R + + E++ D+ + I+
Sbjct: 775 SEIADIPEPKQPAFISRNNVPEAESSENSDLKDSINNVTIT 815
>At1g65800 receptor kinase, putative
Length = 847
Score = 398 bits (1022), Expect = e-111
Identities = 265/770 (34%), Positives = 407/770 (52%), Gaps = 75/770 (9%)
Query: 75 VWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTNDTVATMLDTGNFVL 134
VW+ +R+ P++ + L + + ++ + +R V + A +LD GNFVL
Sbjct: 79 VWVANRDNPLSSSNGTLKISDNNLVIFDQSDRPVWSTNITGGDVRSPVAAELLDYGNFVL 138
Query: 135 QQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTG-HNWSLVSRLAHSLPTPGELSLEW 193
+ N LWQSFD P DTLL MK+G + K+G N L S P+ G+ S +
Sbjct: 139 RDSKNNKPSGFLWQSFDFPTDTLLSDMKMGWDNKSGGFNRILRSWKTTDDPSSGDFSTKL 198
Query: 194 EPKE-GELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYI---IVSNKDEDSFAFEV 249
E I + ++SG N F ++P + YI N + +++ V
Sbjct: 199 RTSGFPEFYIYNKESITYRSGPWLGN-RFSSVPG--MKPVDYIDNSFTENNQQVVYSYRV 255
Query: 250 -KDGKFIRWFISPKGRLIS----DAGST------SNADMCYGYKSDEG---CQVANADMC 295
K + +S G L +A + S D+C YK C + +C
Sbjct: 256 NKTNIYSILSLSSTGLLQRLTWMEAAQSWKQLWYSPKDLCDNYKECGNYGYCDANTSPIC 315
Query: 296 Y---GY----------NSDGGCQKWEEIPNCREPGEVFRKMVGRPNKDNATTDEPANGYD 342
G+ + GC + ++ G V K + P+ + D+ G
Sbjct: 316 NCIKGFEPMNEQAALRDDSVGCVRKTKLSCDGRDGFVRLKKMRLPDTTETSVDKGI-GLK 374
Query: 343 DCKMRCWRNCNCYGFE--ELYSNFTGCIYYSWNSTQDVDLDKKNNFYALVKPTKSPPNSH 400
+C+ RC + CNC F ++ + +GC+ +S + K + +
Sbjct: 375 ECEERCLKGCNCTAFANTDIRNGGSGCVIWSGGLFDIRNYAKGGQDLYVRVAAGDLEDKR 434
Query: 401 GKRRIWIGAAIATALLILCPLILF-LAKKKQKYALQGKKS-----KRKEGKMKDLAE--- 451
K + IG++I ++L+L I+F K+KQK ++ + + ++ M +L +
Sbjct: 435 IKSKKIIGSSIGVSILLLLSFIIFHFWKRKQKRSITIQTPIVDLVRSQDSLMNELVKASR 494
Query: 452 SYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKR 511
SY K+ + D+ ++ + + ++ AT +FS++NKLGQGG+G VYKG+L G+E+AVKR
Sbjct: 495 SYTSKENKTDYL--ELPLMEWKALAMATNNFSTDNKLGQGGFGIVYKGMLLDGKEIAVKR 552
Query: 512 LSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------------------ 553
LSK S QG EF NE+ LI +LQH NLV+LLGCC+ + E++L
Sbjct: 553 LSKMSSQGTDEFMNEVRLIAKLQHINLVRLLGCCVDKGEKMLIYEYLENLSLDSHLFDQT 612
Query: 554 ----LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARM 609
L+W+KR +II GI++GLLYLH+ SR +IIHRDLKASN+LLD+N+ PKISDFGMAR+
Sbjct: 613 RSSNLNWQKRFDIINGIARGLLYLHQDSRCRIIHRDLKASNVLLDKNMTPKISDFGMARI 672
Query: 610 FTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDR 669
F ++E+ NT R+VGTYGYMSPEYAM+GI S KSDV+SFGVLLLEII G++N F++ +R
Sbjct: 673 FGREETEANTRRVVGTYGYMSPEYAMDGIFSMKSDVFSFGVLLLEIISGKRNKGFYNSNR 732
Query: 670 PLNLIGHAWELWNDGEYLQLLDP----SLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMS 725
LNL+G W W +G+ L+++DP +L F E+ RCI +GLLCVQ+ A DRP MS
Sbjct: 733 DLNLLGFVWRHWKEGKELEIVDPINIDALSSEFPTHEILRCIQIGLLCVQERAEDRPVMS 792
Query: 726 DVISMLTNKYKLTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAISTS 775
V+ ML ++ P+RP F + R + +++S D + ++ S
Sbjct: 793 SVMVMLGSETTAIPQPKRPGFCVGRSSLEVDSSSSTQRDDECTVNQVTLS 842
>At4g27300 putative receptor protein kinase
Length = 815
Score = 394 bits (1012), Expect = e-109
Identities = 279/786 (35%), Positives = 408/786 (51%), Gaps = 90/786 (11%)
Query: 34 LKPGDTLNSKSKLCSEQGKFCLYFDSEEAHLVVSSGV--DGAVVWMYDRNQPIAIDSAVL 91
LK GDTL+S ++ + G F L + + H + AVVW+ +RN P+ S L
Sbjct: 34 LKDGDTLSSPDQVF-QLGFFSLDQEEQPQHRFLGLWYMEPFAVVWVANRNNPLYGTSGFL 92
Query: 92 SLDYSGVLKIEFQNRNVPIIIYYSP-----QPTNDTVATMLDTGNFVLQQLHPNGTKSIL 146
+L G L++ F + + S + N+ + + +GN + +G +++L
Sbjct: 93 NLSSLGDLQL-FDGEHKALWSSSSSSTKASKTANNPLLKISCSGNLISS----DGEEAVL 147
Query: 147 WQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEWEPKE-GELNIRKS 205
WQSFD P++T+L MKLG N KT WSL S P+PG+ +L + + +L +RK+
Sbjct: 148 WQSFDYPMNTILAGMKLGKNFKTQMEWSLSSWKTLKDPSPGDFTLSLDTRGLPQLILRKN 207
Query: 206 GKVHWKSGKLKSNGM-FENIPA--KVQRIYQYIIVSNKDEDSFAFEVKDGKFIRWFISPK 262
G + NG+ F PA + ++ Y S+ E ++++ + R ++
Sbjct: 208 GDSSYSYRLGSWNGLSFTGAPAMGRENSLFDYKFTSSAQEVNYSWTPRHRIVSRLVLNNT 267
Query: 263 GRLISDAGSTSNADMCYGYKSDEGCQ---VANADMCYGYNSDG----GC---------QK 306
G+L S N + ++ C + A G NS C +K
Sbjct: 268 GKLHRFIQSKQNQWILANTAPEDECDYYSICGAYAVCGINSKNTPSCSCLQGFKPKSGRK 327
Query: 307 WE----------EIPNCREPGEVFRKMVGRPNKDNATTDEPANG---YDDCKMRCWRNCN 353
W EIP E + F K G D + + A +DCK++C NC+
Sbjct: 328 WNISRGAYGCVHEIPTNCEKKDAFVKFPGLKLPDTSWSWYDAKNEMTLEDCKIKCSSNCS 387
Query: 354 C--YGFEELYSNFTGCIYYSWNSTQDVDLDKKNNFYALVKPTKSPPNSHGKRRIWIGAAI 411
C Y ++ GC+ + + VD+ + ++F V K R +G +
Sbjct: 388 CTAYANTDIREGGKGCLLWFGDL---VDMREYSSFGQDVYIRMGFAKIEFKGREVVGMVV 444
Query: 412 ATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAESYDIKDLENDFKGHDIKVFN 471
+ + I L++ A ++K K R E K + E +DL D+ +F+
Sbjct: 445 GSVVAIAVVLVVVFACFRKKIM----KRYRGENFRKGIEE----EDL-------DLPIFD 489
Query: 472 FTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALIC 531
+I AT DFS N LG+GG+GPVYKG L GQE+AVKRLS SGQG+ EF+NE+ LI
Sbjct: 490 RKTISIATDDFSYVNFLGRGGFGPVYKGKLEDGQEIAVKRLSANSGQGVEEFKNEVKLIA 549
Query: 532 ELQHTNLVQLLGCCIHEEERIL----------------------LDWKKRLNIIEGISQG 569
+LQH NLV+LLGCCI EE +L LDWKKR+NII G+++G
Sbjct: 550 KLQHRNLVRLLGCCIQGEECMLIYEYMPNKSLDFFIFDERRSTELDWKKRMNIINGVARG 609
Query: 570 LLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYM 629
+LYLH+ SRL+IIHRDLKA N+LLD ++NPKISDFG+A+ F +S +TNR+VGTYGYM
Sbjct: 610 ILYLHQDSRLRIIHRDLKAGNVLLDNDMNPKISDFGLAKSFGGDQSESSTNRVVGTYGYM 669
Query: 630 SPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQL 689
PEYA++G S KSDV+SFGVL+LEII G+ N F D LNL+GH W++W + +++
Sbjct: 670 PPEYAIDGHFSVKSDVFSFGVLVLEIITGKTNRGFRHADHDLNLLGHVWKMWVEDREIEV 729
Query: 690 LDPS-LCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAFYI 748
+ L +T V EV RCIHV LLCVQQ DRPTM+ V+ M + L P +P F+
Sbjct: 730 PEEEWLEETSVIPEVLRCIHVALLCVQQKPEDRPTMASVVLMFGSDSSLPH-PTQPGFFT 788
Query: 749 RREIYD 754
R + D
Sbjct: 789 NRNVPD 794
>At1g11350 putative receptor-like protein kinase gi|4008010; similar
to EST gb|AI999419.1
Length = 830
Score = 393 bits (1009), Expect = e-109
Identities = 266/750 (35%), Positives = 383/750 (50%), Gaps = 71/750 (9%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVP----IIIYYSPQPTNDTVATMLDT 129
VVW+ + N PI S ++S+ G L + V +++ P N A +L+T
Sbjct: 71 VVWVANSNSPINDSSGMVSISKEGNLVVMDGRGQVHWSTNVLV---PVAANTFYARLLNT 127
Query: 130 GNFVLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGEL 189
GN VL G + ILW+SF+ P + LPTM L + KTG + L S + P+PG
Sbjct: 128 GNLVLLGTTNTGDE-ILWESFEHPQNIYLPTMSLATDTKTGRSLKLRSWKSPFDPSPGRY 186
Query: 190 SLEWEPKE-GELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYIIVSNKDEDS---- 244
S P EL + K + W+SG F +P RI + + + D
Sbjct: 187 SAGLIPLPFPELVVWKDDLLMWRSGPWNGQ-YFIGLPNMDYRINLFELTLSSDNRGSVSM 245
Query: 245 ----------FAFEVKDGKFIR-----------WFISPKGRLISDAGSTSNADMCYGYKS 283
F + + F R W P + + A A + S
Sbjct: 246 SYAGNTLLYHFLLDSEGSVFQRDWNVAIQEWKTWLKVPSTKCDTYATCGQFASCRFNPGS 305
Query: 284 DEGCQVANADMCYGY------NSDGGCQKWEEIP-NCREPGEVFRKMVG--RPNKDNATT 334
C Y N GC + + R+ + RK G R K
Sbjct: 306 TPPCMCIRGFKPQSYAEWNNGNWTQGCVRKAPLQCESRDNNDGSRKSDGFVRVQKMKVPH 365
Query: 335 DEPANGYD--DCKMRCWRNCNCYGFEELYSNFTGCIYYSWNSTQDVDLDKKNNFYALVKP 392
+ +G + DC C +NC+C + + GC+ +S N D+ ++
Sbjct: 366 NPQRSGANEQDCPESCLKNCSCTAYS--FDRGIGCLLWSGN-LMDMQEFSGTGVVFYIRL 422
Query: 393 TKSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAES 452
S R I I + + ++ K K+ + + ++ +M+ L+ +
Sbjct: 423 ADSEFKKRTNRSIVITVTLLVGAFLFAGTVVLALWKIAKHREKNRNTRLLNERMEALSSN 482
Query: 453 YDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRL 512
L N +K ++ +F F + AT +FS NKLGQGG+G VYKG L G ++AVKRL
Sbjct: 483 DVGAILVNQYKLKELPLFEFQVLAVATNNFSITNKLGQGGFGAVYKGRLQEGLDIAVKRL 542
Query: 513 SKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------------------- 553
S+TSGQG+ EF NE+ +I +LQH NLV+LLG CI EER+L
Sbjct: 543 SRTSGQGVEEFVNEVVVISKLQHRNLVRLLGFCIEGEERMLVYEFMPENCLDAYLFDPVK 602
Query: 554 ---LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMF 610
LDWK R NII+GI +GL+YLH+ SRLKIIHRDLKASNILLDENLNPKISDFG+AR+F
Sbjct: 603 QRLLDWKTRFNIIDGICRGLMYLHRDSRLKIIHRDLKASNILLDENLNPKISDFGLARIF 662
Query: 611 TQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRP 670
E V+T R+VGTYGYM+PEYAM G+ S KSDV+S GV+LLEI+ GR+N+SF++ +
Sbjct: 663 QGNEDEVSTVRVVGTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIVSGRRNSSFYNDGQN 722
Query: 671 LNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISM 730
NL +AW+LWN GE + L+DP + + +E++RC+HVGLLCVQ +ANDRP+++ VI M
Sbjct: 723 PNLSAYAWKLWNTGEDIALVDPVIFEECFENEIRRCVHVGLLCVQDHANDRPSVATVIWM 782
Query: 731 LTNKYKLTTLPRRPAFYIRREIYDGETTSK 760
L+++ P++PAF RR + E++ +
Sbjct: 783 LSSENSNLPEPKQPAFIPRRGTSEVESSGQ 812
>At4g21390 serine/threonine kinase - like protein
Length = 849
Score = 392 bits (1008), Expect = e-109
Identities = 287/818 (35%), Positives = 415/818 (50%), Gaps = 98/818 (11%)
Query: 11 VWIYLWLWWNTTANICVKATSDSLKPGDTLNSKSKLCSEQGKFCLYFDSEEAHLVVSSGV 70
+++Y +L+ ++ A ++ +SL+ G +N K L S Q F L F S + G+
Sbjct: 13 LFLYFFLYESSMAANTIRR-GESLRDG--INHKP-LVSPQKTFELGFFSPGSSTHRFLGI 68
Query: 71 ------DGAVVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPII---IYYSPQPTND 121
D AVVW+ +R PI+ S VL + G L + +N+ + I S N+
Sbjct: 69 WYGNIEDKAVVWVANRATPISDQSGVLMISNDGNLVL-LDGKNITVWSSNIESSTTNNNN 127
Query: 122 TVATMLDTGNFVLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAH 181
V ++ DTGNFVL + T +W+SF+ P DT LP M++ VN +TG N + VS +
Sbjct: 128 RVVSIHDTGNFVLSE---TDTDRPIWESFNHPTDTFLPQMRVRVNPQTGDNHAFVSWRSE 184
Query: 182 SLPTPGELSLEWEPKEG-ELNIRKSGKVH-WKSGKLKSNGMFENIPAK---VQRIYQYII 236
+ P+PG SL +P E+ + + K W+SG+ S +F IP +Y + +
Sbjct: 185 TDPSPGNYSLGVDPSGAPEIVLWEGNKTRKWRSGQWNS-AIFTGIPNMSLLTNYLYGFKL 243
Query: 237 VSNKDEDS---FAFEVKDGKFI--------------RWFISPKGRLISDAGSTSNADMCY 279
S DE F + D + RW + K + S D
Sbjct: 244 SSPPDETGSVYFTYVPSDPSVLLRFKVLYNGTEEELRWNETLKKWTKFQSEPDSECDQYN 303
Query: 280 GYKSDEGCQVANAD-MC---YGY------NSDGGCQ-----KWEEIPNCREPGEVFRKMV 324
C + ++ +C +GY N GC+ K E + E + K V
Sbjct: 304 RCGKFGICDMKGSNGICSCIHGYEQVSVGNWSRGCRRRTPLKCERNISVGEDEFLTLKSV 363
Query: 325 GRPNKDNATTDEPANGYDDCKMRCWRNCNCYGFEELYSNFTGCIYYSWNSTQ-DVDLDKK 383
P D + +DC+ RC RNC+C YS G WN D+ +
Sbjct: 364 KLP--DFEIPEHNLVDPEDCRERCLRNCSC----NAYSLVGGIGCMIWNQDLVDLQQFEA 417
Query: 384 NNFYALVKPTKSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKE 443
++ S + K +I + A+ ++++ L L + K+K + G +
Sbjct: 418 GGSSLHIRLADSEVGENRKTKIAVIVAVLVGVILIGIFALLLWRFKRKKDVSGAYCGKNT 477
Query: 444 GKMKDLAESYDIKDLENDFKGH-------------DIKVFNFTSILEATMDFSSENKLGQ 490
+A+ K+ + F G ++ VF+ +I AT DF EN+LG+
Sbjct: 478 DTSVVVADLTKSKETTSAFSGSVDIMIEGKAVNTSELPVFSLNAIAIATNDFCKENELGR 537
Query: 491 GGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEE 550
GG+GPVYKG+L G+E+AVKRLS SGQG+ EF+NE+ LI +LQH NLV+LLGCC EE
Sbjct: 538 GGFGPVYKGVLEDGREIAVKRLSGKSGQGVDEFKNEIILIAKLQHRNLVRLLGCCFEGEE 597
Query: 551 RIL----------------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKA 588
++L +DWK R +IIEGI++GLLYLH+ SRL+IIHRDLK
Sbjct: 598 KMLVYEYMPNKSLDFFLFDETKQALIDWKLRFSIIEGIARGLLYLHRDSRLRIIHRDLKV 657
Query: 589 SNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSF 648
SN+LLD +NPKISDFGMAR+F ++ NT R+VGTYGYMSPEYAMEG+ S KSDVYSF
Sbjct: 658 SNVLLDAEMNPKISDFGMARIFGGNQNEANTVRVVGTYGYMSPEYAMEGLFSVKSDVYSF 717
Query: 649 GVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIH 708
GVLLLEI+ G++N S + +LIG+AW L+ G +L+DP + T E RCIH
Sbjct: 718 GVLLLEIVSGKRNTSLRSSEHG-SLIGYAWYLYTHGRSEELVDPKIRVTCSKREALRCIH 776
Query: 709 VGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAF 746
V +LCVQ A +RP M+ V+ ML + PR+P F
Sbjct: 777 VAMLCVQDSAAERPNMASVLLMLESDTATLAAPRQPTF 814
>At1g11340 receptor kinase, putative
Length = 901
Score = 389 bits (998), Expect = e-108
Identities = 266/759 (35%), Positives = 387/759 (50%), Gaps = 112/759 (14%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSP-----QPTNDTVATMLD 128
+VW+ +R+ PI S ++ G L + + +I + +PT VAT+ D
Sbjct: 136 IVWVANRDHPINDTSGMVKFSNRGNLSVYASDNETELIWSTNVSDSMLEPT--LVATLSD 193
Query: 129 TGNFVLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGE 188
GN VL P +S W+SFD P DT LP M+LG RK G + SL S +H P G+
Sbjct: 194 LGNLVL--FDPVTGRSF-WESFDHPTDTFLPFMRLGFTRKDGLDRSLTSWKSHGDPGSGD 250
Query: 189 LSLEWEPKE-GELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYIIVSNKDEDSFAF 247
L L E + +L + K W+ G + + I+ V+N+DE SF +
Sbjct: 251 LILRMERRGFPQLILYKGVTPWWRMGSWTGHRWSGVPEMPIGYIFNNSFVNNEDEVSFTY 310
Query: 248 EVKDGKFI---------------------RW---FISPKGRLISDAGSTSNADMCYGYKS 283
V D I RW + PK + + A N GY
Sbjct: 311 GVTDASVITRTMVNETGTMHRFTWIARDKRWNDFWSVPKEQCDNYAHCGPN-----GYCD 365
Query: 284 DEGCQVANADMCYGY-----------NSDGGCQKWEEIPNCREP-GEVFRKMVGRPNKDN 331
+ G+ +S GGC K + C E G V K + P+ +
Sbjct: 366 SPSSKTFECTCLPGFEPKFPRHWFLRDSSGGCTKKKRASICSEKDGFVKLKRMKIPDTSD 425
Query: 332 ATTDEPANGYDDCKMRCWRNCNCYGFEELYSNFT----GCIYYSW---------NSTQD- 377
A+ D +CK RC +NC+C + Y GC+ + NS QD
Sbjct: 426 ASVDMNIT-LKECKQRCLKNCSCVAYASAYHESKRGAIGCLKWHGGMLDARTYLNSGQDF 484
Query: 378 -VDLDKKNNFYA--LVKPTKSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYAL 434
+ +DK++ L++ +SP +S+ + + +LC K A
Sbjct: 485 YIRVDKEDAVEQKWLIREEESPSDSY---QFDCSRNVTNCHFVLCC----------KGAT 531
Query: 435 QGKKSKRKEGKMKDLAESYDIKDL----ENDFKGHDIKVFNFTSILEATMDFSSENKLGQ 490
+ ++ + +D + ++ + ++ +F+ +I+ AT +FSS+NKLG
Sbjct: 532 NAESNRHRSSSANFAPVPFDFDESFRFEQDKARNRELPLFDLNTIVAATNNFSSQNKLGA 591
Query: 491 GGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCI---- 546
GG+GPVYKG+L E+AVKRLS+ SGQG+ EF+NE+ LI +LQH NLV++LGCC+
Sbjct: 592 GGFGPVYKGVLQNRMEIAVKRLSRNSGQGMEEFKNEVKLISKLQHRNLVRILGCCVELEE 651
Query: 547 ------------------HEEERILLDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKA 588
HEE+R LDW KR+ I+ GI++G+LYLH+ SRL+IIHRDLKA
Sbjct: 652 KMLVYEYLPNKSLDYFIFHEEQRAELDWPKRMEIVRGIARGILYLHQDSRLRIIHRDLKA 711
Query: 589 SNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSF 648
SNILLD + PKISDFGMAR+F + T+R+VGT+GYM+PEYAMEG S KSDVYSF
Sbjct: 712 SNILLDSEMIPKISDFGMARIFGGNQMEGCTSRVVGTFGYMAPEYAMEGQFSIKSDVYSF 771
Query: 649 GVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSL-CDTFVPDEVQRCI 707
GVL+LEII G+KN++FH + NL+GH W+LW +GE +++D + +T+ EV +CI
Sbjct: 772 GVLMLEIITGKKNSAFH--EESSNLVGHIWDLWENGEATEIIDNLMDQETYDEREVMKCI 829
Query: 708 HVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAF 746
+GLLCVQ+ A+DR MS V+ ML + P+ PAF
Sbjct: 830 QIGLLCVQENASDRVDMSSVVIMLGHNATNLPNPKHPAF 868
>At4g03230 putative receptor kinase
Length = 852
Score = 385 bits (990), Expect = e-107
Identities = 267/764 (34%), Positives = 395/764 (50%), Gaps = 107/764 (14%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTN-DTVATMLDTGNF 132
VVW+ +R P+ S + ++ G L++ V P + + + ++D GN
Sbjct: 80 VVWVANRESPVLDRSCIFTISKDGNLEVIDSKGRVYWDTGVKPSSVSAERMVKLMDNGNL 139
Query: 133 VLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLE 192
VL + +++WQSF +P DT LP M++ N +L S + + P+ G + +
Sbjct: 140 VL--ISDGNEANVVWQSFQNPTDTFLPGMRMD------ENMTLSSWRSFNDPSHGNFTFQ 191
Query: 193 WEPKEG-ELNIRKSGKVHWKSGK----LKSNGMFENIPAKVQRIYQYIIVSNKDEDSF-- 245
+ +E + I K +WKSG + S+ M I + + + V N
Sbjct: 192 MDQEEDKQFIIWKRSMRYWKSGISGKFIGSDEMPYAISYFLSNFTETVTVHNASVPPLFT 251
Query: 246 ------AFEVKDGKFIRWFISPKGRLISDAGSTSNADMCYGYKSDEG---CQVANADMCY 296
F + ++F R + + D C Y + C N +MC
Sbjct: 252 SLYTNTRFTMSSSGQAQYFRLDGERFWAQIWAEPR-DECSVYNACGNFGSCNSKNEEMCK 310
Query: 297 ---GYNSD-----------GGCQKWEEIPNCREPGEVFRKM--------VGRPNKDNATT 334
G+ + GGC + I C + G V M VG P+ +
Sbjct: 311 CLPGFRPNFLEKWVKGDFSGGCSRESRI--CGKDGVVVGDMFLNLSVVEVGSPD-----S 363
Query: 335 DEPANGYDDCKMRCWRNCNC--YGFEE---LYSNFTGCIYYS-WNSTQDVDLDKKNNFYA 388
A+ +C+ C NC C Y +EE L SN I+ N+ ++ L +N F
Sbjct: 364 QFDAHNEKECRAECLNNCQCQAYSYEEVDILQSNTKCWIWLEDLNNLKEGYLGSRNVFIR 423
Query: 389 LVKPTKSPPNSHGKRRIWIGAAIATALLIL------CPLILFLAKKKQKYALQGKKSKRK 442
+ P G+ R G A +LI+ +++ L+ LQ +K ++
Sbjct: 424 VAVPDIGSHVERGRGRY--GEAKTPVVLIIVVTFTSAAILVVLSSTASYVFLQRRKVNKE 481
Query: 443 EGKM-------------KDLAESYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLG 489
G + K+L ES K ++D +G D+ F +IL AT +FS+ NKLG
Sbjct: 482 LGSIPRGVHLCDSERHIKELIESGRFK--QDDSQGIDVPSFELETILYATSNFSNANKLG 539
Query: 490 QGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEE 549
QGG+GPVYKG+ QE+AVKRLS+ SGQG+ EF+NE+ LI +LQH NLV+LLG C+ E
Sbjct: 540 QGGFGPVYKGMFPGDQEIAVKRLSRCSGQGLEEFKNEVVLIAKLQHRNLVRLLGYCVAGE 599
Query: 550 ERILL----------------------DWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLK 587
E++LL DWK R NII GI++GLLYLH+ SRL+IIHRDLK
Sbjct: 600 EKLLLYEYMPHKSLDFFIFDRKLCQRLDWKMRCNIILGIARGLLYLHQDSRLRIIHRDLK 659
Query: 588 ASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYS 647
SNILLDE +NPKISDFG+AR+F E+ NTNR+VGTYGYMSPEYA+EG+ S KSDV+S
Sbjct: 660 TSNILLDEEMNPKISDFGLARIFGGSETSANTNRVVGTYGYMSPEYALEGLFSFKSDVFS 719
Query: 648 FGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCI 707
FGV+++E I G++N FH+ ++ L+L+GHAW+LW ++LLD +L ++ + +C+
Sbjct: 720 FGVVVIETISGKRNTGFHEPEKSLSLLGHAWDLWKAERGIELLDQALQESCETEGFLKCL 779
Query: 708 HVGLLCVQQYANDRPTMSDVISMLTNKYKLT-TLPRRPAFYIRR 750
+VGLLCVQ+ NDRPTMS+V+ ML + T P++PAF +RR
Sbjct: 780 NVGLLCVQEDPNDRPTMSNVVFMLGSSEAATLPTPKQPAFVLRR 823
>At1g11410 putative brassinosteroid insensitive protein; similar to
EST gb|W43102
Length = 840
Score = 384 bits (985), Expect = e-106
Identities = 278/836 (33%), Positives = 423/836 (50%), Gaps = 108/836 (12%)
Query: 31 SDSLKPGDTLNSKSKLCSEQGKFCLYFDSEEAHLVVSSGV------DGAVVWMYDRNQPI 84
S SLK GD + S+ K +F F S + G+ + +VW+ +R+ PI
Sbjct: 28 SQSLKDGDVIYSEGK------RFAFGFFSLGNSKLRYVGIWYAQVSEQTIVWVANRDHPI 81
Query: 85 AIDSAVLSLDYSGVLKIEFQ-NRNVPI----IIYYSPQPTNDTVATMLDTGNFVLQQLHP 139
S ++ G L + N PI +I +P VA + D GN VL L P
Sbjct: 82 NDTSGLIKFSTRGNLCVYASGNGTEPIWSTDVIDMIQEPA--LVAKLSDLGNLVL--LDP 137
Query: 140 NGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEWEPKE-G 198
KS W+SF+ P +TLLP MK G R++G + + S + P G ++ E +
Sbjct: 138 VTGKSF-WESFNHPTNTLLPFMKFGFTRQSGVDRIMTSWRSPGDPGSGNITYRIERRGFP 196
Query: 199 ELNIRKSGKVHWKSGKLKSNGMFENIPAKVQR-IYQYIIVSNKDEDSFAFEV-------- 249
++ + K + W++G + +P + I+ V+N DE S + V
Sbjct: 197 QMMMYKGLTLWWRTGSWTGQ-RWSGVPEMTNKFIFNISFVNNPDEVSITYGVLDASVTTR 255
Query: 250 ----------------KDGKFIRWFISPKGR--LISDAGSTSNADMCYGYKSDEGC---- 287
+D K+I ++ +P+ + + + G D K + C
Sbjct: 256 MVLNETGTLQRFRWNGRDKKWIGFWSAPEDKCDIYNHCGFNGYCDSTSTEKFECSCLPGY 315
Query: 288 QVANADMCYGYNSDGGCQ--KWEEIPNCREPGEVFRKMVGRPNKDNATTDEPANGYDDCK 345
+ + ++ GC K + I N +E G K V PN D +C+
Sbjct: 316 EPKTPRDWFLRDASDGCTRIKADSICNGKE-GFAKLKRVKIPNTSAVNVDMNIT-LKECE 373
Query: 346 MRCWRNCNCYGFEELY----SNFTGCIYYSWNSTQD-VDLDKKNNFYALVKPTKSPPNSH 400
RC +NC+C + Y GC+ + N L +FY V + +
Sbjct: 374 QRCLKNCSCVAYASAYHESQDGAKGCLTWHGNMLDTRTYLSSGQDFYLRVDKSDAV---E 430
Query: 401 GKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMK----------DLA 450
K RI + C + +++L K++K K DL
Sbjct: 431 WKWRIGEEETCFNSHQFDCSRDV----TTDQFSLLFKEAKTTNTLRKAPSSFAPSSFDLE 486
Query: 451 ESYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVK 510
+S+ +++LE+ + ++ +F ++I AT +F+ +NKLG GG+GPVYKG+L G E+AVK
Sbjct: 487 DSFILEELEDKSRSRELPLFELSTIATATNNFAFQNKLGAGGFGPVYKGVLQNGMEIAVK 546
Query: 511 RLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCI----------------------HE 548
RLSK+SGQG+ EF+NE+ LI +LQH NLV++LGCC+ HE
Sbjct: 547 RLSKSSGQGMEEFKNEVKLISKLQHRNLVRILGCCVEFEEKMLVYEYLPNKSLDYFIFHE 606
Query: 549 EERILLDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMAR 608
E+R LDW KR+ II GI +G+LYLH+ SRL+IIHRDLKASN+LLD + PKI+DFG+AR
Sbjct: 607 EQRAELDWPKRMGIIRGIGRGILYLHQDSRLRIIHRDLKASNVLLDNEMIPKIADFGLAR 666
Query: 609 MFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVD 668
+F + +TNR+VGTYGYMSPEYAM+G S KSDVYSFGVL+LEII G++N++F+ +
Sbjct: 667 IFGGNQIEGSTNRVVGTYGYMSPEYAMDGQFSIKSDVYSFGVLILEIITGKRNSAFY--E 724
Query: 669 RPLNLIGHAWELWNDGEYLQLLDPSLC-DTFVPDEVQRCIHVGLLCVQQYANDRPTMSDV 727
LNL+ H W+ W +GE ++++D + +T+ EV +C+H+GLLCVQ+ ++DRP MS V
Sbjct: 725 ESLNLVKHIWDRWENGEAIEIIDKLMGEETYDEGEVMKCLHIGLLCVQENSSDRPDMSSV 784
Query: 728 ISMLTNKYKLTTLPRRPAFYI--RREIYDGETTSKGPDTDTYSTTAISTSCEVEGK 781
+ ML + P+ PAF RR G ++ P +T ST T +V+G+
Sbjct: 785 VFMLGHNAIDLPSPKHPAFTAGRRRNTKTGGSSDNWPSGETSSTINDVTLTDVQGR 840
>At4g21370 receptor kinase - like protein
Length = 844
Score = 380 bits (975), Expect = e-105
Identities = 276/858 (32%), Positives = 428/858 (49%), Gaps = 112/858 (13%)
Query: 5 KRNKQIVWIYLWLWWNTTANICVKATSDSLKPGDTLNSKSKLCSEQGKFCLYF-----DS 59
K + +++L+ + ++ + + S T++S + S G F L F DS
Sbjct: 8 KHHSYTFFVFLFFFLILFPDLSISVNTLSATESLTISSNKTIVSPGGVFELGFFRILGDS 67
Query: 60 EEAHLVVSSGVDGAVVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPT 119
+ VW+ +R+ P++ +L + + ++ ++ + +V +S T
Sbjct: 68 WYLGIWYKKISQRTYVWVANRDTPLSNPIGILKISNANLVILDNSDTHV-----WSTNLT 122
Query: 120 ----NDTVATMLDTGNFVLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSL 175
+ VA +LD GNFVL+ N + LWQSFD P DTLLP MKLG + K G N +
Sbjct: 123 GAVRSSVVAELLDNGNFVLRGSKINESDEFLWQSFDFPTDTLLPQMKLGRDHKRGLNRFV 182
Query: 176 VSRLAHSLPTPGELSLEWEPK--------EGELNIRKSGKVHWKSGKLKSNGMFENIPAK 227
S + P+ G + E L + +SG W L+ +G+ E +
Sbjct: 183 TSWKSSFDPSSGSFMFKLETLGLPEFFGFTSFLEVYRSGP--WDG--LRFSGILE-MQQW 237
Query: 228 VQRIYQYIIVSNKDEDSFAFEVKD-GKFIRWFISPKGRLISDAGSTSNADM-CYGYKSDE 285
IY + N++E ++ F V D + R I+ GRL + + + + +
Sbjct: 238 DDIIYNF--TENREEVAYTFRVTDHNSYSRLTINTVGRLEGFTWEPTQQEWNMFWFMPKD 295
Query: 286 GCQVANADMCYGY---NSDGGC-----------QKW---EEIPNCREPGEV-------FR 321
C + Y Y ++ C Q W + CR ++ FR
Sbjct: 296 TCDLYGICGPYAYCDMSTSPTCNCIKGFQPLSPQDWASGDVTGRCRRKTQLTCGEDRFFR 355
Query: 322 KMVGRPNKDNATTDEPANGYDDCKMRCWRNCNC--YGFEELYSNFTGCIYY--SWNSTQD 377
M + A + G +C+ +C +CNC Y ++ + +GCI + + ++
Sbjct: 356 LMNMKIPATTAAIVDKRIGLKECEEKCKTHCNCTAYANSDIRNGGSGCIIWIGEFRDIRN 415
Query: 378 VDLDKKNNFYALVKPTKSPPNSHGKRRIW----IGAAIATALLILCPLILFLA-KKKQKY 432
D ++ F L G+RR IG I +L+++ I++ KKKQK
Sbjct: 416 YAADGQDLFVRLAAA------EFGERRTIRGKIIGLIIGISLMLVLSFIIYCFWKKKQKR 469
Query: 433 ALQGKKSKRKEGKMKDLAESYDI-----KDLENDFKGHDIKVFNFTSILEATMDFSSENK 487
A ++++L + + + L + + ++ + F +++ AT +FS N
Sbjct: 470 ARATAAPIGYRDRIQELIITNGVVMSSGRRLLGEEEDLELPLTEFETVVMATENFSDSNI 529
Query: 488 LGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIH 547
LG+GG+G VYK +AVKRLS+ S QG EF+NE+ LI LQH NLV+LL CCI+
Sbjct: 530 LGRGGFGIVYK--------IAVKRLSEMSSQGTNEFKNEVRLIARLQHINLVRLLSCCIY 581
Query: 548 EEERIL-----------------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHR 584
+E+IL L+W+ R +II GI++GLLYLH+ SR KIIHR
Sbjct: 582 ADEKILIYEYLENGSLDSHLFETTQSSNKLNWQTRFSIINGIARGLLYLHQDSRFKIIHR 641
Query: 585 DLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSD 644
DLKASN+LLD+N+ PKISDFGMAR+F + E+ NT ++VGTYGYMSPEYAMEGI S KSD
Sbjct: 642 DLKASNVLLDKNMTPKISDFGMARIFERDETEANTRKVVGTYGYMSPEYAMEGIFSVKSD 701
Query: 645 VYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDT------F 698
V+SFGVL+LEI+ G++N FH+ + NL+G+ WE W +G+ L+++D + D+ F
Sbjct: 702 VFSFGVLVLEIVSGKRNRGFHNSGQDNNLLGYTWENWKEGKGLEIVDSIIVDSSSSMSLF 761
Query: 699 VPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAFYIRREIYDGETT 758
P EV RCI +GLLCVQ+ A DRP MS V+ ML ++ PRRP + +RR D + +
Sbjct: 762 QPHEVLRCIQIGLLCVQERAEDRPKMSSVVLMLGSEKGEYFSPRRPGYCVRRSSLDTDDS 821
Query: 759 SKGPDTDTYSTTAISTSC 776
S + D + S C
Sbjct: 822 SSSTERDNHRRHLWSVLC 839
>At1g11295 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 377 bits (969), Expect = e-104
Identities = 264/739 (35%), Positives = 377/739 (50%), Gaps = 86/739 (11%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPT-NDTVATMLDTGNF 132
V+W+ +++ PI S V+S+ G L + R V S + + N TVA +L++GN
Sbjct: 76 VIWVANKDTPINDSSGVISISEDGNLVVTDGQRRVLWSTNVSTRASANSTVAELLESGNL 135
Query: 133 VLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGH-NWSLVSRLAHSLPTPGE--- 188
VL+ + T + LW+SF P D+ LP M +G N +TG N ++ S S P+PG
Sbjct: 136 VLKDAN---TDAYLWESFKYPTDSWLPNMLVGTNARTGGGNITITSWTNPSDPSPGSYTA 192
Query: 189 -LSLEWEPKEGELNIRKSGKVHWKSGKLKSNG-MFENIPAKVQRIYQYIIVSNKDEDSFA 246
L L P+ N + W+SG NG MF +P ++ Y N D + A
Sbjct: 193 ALVLAPYPELFIFNNNDNNATVWRSGPW--NGLMFNGLPDVYPGLFLYRFKVNDDTNGSA 250
Query: 247 FE--VKDGKFIRWFISPKGRLI----SDA------GSTSNADMCYGYKSD---------- 284
D ++ +G I S+A GS A C Y
Sbjct: 251 TMSYANDSTLRHLYLDYRGFAIRRDWSEARRNWTLGSQVPATECDIYSRCGQYTTCNPRK 310
Query: 285 -------EGCQVANADMCYGYNSDGGCQKWE----EIPNCREPGEVFRKMVGRPNKDNAT 333
+G + N N GGC + E N + + F K+ D A
Sbjct: 311 NPHCSCIKGFRPRNLIEWNNGNWSGGCIRKLPLQCERQNNKGSADRFLKLQRMKMPDFAR 370
Query: 334 TDEPANGYDDCKMRCWRNCNCYGFEELYSNFTGCIYYSWN-STQDVDLDKKNNFYALVKP 392
E + +C M C ++C+C F + GC+ WN S D + + ++
Sbjct: 371 RSEASE--PECFMTCLQSCSCIAFA--HGLGYGCMI--WNRSLVDSQVLSASGMDLSIRL 424
Query: 393 TKSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAES 452
S + +R I IG ++A + ++ +L + K K++K+K + + +
Sbjct: 425 AHSEFKTQDRRPILIGTSLAGGIFVVATCVLLARRIVMK-----KRAKKKGTDAEQIFKR 479
Query: 453 YDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRL 512
+ + K ++ +F F + AT +FS NKLGQGG+GPVYKG+L GQE+AVKRL
Sbjct: 480 VEALAGGSREKLKELPLFEFQVLATATDNFSLSNKLGQGGFGPVYKGMLLEGQEIAVKRL 539
Query: 513 SKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------------------- 553
S+ SGQG+ E E+ +I +LQH NLV+L GCCI EER+L
Sbjct: 540 SQASGQGLEELVTEVVVISKLQHRNLVKLFGCCIAGEERMLVYEFMPKKSLDFYIFDPRE 599
Query: 554 ---LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMF 610
LDW R II GI +GLLYLH+ SRL+IIHRDLKASNILLDENL PKISDFG+AR+F
Sbjct: 600 AKLLDWNTRFEIINGICRGLLYLHRDSRLRIIHRDLKASNILLDENLIPKISDFGLARIF 659
Query: 611 TQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRP 670
E NT R+VGTYGYM+PEYAM G+ S KSDV+S GV+LLEII GR+N+
Sbjct: 660 PGNEDEANTRRVVGTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIISGRRNSH------- 712
Query: 671 LNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISM 730
L+ H W +WN+GE ++DP + D E+++C+H+ LLCVQ ANDRP++S V M
Sbjct: 713 STLLAHVWSIWNEGEINGMVDPEIFDQLFEKEIRKCVHIALLCVQDAANDRPSVSTVCMM 772
Query: 731 LTNKYKLTTLPRRPAFYIR 749
L+++ P++PAF R
Sbjct: 773 LSSEVADIPEPKQPAFMPR 791
>At1g61480 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 809
Score = 360 bits (924), Expect = 2e-99
Identities = 259/762 (33%), Positives = 385/762 (49%), Gaps = 91/762 (11%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTNDTVATMLDTGNFV 133
VVW+ +R +P+ +A L++ +G L + +N +V I + +N + A + D GN V
Sbjct: 72 VVWVANREKPVTDSAANLTISSNGSLLLFNENHSVVWSIGET-FASNGSRAELTDNGNLV 130
Query: 134 LQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEW 193
+ N + LW+SF+ DT+LP L N TG L S +H+ P+PG+ +++
Sbjct: 131 VID---NNSGRTLWESFEHFGDTMLPFSNLMYNLATGEKRVLTSWKSHTDPSPGDFTVQI 187
Query: 194 EPK-EGELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYIIVSNKDED---SFAFEV 249
P+ + + K +W+SG F IP + Y +D + SF +
Sbjct: 188 TPQVPSQACTMRGSKTYWRSGPWAKT-RFTGIPV-MDDTYTSPFSLQQDTNGSGSFTYFE 245
Query: 250 KDGKFIRWFISPKGRL-----------ISDAGSTSNADMCYGYKSDEG-CQVANADMCYG 297
++ K I+ +G L ++ ++ D+ YG+ G C ++ C
Sbjct: 246 RNFKLSYIMITSEGSLKIFQHNGMDWELNFEAPENSCDI-YGFCGPFGICVMSVPPKCKC 304
Query: 298 Y--------------NSDGGCQKWEEIPNCREPGEVFRKMVGRPNKDNATTDEPANGYD- 342
+ N GC + E+ +C+ G K V + +P + Y+
Sbjct: 305 FKGFVPKSIEEWKRGNWTDGCVRHTEL-HCQ--GNTNGKTVN--GFYHVANIKPPDFYEF 359
Query: 343 -------DCKMRCWRNCNCYGFEELYSNFTGCIYYSWNSTQDVDLDKKNNFYALVKPTKS 395
C C NC+C F Y N GC+ ++ + V + ++ S
Sbjct: 360 ASFVDAEGCYQICLHNCSCLAFA--YINGIGCLMWNQDLMDAVQFSAGGEILS-IRLASS 416
Query: 396 PPNSHGKRRIWIGAAIATALLILCPLILF-LAKKKQKYALQGKKSK--RKEGKMKDLAES 452
+ + +I + + ++ +L ++ F + K K+ + K SK KE DL E
Sbjct: 417 ELGGNKRNKIIVASIVSLSLFVILAFAAFCFLRYKVKHTVSAKISKIASKEAWNNDL-EP 475
Query: 453 YDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRL 512
D+ L K F +I AT +FS NKLGQGG+G VYKG L G+E+AVKRL
Sbjct: 476 QDVSGL---------KFFEMNTIQTATDNFSLSNKLGQGGFGSVYKGKLQDGKEIAVKRL 526
Query: 513 SKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------------------- 553
S +SGQG EF NE+ LI +LQH NLV++LGCCI EER+L
Sbjct: 527 SSSSGQGKEEFMNEIVLISKLQHKNLVRILGCCIEGEERLLVYEFLLNKSLDTFLFDSRK 586
Query: 554 ---LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMF 610
+DW KR NIIEGI++GL YLH+ S L++IHRDLK SNILLDE +NPKISDFG+ARM+
Sbjct: 587 RLEIDWPKRFNIIEGIARGLHYLHRDSCLRVIHRDLKVSNILLDEKMNPKISDFGLARMY 646
Query: 611 TQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRP 670
E NT R+ GT GYM+PEYA G+ S KSD+YSFGV+LLEII G K + F +
Sbjct: 647 QGTEYQDNTRRVAGTLGYMAPEYAWTGMFSEKSDIYSFGVILLEIITGEKISRFSYGRQG 706
Query: 671 LNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISM 730
L+ +AWE W + + LLD + D+ P EV+RC+ +GLLCVQ DRP +++SM
Sbjct: 707 KTLLAYAWESWCESGGIDLLDKDVADSCHPLEVERCVQIGLLCVQHQPADRPNTMELLSM 766
Query: 731 LTNKYKLTTLPRRPAFYIRREIYDGETTSKGPDTDTYSTTAI 772
LT LT+ P++P F + D E+ S+G T T ++
Sbjct: 767 LTTTSDLTS-PKQPTFVVHTR--DEESLSQGLITVNEMTQSV 805
>At1g61440 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 792
Score = 330 bits (847), Expect = 1e-90
Identities = 238/733 (32%), Positives = 358/733 (48%), Gaps = 90/733 (12%)
Query: 74 VVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTNDTVATMLDTGNFV 133
VVW+ +R +P+ +A L + SG L + ++ + + + A + D GN +
Sbjct: 65 VVWVANREKPVTDSAANLVISSSGSLLL-INGKHDVVWSTGEISASKGSHAELSDYGNLM 123
Query: 134 LQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELSLEW 193
++ N T LW+SF+ +TLLP + N TG L S +++ P+PG+ ++
Sbjct: 124 VKD---NVTGRTLWESFEHLGNTLLPLSTMMYNLVTGEKRGLSSWKSYTDPSPGDFWVQI 180
Query: 194 EPKEGELNIRKSGKVHWKSGKLKSNGMFENIPAKVQRIYQYIIVSNKDEDS---FAFEVK 250
P+ G + + + IP ++ Y ++D + F++ +
Sbjct: 181 TPQVPSQGFVMRGSTPYYRTGPWAKTRYTGIP-QMDESYTSPFSLHQDVNGSGYFSYFER 239
Query: 251 DGKFIRWFISPKGRL-----------ISDAGSTSNADMCYGYKSDEG-CQVANADMCYGY 298
D K R ++ +G + S G ++ D+ YG G C +++ C +
Sbjct: 240 DYKLSRIMLTSEGSMKVLRYNGLDWKSSYEGPANSCDI-YGVCGPFGFCVISDPPKCKCF 298
Query: 299 --------------NSDGGCQKWEEIPNCR-----EPGEVFRKMVGRPNKDNATTDEPAN 339
N GC + E+ +C+ + VF + PN E AN
Sbjct: 299 KGFVPKSIEEWKRGNWTSGCARRTEL-HCQGNSTGKDANVFHTV---PNIKPPDFYEYAN 354
Query: 340 GYD--DCKMRCWRNCNCYGFEELYSNFTGCIYYSWNSTQDVDLDKKNNFYALVKPTKSPP 397
D C C NC+C F Y GC+ +S + + + ++ S
Sbjct: 355 SVDAEGCYQSCLHNCSCLAFA--YIPGIGCLMWSKDLMDTMQFSAGGEILS-IRLAHSEL 411
Query: 398 NSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAESYDIKD 457
+ H ++ + + ++ L ++ F + K + DL +S D+
Sbjct: 412 DVHKRKMTIVASTVSLTLFVILGFATF--------GFWRNRVKHHDAWRNDL-QSQDVPG 462
Query: 458 LENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSG 517
LE F +I AT +FS NKLG GG+G VYKG L G+E+AVKRLS +S
Sbjct: 463 LE---------FFEMNTIQTATSNFSLSNKLGHGGFGSVYKGKLQDGREIAVKRLSSSSE 513
Query: 518 QGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL----------------------LD 555
QG EF NE+ LI +LQH NLV++LGCC+ +E++L LD
Sbjct: 514 QGKQEFMNEIVLISKLQHRNLVRVLGCCVEGKEKLLIYEFMKNKSLDTFVFGSRKRLELD 573
Query: 556 WKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQES 615
W KR +II+GI +GLLYLH+ SRL++IHRDLK SNILLDE +NPKISDFG+AR+F +
Sbjct: 574 WPKRFDIIQGIVRGLLYLHRDSRLRVIHRDLKVSNILLDEKMNPKISDFGLARLFQGSQY 633
Query: 616 IVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIG 675
T R+VGT GYMSPEYA G+ S KSD+YSFGVLLLEII G K + F + L+
Sbjct: 634 QDKTRRVVGTLGYMSPEYAWTGVFSEKSDIYSFGVLLLEIISGEKISRFSYGEEGKALLA 693
Query: 676 HAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKY 735
+ WE W + + LLD +L D+ P EV RC+ +GLLCVQ DRP +++SMLT
Sbjct: 694 YVWECWCETRGVNLLDQALDDSSHPAEVGRCVQIGLLCVQHQPADRPNTLELLSMLTTTS 753
Query: 736 KLTTLPRRPAFYI 748
L LP++P F +
Sbjct: 754 DL-PLPKQPTFAV 765
>At4g23180 receptor-like protein kinase 4 (RLK4)
Length = 669
Score = 330 bits (846), Expect = 2e-90
Identities = 182/397 (45%), Positives = 252/397 (62%), Gaps = 43/397 (10%)
Query: 392 PTKSPPNS--HGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDL 449
P +PP S G ++ + IA + I+ ++LF+A Y +++++
Sbjct: 271 PVSAPPRSGKDGNSKVLV---IAIVVPIIVAVLLFIAG----YCFLTRRARKSYYTPSAF 323
Query: 450 AESYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAV 509
A +D D ++ +I AT DF NK+GQGG+G VYKG L+ G EVAV
Sbjct: 324 AG--------DDITTADSLQLDYRTIQTATDDFVESNKIGQGGFGEVYKGTLSDGTEVAV 375
Query: 510 KRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---------------- 553
KRLSK+SGQG VEF+NE+ L+ +LQH NLV+LLG C+ EER+L
Sbjct: 376 KRLSKSSGQGEVEFKNEVVLVAKLQHRNLVRLLGFCLDGEERVLVYEYVPNKSLDYFLFD 435
Query: 554 ------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMA 607
LDW +R II G+++G+LYLH+ SRL IIHRDLKASNILLD ++NPKI+DFGMA
Sbjct: 436 PAKKGQLDWTRRYKIIGGVARGILYLHQDSRLTIIHRDLKASNILLDADMNPKIADFGMA 495
Query: 608 RMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDV 667
R+F ++ NT+RIVGTYGYMSPEYAM G S KSDVYSFGVL+LEII G+KN+SF+
Sbjct: 496 RIFGLDQTEENTSRIVGTYGYMSPEYAMHGQYSMKSDVYSFGVLVLEIISGKKNSSFYQT 555
Query: 668 DRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDV 727
D +L+ +AW LW++G L+L+DP++ + +EV RC+H+GLLCVQ+ +RPT+S +
Sbjct: 556 DGAHDLVSYAWGLWSNGRPLELVDPAIVENCQRNEVVRCVHIGLLCVQEDPAERPTLSTI 615
Query: 728 ISMLTNKYKLTTLPRRPAFYIRREI----YDGETTSK 760
+ MLT+ +PR+P + + I D +TTSK
Sbjct: 616 VLMLTSNTVTLPVPRQPGLFFQSRIGKDPLDTDTTSK 652
>At4g11470 serine/threonine kinase-like protein
Length = 666
Score = 323 bits (829), Expect = 2e-88
Identities = 178/382 (46%), Positives = 234/382 (60%), Gaps = 47/382 (12%)
Query: 396 PPNSHGKRRIWIGAAIATAL-----LILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLA 450
PP ++I G +A + ++L L L + K++Q Y K L
Sbjct: 267 PPRDPDGKKISTGVIVAIVVSAVIFVVLVALGLVIWKRRQSY--------------KTLK 312
Query: 451 ESYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVK 510
D +D F+FT+I AT +FS NKLGQGG+G VYKG+L E+AVK
Sbjct: 313 YHTD-----DDMTSPQSLQFDFTTIEVATDNFSRNNKLGQGGFGEVYKGMLPNETEIAVK 367
Query: 511 RLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL----------------- 553
RLS SGQG EF+NE+ ++ +LQH NLV+LLG CI +E+IL
Sbjct: 368 RLSSNSGQGTQEFKNEVVIVAKLQHKNLVRLLGFCIERDEQILVYEFVSNKSLDYFLFDP 427
Query: 554 -----LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMAR 608
LDWK+R NII G+++GLLYLH+ SRL IIHRD+KASNILLD ++NPKI+DFGMAR
Sbjct: 428 KMKSQLDWKRRYNIIGGVTRGLLYLHQDSRLTIIHRDIKASNILLDADMNPKIADFGMAR 487
Query: 609 MFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDV- 667
F ++ T R+VGT+GYM PEY G STKSDVYSFGVL+LEI+CG+KN+SF +
Sbjct: 488 NFRVDQTEDQTGRVVGTFGYMPPEYVTHGQFSTKSDVYSFGVLILEIVCGKKNSSFFQMD 547
Query: 668 DRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDV 727
D NL+ H W LWN+ L L+DP++ +++ DEV RCIH+G+LCVQ+ DRP MS +
Sbjct: 548 DSGGNLVTHVWRLWNNDSPLDLIDPAIKESYDNDEVIRCIHIGILCVQETPADRPEMSTI 607
Query: 728 ISMLTNKYKLTTLPRRPAFYIR 749
MLTN +PR P F+ R
Sbjct: 608 FQMLTNSSITLPVPRPPGFFFR 629
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,558,176
Number of Sequences: 26719
Number of extensions: 923940
Number of successful extensions: 5325
Number of sequences better than 10.0: 1005
Number of HSP's better than 10.0 without gapping: 843
Number of HSP's successfully gapped in prelim test: 162
Number of HSP's that attempted gapping in prelim test: 1967
Number of HSP's gapped (non-prelim): 1287
length of query: 782
length of database: 11,318,596
effective HSP length: 107
effective length of query: 675
effective length of database: 8,459,663
effective search space: 5710272525
effective search space used: 5710272525
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC140024.3


